Abstract
RNA interference (RNAi)-based transgenic technologies have evolved as potent biochemical tools for silencing specific genes of plant pathogens and pests. The approach has been demonstrated to be useful in silencing genes in insect species. Here, we report on the successful construction of RNAi-based plasmid containing an interfering cassette designed to generate dsRNAs that target a novel v-ATPase transcript in whitefly (Bemisia tabaci), an important agricultural pest in tropical and sub-tropical regions. The presence of the transgene was confirmed in T0 and T1 generations of transgenic lettuce lines, segregating in a Mendelian fashion. Seven lines were infested with whiteflies and monitored over a period of 32 days. Analysis of mortality showed that within five days of feeding, insects on transgenic plants showed a mortality rate of 83.8–98.1%. In addition, a reduced number of eggs (95 fold less) was observed in flies feeding on transgenic lettuce plants than insects on control lines. Quantitative reverse transcription PCR showed decreased expression level of endogenous v-ATPase gene in whiteflies feeding on transgenic plants. This technology is a foundation for the production of whitefly-resistant commercial crops, improving agricultural sustainability and food security, reducing the use of more environmentally aggressive methods of pest control.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Whitefly (Bemisia tabaci Gennadius) is a highly polyphagous insect pest that causes great damage in several crops with a host range of over 600 plant species, including cotton, soybean, potato, common bean, cassava, eggplant, cucumber, pepper and tomato (CABI 2017). Besides sucking plant nutrients from phloem, which results in accumulation of toxic molecules leading to plant breakdown, B. tabaci transmits several viruses to plant species (Li et al. 2011; Brown et al. 2015). Crop yield losses due to whitefly-transmitted begomoviruses alone are between 20 and 100% (Brown and Bird 1992; Cathrin and Ghanim 2014).
The most widely used cultural practice of controlling the insect is application of insecticides (Karatolos et al. 2010). The activities of natural enemies of B. tabaci have been employed as a means of biological control, leading to the production of parasitoids (Rivnay and Gerling 1987; Gerling et al. 2001). Furthermore, pathogenic fungi have been used to generate mycoinsecticide (Faria and Wraight 2001; Cuthbertson and Walters 2005; Cuthbertson et al. 2005). Despite the progress made in developing alternative methods of control, the use of insecticides remains the common practice. However, due to its rapid reproduction lifecycle, the pest can easily overwhelm crops prompting farmers to applying high doses of insecticides even when the number of flies per plant is low. This has, over the years, led to the emergence of resistance (Wang et al. 2010), which underscores the need to develop new methods for control, including the use of RNA interference technologies. The demonstration that double-stranded RNAs (dsRNAs) supplied in artificial diet trigger RNA interference (RNAi) in several coleopteran species, resulting in larval stunting and mortality, is an indication of the potential RNAi holds (Baum et al. 2007; Scott et al. 2013). RNA interference has evolved as a natural cellular defense mechanism in eukaryotes against viruses and genomic confinement of retrotransposons (Fire et al. 1998; Ibrahim and Aragão 2015). It is an effective tool for silencing genes in various organisms, including insects. Its usefulness in insect pest control was highlighted with the demonstration of gene silencing in tritomid bug (Rhodnius prolixus) (Araujo et al. 2006) and light brown apple moth (Epiphyrsu postvittana) (Turner et al. 2006). Transgenic plant-mediated approaches using RNAi to target insect genes have been reported in experiments showing silencing of genes in western corn rootworm (Diabrotica virgifera) (Baum et al. 2007), cotton bollworm (Helicoverpa armigera) (Mao et al. 2007), brown plant hopper (Nilaparvara lugens) (Zha et al. 2011) and tobacco hornworm (Manduca sexta) (Kumar et al. 2012) among others. The demonstration that tissue-specific mRNAs of different genes in whitefly may be preferentially silenced, or the expression thereof reduced, using RNAi-based strategy has provided evidence of its effectiveness in controlling the insect (Ghanim et al. 2007; Yu et al. 2012). The development of a method for the delivery of dsRNA and siRNA into whitefly through oral route has led to silencing and knocking down the expression of its important genes (Upadhyay et al. 2011). Recently, researchers have reported the use of transgenic tobacco, (Nicotiana tabacum) to generate dsRNAs against aquaporin and alpha glucosidase genes, responsible for osmoregulation, which when silenced in whitefly led to 70% mortality (Raza et al. 2016). Similarly, acetylcholinesterase (AChE) and ecdysone receptor (EcR) genes of whitefly were used to induce mortality of up to 90% in tobacco (Malik et al. 2016). Of the candidate genes targeted for silencing in insects, v-ATPase has consistently featured among the most potent (Upadhyay et al. 2011; Thakur et al. 2014). Since v-ATPase affects such diverse cellular processes as intracellular membrane transport and the processing and transport of neurotransmitters, as well as regulation of the entry of microorganisms like viruses, it is expected that silencing its gene will be lethal (Davies et al. 1996; Dow et al. 1997; Beyenbach and Wieczorek 2006; Forgac 2007).
