Abstract
Whitefly (Bemisia tabaci) is a polyphagous insect that causes huge damage in several horticultural crops, including tomato, by sucking nutrients from the phloem and transmitting viruses. Whiteflies are particularly difficult to manage and the use of chemicals remains the common practice, which causes the development of insecticidal resistance. Thus, there is considerable interest in the introduction of resistance to whitefly by classical and molecular breeding. Here, we explored the concept of using an RNA interference construct to silence a v-ATPase gene in whiteflies interacting with transgenic tomato plants that express siRNA molecules corresponding to a fragment from the B. tabaci vATPase. PCR analyses revealed the presence of both ΔATPase and nptII transgenes in all transgenic lines. siRNA expressing lines were challenged against whitefly and revealed a mortality rate of 57.1% in transgenic line 4.4.1, while in the control the mortality was 7.6%. Mortality of 2nd instar nymphs was higher on the transgenic plants and the development of 3rd instar nymphs was slightly longer than on the control plants. Although the attraction of insects was not significantly different between treatments, the number of eggs laid by the insects on the transgenic plants was significantly lower, compared to the controls. RT-qPCR revealed a decreased expression level of endogenous v-ATPase gene in whiteflies feeding on transgenic plants. No unexpected effect was observed on the non-target insects Myzus persicae or Tuta absoluta. Results presented here may form the foundation for the generation of elite tomato varieties resistant to whitefly, a devastating insect pest.
Key message
Genetically modified tomato was generated to express a vATPase small interfering RNA. Transgenic plants were tolerant to whiteflies (Bemisia tabaci) and revealed no effect on Myzus persicae and Tuta absoluta.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tomato (Solanum lycopersicum) is one of the most important vegetable plants in the world, with global production estimated as 180 million tons (FAOSTAT 2019, http://www.fao.org/faostat/); due to its nutritional importance as food and source of health-promoting compounds, and to the balanced mixture of minerals and antioxidants (Dorais et al. 2008). However, its production is affected by a diversity of insect pests and diseases. Among them, whiteflies (Bemisia tabaci) and whitefly-transmitted viruses present some of the most important constraints worldwide.
Whitefly is recognized as a complex of cryptic species. It is a polyphagous insect that causes huge damage in hundreds of host species, including horticultural crops, such as tomato, lettuce, eggplant, cauliflower, and cucumber (Greathead 1986; Oliveira et al. 2001; Shah and Liu 2013). Besides sucking nutrients from the phloem, which results in accumulation of toxic molecules leading to plant breakdown, B. tabaci transmits several plant viruses, such as begomoviruses (Geminiviridae), criniviruses (Closteroviridae), and torradoviruses (Secoviridae) (Jones 2003; Oliveira et al. 2001; Navas-Castillo et al. 2011) and these then cause yield losses, even with a low insect population. In addition, whitefly excretes honeydew on the leaf surface, which promotes its colonization by Ascomycete fungi, generally called “sooty mold”, which reduce photosynthesis efficiency (Byrne and Bellows Junior 1991; Perring et al. 2018). Whiteflies are particularly difficult to manage, due to their diversity, adaptability, rapid reproduction lifecycle, extensive host range, and the ability to quickly select populations resistant to insecticides, and also to transmit several viruses. Numerous management methods have been employed, such as biological control and agricultural practice strategies. However, the use of chemicals remains the common practice, which causes the development of insecticidal resistance. Consequently, there is considerable interest in the introduction of whitefly resistance into plants by classical and molecular breeding.
RNA interference (RNAi) has evolved as an important defense mechanism in eukaryotes against viruses and transposable elements (Obbard et al. 2009). In insects, RNAi have been reported in species from Lepidoptera, Coleoptera, Diptera, Hemiptera, Hymenoptera, Dictyoptera, Isoptera and Orthoptera orders (Swevers and Smagghe 2012). Three RNAi pathways have been identified mediated by different small RNA molecules playing a role in defense against viruses and transposons (via siRNA), regulating gene expression (via miRNA) and suppression of germ-line transposon expression (via piRNA) (Yan et al. 2020).