Here we report the cloning of a novel v-ATPase gene fragment from whitefly that was used to generate an intron-hairpin construct, and was expressed in genetically engineered lettuce, an important leafy crop. We hypothesized that the expression of this construction, with the consequent production of siRNA, would silence the whitefly endogenous gene, interfering in the insect life cycle and generating resistant plants.
Materials and methods
Plant and insect culture
Whiteflies (Biotype B) were kindly provided by Dr. Josias Correa de Faria of Embrapa Arroz e Feijão (Santo Antônio de Goiás, Brazil), from a known non-viruliferous colony maintained in greenhouse. The insects were released on potted cotton plants in Embrapa Recursos Genéticos e Biotecnologia (Brasília, Brazil).
RNAi construct
A 647 bp sequence corresponding to a fragment from whitefly v-ATPase gene was amplified by PCR from genomic DNA using the primers ATPaF1 (5′-GAGGGTGACATGGCCACCATCCAGGT-3′) and ATPaR1 (5′-GACRTCRGAGTTGGAGTACTTGGACAG-3′), and cloned in the pGEMTEasy vector (Supporting Information). These primers were based on conserved regions of available insect v-ATPase genes from the related species: cotton bollworm (Helicoverpa zea), silkworm (Bombyx mori), Culex quinquefascitatus, Asian corn borer (Ostrinia furnacalis), tobacco hornworm (Manduca sexta) and cotton leaf worm (Spodoptera littoralis). The 647 bp fragment was used as template for amplification of a sequence of 545 bp using the primers ATPXS (5′-TTCTA GA GCTCGCATCCGAAAGCGCCGGGAATG-3′; including the sites for Xba I [underlined] and Sac I [in bold]) and ATPSK (5′-GGGTACC ACTAGTCGGCGACCCTGTACAGCGAAC-3′; including the sites for Kpn I [underlined] and Spe I [in bold]). PCR was carried out according to Bonfim et al. (2007). This fragment was cloned in sense and antisense orientations in the vector pSIU (Tinoco et al. 2010), generating the vector pBtATPase. The interfering cassette was removed with Hind III and Eco RI from the vector pBtATPase and cloned into the pCAMBIA3300 vector, which contains the bar gene (that confers tolerance for the herbicide glufosinate ammonium), generating the vector pBtATPaseC3300. The interfering cassette containing the v-ATPase sequence hairpin will be referred to hereafter as ΔATPase (Fig. 1a).
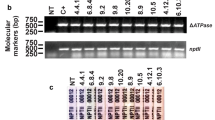
a Diagram of the vector pBtATPaseC3300 showing the ΔATPase interfering cassette The 545 bp fragment of the whitefly v-ATPase gene was directionally cloned in order to generate sense and antisense arms flanking the malate synthase gene intron 3 from Arabidopsis thaliana (Atms-i3) The cassette was cloned into the pCAMBIA3300 vector, generating the pBtATPaseC3300, which was used to transform lettuce. b PCR analysis of 17 transformed lines for detection of the ΔATPase (upper gel) and the bar gene (lower gel), used for selection of transformed plants with glufosinate ammonium) C1 is a non-transgenic plant and C+ is the positive control (vector). c Immuno-chromatographic analyses of PCR-positive plants for the expression of the bar gene (presence of PAT protein) showing plants displaying strong (++ and +++) and weak (+) signals C is a non-transgenic plant
Lettuce genetic transformation
The plasmid vector pBtATPaseC3300 was transformed into Agrobacterium tumefaciens strain EHA105 which was then co-cultured with cut pieces of lettuce cotyledons, according to the method of Dias et al. (2006). Briefly, lettuce seeds (cv Verônica) were surface sterilized by immersion in Tween 20 0.01% followed by NaOCl 2.5% for 7 min. The seeds were then rinsed three times in sterile distilled water and germinated in a Petri dish containing MS media solidified with agar 0.8% for 48 h. Cotyledons cut from seedlings were co-cultured with bacterial suspension (O D600 of 0.5–1.0) in MS media containing acetosyringone 0.5 M and glucose 3% for 15 min. The explants were dried on autoclaved filter paper and incubated in an MS medium containing sucrose 3%, naphthalene acetic acid 0.05 mg/L (NAA) and 6-benzylaminopurine 0.2 mg/L (BAP) at 25 ± 2 °C for 48 h. The explants were then sub-cultured into the same medium with addition of glufosinate ammonium 4 mg/L. Proliferating calli arising from the explants were subcultured into the same fresh medium every week. Emerging shoots were individualized and cultured on MS medium containing sucrose 3%, kinetin 0.5 mg/L, zeatin 0.5 mg/L and glufosinate ammonium 4 mg/L. The explants were sub-cultured on a weekly basis until they were fully grown, and then they were transferred to rooting medium containing MS basal medium and glufosinate ammonium 4 mg/L. With the development of roots, explants were acclimatized in plastic cups containing 50:50 ratio of soil and vermiculate in a humidity chamber made of a plastic sac and rubber band. When fully hardened, the plants were transferred to large pots. With the emergence of seeds, these were collected and planted in plastic cups for further screening and analyses.
Screening of transgenic plants
Regenerated plants were first screened using the immunochromatographic strip test, method of TraitChek™ (Romer Labs) based on the manufacturer’s recommendation for the detection of PAT protein. DNA from leaves was isolated using a modified version of Doyle and Doyle (1987), and used for PCR analysis using the primer pairs ATPXS/ATPSK for the detection of ΔATPase. For the detection of bar gene, the primers BarF (5′-AAACCCACGTCATGCCAGTT-3′)/BarR (5′-CATCGAGACAAGCACGGTCA-3′) were used PCR conditions were set according to Bonfim et al. (2007).
Progeny analysis
Seeds of the first generation (T1) of self-pollinated plants were germinated and analyzed for the presence of ΔATPase by PCR, as previously described. Pearson’s Chi squared (χ2) was used to determine whether or not the observed segregation ratio was consistent with a Mendelian ratio (3:1 or 15:1), at 95% level of confidence. For Lines 1, 3, 4, 6, 19 and 25, the sample sizes were 30 each. For Line 31, where the sample size small, Yates’s correction factor was used.
Whitefly toxicity assay
The toxicity assay was performed using 20 emerging (synchronized) young whiteflies inoculated on 4-week-old potted T1 plants in plastic cups, which were then transferred into plastic jars with cover. The covers were perforated at the top and the hole sealed with cotton wool to allow air circulation. The population of the flies was monitored on a daily basis. The number of whiteflies per plant was recorded over a period of 2 weeks until all the whiteflies had died off. During the third week after inoculation, emerging eggs, crawlers (nymphs), pupae and adults were recorded on the leaves by visualizing individual leaves of each plant under stereomicroscope. Two types of controls were used; transgenic plants expressing bar gene with no interfering cassette, and non-transgenic plants. Emergence of eggs, crawlers (nymphs, all instar stages), pupae and adults was monitored during a cycle of 32 days. The system was kept in a 16 h photoperiod and 25 ± 2 °C. Plants were irrigated by means of hypodermic syringe and carefully removed using a metal grip for counting of eggs, crawlers, pupae and adults after every 4 days, starting 11 days after inoculation. Day 11 after inoculation was considered day 1 for egg to adult analysis. Seven transgenic lines containing the v-ATPase-LEG fragment were used for the bioassay in addition to the two control lines. For each line, 12 biological repetitions were used and their mean values used for Tukey analysis using Prism software, version 5.0.
Analysis of siRNAs
Total RNA from lettuce was isolated using miRNeasy mini kit (Quiagen) as recommended by the manufacturer. The siRNA analysis was carried out as described by Bonfim et al. (2007). Hybridization was performed using a DNA probe corresponding to the v-ATPase PCR fragment amplified using the primer pair ATPXS/ATPSK labeled with α32P dCTP and a random primer DNA labeling kit (Amersham Pharmacia Biotech.), according to the manufacturer’s instructions. Hybridization and post-hybridization washes were conducted as described by Yoo et al. (2004). Oligomers (18 and 24 nucleotides) were used as molecular size markers. The bands were visualized with a fluorescent image analyzer (FLA-3000; FUJIFILM). Three independent autoradiographs, corresponding to three biological replicates (individual plants) were used.
Quantitative real-rime PCR
The transcription level of the endogenous v-ATPase gene in B. tabaci interacting with transgenic lettuce leaves (T1 generation) was determined by quantitative reverse transcription. RNA isolation and qRT-PCR were performed as described by Andrade et al. (2016). Fifty (50) whiteflies were released into bioassay systems containing lettuce lines 1, 3, 4, 6, 19, 25, 31 and control as described above (whitefly toxicity assay). After 48 h, total RNA was extracted from the surviving insects, of which 200 ng from each of the samples was used to synthesize cDNA using Promega GoScript™ Reverse Transcription System (Promega, A5000) as recommended by the manufacturer. Three biological repetitions of each of the lines were used for the cDNA synthesis. The primers for the endogenous v-ATPase (TCTTTCCCTCAAACTCCAGC and AGGCACGATCACCAAGATG) and actin (GACCAGCCAAGTCCAAACGA and CCTTTGTGGTAGAGGTCTCAGTT) genes were designed using the PrimerQuest Tool (IDT Integrated DNA Technologies, Inc.). Products from the PCR were separated by agarose gel electrophoresis and sequenced to confirm their identities. The relative v-ATPase transcription levels in different RNA samples were normalized with respect to the internal standard actin gene. Triplicate quantitative assays were performed on each cDNA sample. The relative level of the expression was calculated using the Livak method (Livak and Schmittgen 2001). Results were compared by one-way ANOVA with Dunnett’s posttest (α = 0.05) to verify differences between insect groups interacting with each RNAi expressing lettuce line compared to the control using Prism software, version 5.0.
Results
Generation of transgenic lettuce lines
In order to assess the effectiveness of RNAi technology in generating lettuce plants resistant to whitefly, we designed and engineered an intron-hairpin construct (ΔATPase) into which a fragment of a novel v-ATPase gene from the insect was directionally cloned to produce sense and antisense arms (Fig. 1a).