Although some insect species are insensitive to RNAi through feeding, gene silencing is usually triggered by supplying exogenous dsRNA by topical applications, such as foliar spray, micro-injection, root dipping and seed treatment, and expression of dsRNA in transgenic plants (for a review see Yan et al. 2020). The effectiveness of RNAi as a tool to generate transgenic crops resistant to insect pests has been demonstrated (Baum et al. 2007; Mao et al. 2007; Gordon and Waterhouse 2007) and is now a reality with the development of some commercial products (Head et al. 2017).
RNAi-mediated gene silencing has been demonstrated in whiteflies. For example, silencing of genes (Cyp315a1, Cyp18a1, EcR and E75) from the ecdysone pathway through leaf-mediated dsRNA feeding assays had a limited effect on the survival and fecundity of adult whiteflies, while in nymphs, gene silencing reduced survival and delayed development (Luan et al. 2013). Raza et al. (2016) reported 70% insect mortality in transgenic tobacco (Nicotiana tabacum) engineered to express dsRNAs for silencing insect aquaporin and alpha glucosidase genes. Similarly, acetylcholinesterase (AChE) and ecdysone receptor (EcR) genes from whitefly were used to induce mortality of up to 90% in tobacco (Malik et al. 2016). Thakur et al. (2014) showed mortality of 34 to 85% among insects feeding on transgenic tobacco leaf discs expressing a v-ATPase gene target. Experiments to express a 370-bp fragment from the green fluorescent protein gene (dsGFP) in transgenic tomato plants revealed its partial non-specific degradation when ingested by the insects (Luo et al. 2017). However, most of the degradation of dsGFP was when it was orally co-administrated with dsRNA against insect dsRNases. In addition, the insect mortality reached 50% over 6 days on artificial diets stacking dsRNA against B. tabaci dsRNases, and two insect coding genes, aquaporin (AQP1) and sucrase (SUC1) (Luo et al. 2017). Recently, Xia et al. (2021) reported resistance in transgenic tomato plants modified to silence the BtPMaT1 gene by impairing whitefly ability to detoxicate plant phenolic glucosides.
We have previously shown mortality of 83–98 % in adult whiteflies and 95% reduction in the number of eggs of whiteflies feeding on transgenic lettuce expressing dsRNA corresponding to a vATPase gene (Ibrahim et al. 2017). Since transmembrane ATPases are crucial for several functions in the cell metabolism, intracellular membrane transport, and their processing, and transport of neurotransmitters, they seem to be promising candidates for engineering RNAi-based resistance to whiteflies in transgenic plants (Beyenbach and Wieczorek 2006; Upadhyay et al. 2011; Thakur et al. 2014; Ibrahim et al. 2017). However, since vATPase genes present some similarities among insect species, the effect of gene silencing on non-target organisms might be a constraint that should be investigated. Here we report the generation of transgenic tomato lines engineered to express dsRNA molecules targeting a B. tabaci vATPase gene. We hypothesized that the expression of this vATPase intron-hairpin construction would interfere in insect survival, generating tolerant tomato plants.
Materials and methods
RNAi construct
The RNAi cassette was cut from the vector pBtATPaseC3300 (Ibrahim et al. 2017) with Hind III and Eco RI and transferred to pCambia2300, generating pC2300ATPase (Fig. 1a), used to transform tomato. In addition, pC2300ATPase has the nptII gene that confers tolerance to kanamycin, used to select the transgenic plants. The interfering cassette containing the v-ATPase sequence hairpin will be referred to hereafter as ΔATPase (Fig. 1a). It is composed of a fragment of 545 bp from the B. tabaci vacuolar ATP synthase catalytic subunit A gene (Ibrahim et al. 2017) cloned in sense and antisense orientations into the vector pSIU (Tinoco et al. 2010), flanked by the intron 3 of the malate synthase gene from Arabidopsis thaliana (Atms-i3) and driven by a double 35 S cauliflower mosaic virus (35SCaMV) promoter plus a leader sequence from Alfalfa mosaic virus (AMV) (Datla et al. 1991).
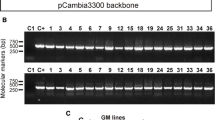
Molecular analyses of transgenic lines. a Schematic representation of the T-DNA region of the pC2300ATPase vector containing the RNAi constructs for sequences specific to the whitefly vATPase gene (ΔATPase), and the nptII gene, which encodes for the enzyme NPTII (neomycin phosphotransferase II) and confers resistance to kanamycin. The backbone is from pCambia2300. b PCR analysis of 9 transformed lines for detection of the ΔATPase and nptII transgenes. C+ is the positive control (vector pC2300ATPase). c ImmunoStrip test of PCR-positive plants for detection of neomycin phosphotransferase II. NT are non-transgenic plants
Tomato genetic transformation
Plant genetic transformation was carried out according to Sun et al. (2006) with modifications. Mature tomato seeds (cv. MicroTom) were surface disinfested at 70% ethanol for 1 min and 2% sodium hypochlorite for 5 min, followed by five rinses with autoclaved distilled water. Seeds were then dried out on filter paper and transferred to half-strength MS medium. Germination was carried out for a period of 6 days under dark conditions, followed by 2 to 4 days at 16 h-photoperiod at 25±2 °C. Cotyledons from germinated seedlings were transversely cut in two halves, and the explants (distal and proximal) were immersed in an agrobacterial suspension for 30 min. Explants were dried out on filter paper and transferred to co-culture medium (MS medium salts, B5 medium vitamins, 200 mg/L PVP, 200 mg/L MES, 5 µM BAP, 0.1 µM IAA, 200 µM acetosyringone, 400 mg/L cysteine, 3% sucrose, 0.6% agar, pH 5.2) with the abaxial surface facing downwards, for 48 h in the dark at 25±2 °C. After two days, explants were transferred to MS medium supplemented with 200 mg/L PVP, 200 mg/L MES, 2 mg/L silver nitrate, 5 µM zeatin, 0.1 µM IAA, 2% sucrose, B5 medium vitamins, 100 mg/L Timentin, 200 mg/L cefotaxime, 50 mg/L kanamycin and 0.6% agar, pH 5.7. After 3 to 4 weeks, the concentration of kanamycin was reduced to 25 mg/L and the explants were sub-cultured every 15 days, until formation of plantlets. Emerging shoots were transferred to an elongation medium (MS medium salts supplemented with 3% sucrose, B5 medium vitamins, 100 mg/L Timentin, 200 mg/L cefotaxime, 25 mg/L kanamycin and 0.6% agar, pH 5.7). Well-developed and elongated shoots were transferred to a rooting medium (MS medium salts, B5 medium vitamins, 1 mg/L IBA, 100 mg/L Timentin, 200 mg/L cefotaxime, 25 mg/L kanamycin and 3% sucrose, 0.6% agar, pH 5.7). With the full development of the roots, plants were acclimated in a greenhouse and allowed to set seeds. Agrobacterial suspension was prepared cultivating Agrobacterium tumefaciens EHA105 on solid AB medium (Chilton et al. 1974) containing 100 mg/L rifampicin and 100 mg/L kanamycin for 2 to 3 days at 28 °C. Then bacterial culture was suspended in liquid co-culture medium and diluted to OD600 = 0.5.
Screening of transgenic plants
Regenerated plants were screened for the presence of the neomycin phosphotransferase II (NPTII) using the ImmunoStrip (Agdia, USA), according to the manufacturer’s recommendations. DNA from leaves was isolated using a modified version of Doyle and Doyle (1987) and amplified by PCR analyses using the primer pairs ATPXS1 (TTCTAGAGCTCTATCACACTATCTGAGTAC) / ATPSK1 (GGTACCACTAGTGGGAAGTTTTTATCGTAG) for the detection of ΔATPase and NPTII 60 (GAGGCTATTCGGCTATGACTG) / NPTII 470 (TCGACAAGACCGGCTTCCATC) for the detection of the nptII gene. PCR was carried out according to Bonfim et al. (2007). The reaction was subjected to 95 °C (5 min) for denaturation, and amplified by 35 cycles (95 °C for 1 min, 55 °C for 1 min, 72 °C for 2 min) with a final elongation cycle of 7 min at 72 °C.