A total of 17 independent lines of transgenic lettuce plants were obtained. All primary transformants (T0) appeared normal, and PCR analysis confirmed the presence of both ΔATPase and bar cassettes (Fig. 1b). Progenies of the transgenic lines were analyzed for determination of the segregation pattern screened for the presence of PAT protein, using the immunochromatographic method. Seven lines revealed a Mendelian segregation pattern of 3:1 (Line 1: χ2 = 0.2, P = 0.61; Line 3: χ2 = 0.5, P = 0.60; Line 4: χ2 = 1.6, P = 0.2; Line 6: χ2 = 0.5, P = 0.48; Line 1.9: χ2 = 0.5, P = 0.49; Line 25: χ2 = 0.2, P = 0.62 and Line 31: χ2 = 0.7, P = 0.41; 1 df); and showed a strong PAT protein signals (Fig. 1c). Based on these results, these seven lines were selected for further analyses.
Total RNA from the seven lines was used for northern blot analysis to detect the transgene-derived siRNA in leaves (Fig. 2). Results revealed siRNA strong bands of expected size range in all transgenic plants. There was no siRNA signal in the non-transgenic plants.
Transgenic lettuce plants expressing v-ATPase siRNA induced high mortality in whitefly
A feeding experiment was performed in which transgenic lettuce plants expressing v-ATPase siRNA were challenged with newly emerged adult whiteflies and their mortality monitored over a period of 32 days. Within the first three days, there was a sharp decrease in the number of whiteflies feeding on all transgenic lines (Fig. 3). This decrease was significantly lower and slower in the two controls used (P < 0.05). On day five, several flies were visible on the control, while there were only a few on the transgenic lines. Whiteflies fed with transgenic plants presented statistically significant higher mortality rate when compared to insects fed with non-transgenic plants or with transgenic plants expressing only the bar gene (P < 0.05). Death by ingestion of the siRNA from the leaves appears to start from the first three days after interacting with transgenic plants, when more than 50% of the earlier inoculated adults were dead, whereas only 5% of the population was reduced in control lines (Fig. 3). On day three, there were between 15 and 17 insects (corresponding to 76.3–85.0%) on the control lines, while there were between 6 and 11 (30–546%) insects on the test plants. This may be translated to a mortality of between 56.4 and 70% in transgenic plants within the first three days. On day five, this numerical relationship stood at 11 and 12 insects (56.3–56.7%) on the control lines and 2 and 3 (9.6–16.3%) on the transgenic plants, corresponding to a mortality of between 83.8 and 98.1% on the test plants (Fig. 3). By the tenth day, the number of whiteflies was near zero on all the transgenic plants expressing v-ATPase siRNAs, while control lines still had a little below 5% of the original population.
Toxicity of transgenic lettuce plants producing v-ATPase siRNAs against whiteflies All ΔATPase-transgenic plants (Lines 1, 3, 4, 6, 19, 25 and 31) induced higher mortality than internal control [non-transgenic plants (C1) and plants transformed with the vector pCAMBIA3300, without the ΔATPase cassette (C2)] All transgenic lines were similar and statistically different from controls in all days (*P < 0.05, n = 12) Bars represent ± SD
v-ATPase siRNAs in transgenic lettuce promote low oviposition and delay pupation in whitefly
The cycle of the whiteflies feeding on lettuce plants was evaluated to monitor the emergence of eggs, crawlers, pupae and adults. Generally, lower number of all stages was recorded in flies feeding on the test plants than on the control lines. Monitoring oviposition by 20 whiteflies feeding on transgenic lettuce plants showed that flies on the test plants produced a lower number of eggs than both controls (P < 0.05). On the 12th day (day 1 of this count) after inoculation, the 20 whiteflies deposited between 227 and 231 eggs and produced between 107 and 125 crawlers in control lines; while those feeding on the transgenic test plants produced between 24 and 66 eggs and between 13 and 52 crawlers, respectively (Fig. 4a). The number of eggs fell in all transgenic lines and in the control throughout the 32 days of analysis. However, in the control lines, this fall was sharper within the first nine days from the counting of eggs. It then slowed down and fell sharply by the 17th day, when it suddenly started to rise again. This new rise correlates with the increasing number of emerging adults. In the test plants however, this fall was much slower. Although insects in lines 3 and 31 appeared to stabilize in number of eggs over the first week, they all tended to converge with the remaining transgenic test lines to show a steep fall by day 9 through day 13. In addition, several eggs appeared to have been aborted.
Effect of ΔATPase dsRNA on oviposition (a), emergence of crawlers (nymphs) (b), emergence of pupa (c) and re-emergence of adult whiteflies (d) derived from the initial population of 20 whiteflies feeding on transgenic lettuce lines Bars represent ± SD *P < 0.05, n = 12, related to controls (C1 = non-transgenic plants; C2 = plants transformed with the vector pCAMBIA3300, without the ΔATPase cassette
The emergence of crawlers followed a similar pattern to the deposition of eggs, with the number of crawlers being much lower (Fig. 4b). The difference between the control and the test lines in terms of population of nymphs becomes very clear when we consider the fact that day 1 is the peak day for their production. Whereas the control lines recorded between 107 and 125 crawlers, the highest number of crawlers generated on the transgenic lines on this day was 48 (Line 4), with the lowest being 14 (Line 6).
Both controls produced pupae earlier and at higher rates than test plants (Fig. 4c) On day 5, between 5 and 6 pupae emerged on the control lines, whereas there was no evidence of such an emergence on any of the transgenic lines. The peak day for the emergence of pupa on both control and tests plants was day 13. However, whereas there were between 24 and 25 on controls, the highest number of pupae from transgenic plants was 15 (line 4) with the lowest (≤2) being common to Lines 6 and 19.
New adults started to emerge by the 18th day after infestation in all the control lines. However, this emergence was delayed by at least 2 days in the transgenic systems monitored. While the peak for the emergence of adults for all control lines was on day 17 (with 11 to 13 flies on the day), the peak for the test plants was on day 13, with the highest number of adults being 2 only. From then on, adult emergence diminished on all the transgenic lines (Fig. 4d). This places emergence in the transgenic line at about 78% lower than in control. On any given day, there were more flies per leaf of each of the control lines than in the transgenic lines. While counting of adult flies was done at intervals of four days, emerging adults sometimes died before they were counted. Carcasses of the flies were clearly visible at different stages over the period of 32 days.
Again, at the level of eggs, several aborted eggs were observable. It is not clear if the feeding triggered the systemic RNAi pathway and was responsible for this abortion, or indeed if the molecules acted at different stages in the development of the whitefly. What became apparent was that the death of emerging whiteflies often took place 3 days after they started to feed on the crop.
Bt v-ATPase was down-regulated in insects feeding on transgenic lettuce lines
Analyses were carried out to quantify the transcription level of the endogenous v-ATPase gene in B. tabaci interacting with transgenic lettuce leaves from seven different lines (T1 generation). Results showed that it was significantly lower in all the transgenic events expressing v-ATPase-related siRNA, when compared to the two types of controls (Fig. 5). There were no statistical differences among transgenic lines (P < 0.05). Insects interacting with both control lines (non-transgenic and transformed with the backbone vector) presented higher expression levels of the endogenous v-ATPase when compared with RNAi expressing lettuce lines.
Relative expression of endogenous B tabaci v-ATPase in insects feeding on leaves of transgenic lettuce lines for 48 h, determined by quantitative real-time RT-PCR Data represent means of three replications (± SD) *P < 0.05, n = 9, related to controls (C1 = non-transgenic plants; C2 = plants transformed with the vector pCAMBIA3300, without the ΔATPase cassette
Discussion
The impetus for the development of whitefly-resistant plants derives from early reports showing that siRNAs can effectively interfere with the physiology of insects leading to impaired growth of larvae, disruption of feeding habit and ultimately death (Baum et al. 2007; Mao et al. 2007). In the pioneering work of Baum et al. (2007), 14 genes were identified and knocked out using low concentrations of dsRNA, leading to death of Western corn rootworm larvae. With the demonstration that tissue-specific mRNAs of different genes of whitefly could be preferentially silenced by up to 70%, a critical point was reached in the development of whitefly-resistant plants (Ghanim et al. 2007). However, any method that seeks to control whitefly via RNAi must take into account the means by which siRNAs and/or dsRNA are delivered. Since it is impracticable to inject every whitefly in the field, oral delivery and uptake of dsRNA in the gut have become imperative (Upadhyay et al. 2011). Indeed, the use of this approach led to significant mortality of whiteflies based on artificial diet prepared from dsRNA corresponding to five different genes including v-ATPase Although Upadhyay et al. (2011) reported a similarity in the pattern of toxicity of siRNA diets based on the 5 genes, dsRNA from the A subunit of ATPase was found to be most effective, with a mortality of more than 80% over 6 days. In the present work, insect mortality of between 83.8 and 98.1% was observed within 5 days of feeding on transgenic lettuce expressing v-ATPase siRNA (Fig. 3). This represents an improvement on all the reports so far on silencing of v-ATPase gene. The improvement is indeed further enhanced because the method developed in this study does not only dispense with the preparation of artificial diets containing siRNA/dsRNA, but also eliminates the necessity of monitoring mortality under conditions that may not be easily controlled and therefore require the use of complex population dynamic parameters for analysis. The effect of siRNA v-ATPase on a bioassay employing the use of leaf discs from transgenic tobacco showed a mortality rate of between 15 and 38% on day 2 and between 34 and 85% on day 6 following infestation (Thakur et al. 2014). A feeding experiment using dsRNAs that targeted genes within the molting hormone-ecdysone synthesis and signaling pathway showed low survival and delayed development of whitefly during nymphal stages (Luan et al. 2013). An alternative strategy in which glutathione synthase transferase, coding for GST, an enzyme known to detoxify insecticides, might be silenced by siRNAs designed to block its synthesis is quite attractive. Indeed, it has recently been demonstrated that dsRNAs synthesized based on whitefly GST, and fed to the insect in dietary form, resulted in a mortality rate ranging from 40 to 77.43% over a period of 72 h (Asokan et al. 2015). In addition, dsRNAs that target aquaporin and alpha glucosidase genes have been reported to cause 70% mortality (Raza et al. 2016). The same group showed a 90% mortality when dsRNAs designed to target AChE and EcR genes of B. tabaci were expressed in tobacco (Malik et al. 2016).
Although the methods employed in evaluating the effect of siRNA on mortality of whiteflies by Upadhyay et al. (2011) and Thakur et al. (2014) are different from that used in this study, insect mortality in the three studies may be averaged within the range of 75–85%. In all cases, mortality was recorded in the first 10 days of inoculation, irrespective of the number of whiteflies inoculated or the bioassay system used. However, the method developed in this work may be considered superior to the previous two studies in two aspects: (1) For the first time, RNAi-based strategy that employs a transgenic crop was used to silence v-ATPase; (2) it led to the development of a bioassay system using simple, cheap and reproducible materials, without the need either to synthesize siRNA/dsRNA for use in feeding experiment or to prepare diets for the same purpose.
A key factor in monitoring the mortality of whiteflies as developed in this method is the heterogeneity of the freshly emerged adults. While care was taken in introducing relatively synchronized and only emerging adult whiteflies of approximately the same age into the bioassay system, some factors may have interfered with the overall toxicity of the siRNAs. For example, physiological barriers, behavioral factors and geographical orientation of individual whiteflies on the leaves may have influenced the overall toxic effect of the siRNA consumed by the insects. In almost all of the bioassay units used in this study, the whiteflies tended to colonize the lower leaf of the host plant with the remaining leaves left non-colonized. With the subsequent emergence of crawlers and new adults, they migrated to the youngest available leaf, to repeat the cycle all over again. Although previous experience of whiteflies may vary among populations, behavioral changes in terms of host preference, oviposition and feeding may arise depending on the host plant (Shah and Liu 2013). At any rate, this observation may lead to the use of lower leaves as baits in a similar bioassay or diet-based analysis to allow for more stringent control and monitoring of the whitefly cycle.
The presence of enzymes in insect gut, including v-ATPase, may also have determined the toxicity of the siRNAs. For example, the siRNAs may be degraded in the gut before they can exert their toxic effect, since there are detoxifying nucleases that may reduce their availability for gene silencing (Katoch and Thakur 2012). While in situ studies are required to study the cellular location and physiological effect(s) of the siRNAs derived in this study, the pattern of death, which often took place by the third day of contact with lettuce plants. It suggests that silencing may be in the gut of B. tabaci, and it is observable at the adult stage within the first three days of feeding. Clearly, the dramatic silencing effect leading to the death of insects is intimately related to the transgenic plant’s ability to express hairpin RNA that targets gut v-ATPase at fast rates (Gordon and Waterhouse 2007). This dosage and time-dependent effect may explain why some of the whiteflies reached the next generation unharmed and were able to lay eggs.
Moving forward, a siRNA profile of the entire lines generated in this study will provide clues to the level of the molecules within a large population of individuals from different lines. In addition, microscopic analysis of the insect gut within the first few days of feeding may furnish valuable physiological information on the effect of the siRNA produced by the transgenic plants. Experiments targeting acetycholinesterase (AchE) gene in cotton bollworm using dietary siRNA reported reduction in larval growth, pupal weight loss and malformation and death (Kumar et al. 2009). Similarly, RNAi was used to express TLR7 dsRNA of whitefly in recombinant entomopathogenic fungi Isaria fumosorossea on China rose (Hibiscus rosasinensis) knocking down the gene and leading to mortality of over 90% in nymphs (Chen et al. 2015). At the level of gene expression, a decrease in target mRNA levels was reported in light brown apple moth (Epiphyas postvittana) (Turner et al. 2006) and red flour beetle (Tribolium castaneum) (Tomoyasu et al. 2008). A 62% decrease in the expression level of ATPase RNA was reported following feeding with transgenic tobacco plants (Thakur et al. 2014).
How soon RNAi technology may emerge as an industrial-scale crop protection strategy against sap-sucking hemipteran pests such as whiteflies, aphids and leafhoppers, may ultimately depend on the speed with which similar approaches to the one described in this study are optimized (Price and Gatehouse 2008; Upadhyay et al. 2011). Crucial optimization parameters include such vital components as the siRNA delivery system as well as the enhancement of the tissue accumulation of miRNA, siRNA and dsRNA in sieve tubes by designing transformation vectors with phloem specific promoters (Buhtz et al. 2008; Kehr and Buhtz 2008; Pant et al. 2008; Zhang et al. 2013). In addition, the mechanisms by which processed and unprocessed dsRNA and ssRNA are taken up by insect cells need to be studied and understood further. This knowledge is crucial for anticipating possible mechanisms of resistance and suppression that may arise under natural field conditions. Moreover, new candidate genes need to be discovered and their amenability to the approach tested RNAi strategies could be combined with other biotechnological approaches, such as the use of bacteria or viruses that might interfere with the viability of whitefly reproduction (Faria et al. 2016).
In conclusion, 17 transgenic lines of lettuce expressing siRNAs derived from v-ATPase were generated and screened. Northern analysis showed the presence of siRNA specific to the v-ATPase hairpin-forming sequence in seven of the lines. When subjected to bioassay, these lines had protective effects against whitefly by the induction of high mortality in adults, reduced oviposition and delayed conversion of pupae to adults. A quantitative RT-PCR analysis established the cause–effect relationship between silencing of the v-ATPase gene induced by ATPase-specific siRNA produced by the host and impairment of the insect life cycle. Silencing of genes for the control of whitefly may form the foundation for a field trial with the ultimate aim of breeding elite lines of the crops for protection against whitefly and other sap-sucking insects.
References
Andrade CM, Tinoco MLP, Rieth AF, Maia FCO, Aragão FJL (2016) Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol 65:626–632
Araujo RN, Santos A, Pinto FS, Gontijo NF, Lehane MJ, Pereira MH (2006) RNA interference of the salivary gland nitrophorin 2 in the triatomine bug Rhodnius prolixus (Hemiptera: Reduviidae) by dsRNA ingestion or injection. Insect Biochem Mol Biol 36:683–693
Asokan R, Rebijith KB, Roopa HK, Kumar NKK (2015) Non-invasive delivery of dsGST is lethal to the sweet potato whitefly, Bemisia tabaci (G) (Hemiptera: Aleyrodidae). Appl Biochem Biotechnol 175:2288–2299
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleasu M, Vaughn T, Roberts J (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326
Beyenbach KW, Wieczorek H (2006) The V-type H + ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol 209:577–589
Bonfim K, Faria JC, Nogueira EOPL, Mendes EA, Aragão FJL (2007) RNAi-mediated resistance to bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant Microbe Interact 20:717–726
Brown JK, Bird J (1992) Whitefly-transmitted geminiviruses and associated disorders in the Americas and the Caribbean Basin. Plant Dis 76:220–225
Brown JK, Zerbini FM, Navas-Castillo J, Moriones E, Ramos Sobrinho R, Silva JCF, Briddon RW, Hernandez-Zepeda C, Idris AM, Malathi VG, Martin DP, Rivera-Bustamante R, Ueda S, Varsani A (2015) Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch Virol 160:1593–1619
Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J (2008) Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J 53:739–749
CABI (2017) Invasive species compendium. Bemisia tabaci (tobacco whitefly) Wallingford, UK: CAB International. http://www.cabi.org/isc/datasheet/8927. Accessed 26 June 2017
Cathrin PB, Ghanim M (2014) Recent advances on interactions between the whitefly Bemisia tabaci and begomoviruses, with emphasis on Tomato yellow leaf curl virus. In: Gaur RK, Hohn T, Sharma P (eds) Plant Virus-Host Interaction: Molecular Approaches and Viral Evolution. Elsevier, Amsterdam, pp 79–103
Chen X, Li L, Hu Q, Zhang B, Wu W (2015) Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol 15:64
Cuthbertson AGS, Walters KFA (2005) Pathogenicity of the entomopathogenic fungus, Lecanicillium muscarium, against the sweet potato whitefly Bemisia tabaci, under laboratory and glasshouse conditions. Mycopathologia 160:315–319
Cuthbertson AGS, Walters KFA, Northing P (2005) The susceptibility of immature stages of Bemisia tabaci to the entomopathogenic fungus Lecanicillium muscarium on tomato and verbena foliage. Mycopathologia 159:23–29
Davies SA, Goodwin SF, Kelly DC, Wang Z, Sozen MA, Kaiser K, Dow JAT (1996) Analysis and inactivation of vha55, the gene encoding the vacuolar ATPase B-subunit in Drosophila melanogaster reveals a larval lethal phenotype. J Biol Chem 271:30677–30684
Dias BBA, Cunha WG, Morais LS, Vianna GR, Rech EL, Capdeville G, Aragão FJL (2006) Expression of an oxalate decarboxylase gene from Flammulina sp in transgenic lettuce (Lactuca sativa) plants and resistance to Sclerotinia sclerotiorum. Plant Pathol 55:187–193
Dow JAT, Davies SA, Gua Y, Graham S, Finbow ME, Kaiser K (1997) Molecular genetic analysis of v_ATPase function in Drosophila melanogaster. J Exp Biol 200:237–245
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Faria M, Wraight SP (2001) Biological control of Bemisia tabaci with fungi. Crop Prot 20:767–778
Faria JC, Aragão FJL, Souza T, Quintela E, Kitajima EW, Ribeiro SG (2016) Golden mosaic of common beans in Brazil: management with a transgenic approach. APS Features 10:1094
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998) Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806–811
Forgac M (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat Rev Mol Cell Biol 8:917–929
Gerling D, Alomar O, Arnó J (2001) Biological control of Bemisia tabaci using predators and parasitoids. Crop Prot 20:779–799
Ghanim M, Kontsedalov S, Czosnek H (2007) Tissue-specific gene silencing by RNA interference in the whitefly Bemisia tabaci (Gennadius). Insect Biochem Mol Biol 37:732–738
Gordon KHJ, Waterhouse PM (2007) RNAi for insect-proof plants. Nat Biotechnol 25:1231–1232
Ibrahim AB, Aragão FJL (2015) RNAi-mediated resistance to viruses in genetically engineered plants. Methods Mol Biol 1287:81–92
Karatolos N, Denholm I, Williamson M, Nauen R, Gorman K (2010) Incidence and characterization of resistance to neonicotinoid insecticides and pymetrozine in the greenhouse whitefly, Trialeurodes vaporariorum Westwood (Hemiptera: Aleyrodidae). Pest Manag Sci 66:1304–1307
Katoch R, Thakur N (2012) Insect gut nucleases: a challenge for RNA interference mediated insect control strategies. Int J Biochem Biotechnol 1:198–203
Kehr J, Buhtz A (2008) Long distance transport and movement of RNA through the phloem. J Exp Bot 59:85–92
Kumar M, Gupta GP, Rajam MV (2009) Silencing of acetylcholinesterase gene of Helicoperva armigera by siRNA affects larval growth and its life cycle. J Insect Physiol 55:273–278
Kumar P, Pandit SS, Baldwin IT (2012) Tobacco rattle virus vector: a rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS ONE 7:e31347
Li SJ, Xue X, Ahmed MZ, Ren S-X, Du Y-Z, Wu J-H, Cuthbertson AGS, Qui B-L (2011) Host plants and natural enemies of Bemisia tabaci (Hemiptera: Aleyrodidae) in China. Insect Sci 18:101–120
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the \(2^{- \Delta \Delta {\rm C}_{\rm T}}\) method. Methods 25:402–408. doi:10.1006/meth.2001.1262
Luan JB, Ghanim M, Liu SS, Czosnek H (2013) Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem Mol Biol 43:740–746
Malik HJ, Raza A, Amin I, Scheffler JA, Scheffler EB, Brown JK, Mansoor S (2016) RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco. Plant Sci Rep 6:38469
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25:1307–1313
Pant BD, Buhtz A, Kehr J, Scheible WR (2008) MicroRNA399 is a long distance signal for the regulation of plant phosphate homeostasis. Plant J 53:731–738
Price DRG, Gatehouse JA (2008) RNAi-mediated crop protection against insects. Trends Biotechnol 26:393–400
Raza A, Malik HJ, Shafiq M, Amin I, Scheffler JA, Scheffler EB, Mansoor S (2016) RNA interference based approach to down regulate osmoregulators of whitefly (Bemisia tabaci): potential technology for the control of whitefly. PLoS ONE 11:e0153883
Rivnay T, Gerling D (1987) Aphelinidae parasitoids (Hymenoptera: Chalcidoidea) of whiteflies (Hemiptera: Aleyrodidae) in Israel, with description of three new species. Entomophaga 32:463–475
Scott JG, Michel K, Bartholomay LC, Siegfried BD, Hunter WB, Smagghe G, Zhu KY, Douglas AE (2013) Towards the elements of successful insect RNAi. J Insect Physiol 59:1212–1221
Shah MMR, Liu T-X (2013) Feeding experience of Bemisia tabaci (Hemiptera: Aleyrodidae) affects their performance on different host plants. PLoS ONE 8(10):e77368
Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase a gene. PLoS ONE 9:e87235
Tinoco MLP, Dias BBA, Dall’Astta RC, Pamphile JA, Aragão FJL (2010) In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol 8:27
Tomoyasu Y, Miller SC, Tomita S, Schoppmeier M, Grossmann D, Bucher G (2008) Exploring systemic RNA interference in insects: a genome-wide survey for RNAi genes in Tribolium. Genome Biol 9:R10
Turner CT, Davy MW, MacDiarmid RM, Plummer KM, Birch NP, Newcomb RD (2006) RNA interference in the light brown apple moth, Epiphyas postvittana (Walker) induced by double-stranded RNA feeding. Insect Mol Biol 15:383–391
Upadhyay SK, Chandrashekar K, Thakur N, Verma PC, Borgio JF, Singh PK, Tuli R (2011) RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36:153–161
Wang Z, Yan H, Yang Y, Wu Y (2010) Biotype and insecticide resistance status of the whitefly (Bemisia tabaci) from China. Pest Manag Sci 66:1360–1366
Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16:1979–2000
Yu N, Christiaens O, Liu J, Niu J, Cappelle K, Caccia S, Huvenne H, Smagghe G (2012) Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 20:4–14
Zha W, Peng X, Chen R, Du B, Zhu L, He G (2011) Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the Hemipteran insect Nilaparvata lugens. PLoS ONE 5:e20504
Zhang H, Li HC, Miao XX (2013) Feasibility, limitation and possible solutions of RNAi-based technology for insect pest control. Insect Sci 20:15–30
Acknowledgements
We gratefully acknowledge the financial support of CNPq (Brazil). AB Ibrahim was supported by a fellowship from CAPES (Brazil). We are thankful to Dr. Josias Faria (Embrapa Arroz e Feijão) for providing whiteflies and Dr. Mirella Pupo (UFRJ) for assisting with statistical analyses.
Funding
Funding was provided by Conselho Nacional de Desenvolvimento Científico e Tecnológico and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ibrahim, A.B., Monteiro, T.R., Cabral, G.B. et al. RNAi-mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res 26, 613–624 (2017). https://doi.org/10.1007/s11248-017-0035-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-017-0035-0