Progeny analysis
Seeds of the first generation (T1) of self-pollinated plants were germinated and analyzed for the presence of ΔATPase by PCR, as described. Pearson’s Chi squared (Χ2) was used to determine whether the observed segregation ratio was consistent with a Mendelian ratio of 3:1, at 95% level of confidence. Yates’s correction factor was used when one of the frequencies was less than 5, or when the total value of plants evaluated was less than 20.
Analysis of siRNAs
Total RNA from tomato transgenic (in homozygosis) and non-transgenic lines was extracted with Trizol (Invitrogen) as recommended by the manufacturer. The siRNA analysis was carried out as described by Bonfim et al. (2007). Hybridization was performed using a DNA probe corresponding to the vATPase PCR fragment amplified using the primer pair ATPXS1/ATPSK1 labeled with α32P dCTP and the DecaLabel DNA Labeling Kit (Thermo Fisher Scientific), according to the manufacturer’s instructions. Hybridization and post-hybridization washes were conducted as described by Yoo et al. (2004). Oligomers (18, 25 and 39 nucleotides) were used as molecular size markers. The bands were visualized with a fluorescent image analyzer (FLA-3000; Fujifilm).
Whitefly mortality, development and preference assays
Bemisia tabaci colonies have been maintained in common beans in a screenhouse at Embrapa Arroz e Feijão, in Santo Antonio de Goias, Goias state, Central Brazil, for over ten years. Care has been taken to avoid inbreeding depression by introducing new specimens regularly into the colony. The whitefly population in our colony was identified as B. tabaci MEAM1 (=biotype B) and it has been monitored by sequencing the mitochondrial cytochrome oxidase I (mtCOI) gene.
Whitefly mortality was evaluated on 4-week-old plants from the following transgenic tomato lines: 4.4.1, 6.8.4, 9.2 and control (non-transgenic plants). Three plants of each line were submitted to whitefly oviposition for 2 h. After that, the adult whiteflies were removed and plants were isolated in individual cages (68 cm x 27 cm x 27 cm) in the greenhouse. Insects developed from eggs into adults, and adult mortality was registered after 5 to 7 days. Plants were placed over a black plastic canvas, so dead insects were easily found inside the cage. On the seventh day, the number of living and dead insects was counted, as the insects were removed from the cages using an insect aspirator. Then, the number of empty pupa cases on the leaves was counted under stereomicroscope. To calculate the percentage of mortality, the number of empty pupa cases was used as the total number of adult insects in each replicate. This experiment was repeated twice.
Insect development from egg to adult was evaluated on 10 individual eggs per plant on tomato line 4.4.1 and the control (non-transgenic plants), with 5-8 replicates. For that, 4-week-old plants were exposed to whitefly oviposition for 1 h, the adults were removed. When the 1st instar nymphs became sessile, their spots on the leaf were circled and numbered using a permanent marker, taking care not to damage the plant or the insect. Plants were maintained at 25 ± 1°C, with 70–80 % relative humidity and 16 h/8 h light/dark conditions. The nymphal stage of each individual was recorded daily, until adult emergence, using a stereomicroscope. Mortality in each stage was also recorded.
A free-choice experiment was carried out to evaluate preference and attraction of adult whiteflies to the transgenic tomato lines. For that, 4-week-old plants of the line 4.4.1 and control plants (10 replicates per treatment) were intercalated in a circle, in a whitefly-free screenhouse and then, 300 adult insects were released in the center of the circle. After 48 h, the number of adults in each plant was counted, by carefully looking at the leaves. After five days, the number of eggs per plant was counted using a stereomicroscope.
Quantitative real-time PCR
The transcription level of the endogenous v-ATPase gene in B. tabaci interacting with transgenic line 4.4.1 and non-transgenic tomato leaves was determined by quantitative reverse transcription. RNA isolation and RT-qPCR were performed as described by Andrade et al. (2016). Whiteflies were released into bioassay systems containing tomato plants as described above for the whitefly mortality assays. After 48 h, total RNA was extracted from 150 surviving insects per replicate, of which 200 ng from each of the samples was used to synthesize cDNA using Promega GoScript Reverse Transcription System (Promega) as recommended by the manufacturer. Reactions were performed using the StepOnePlus Real-Time PCR System (Thermo Fisher Scientific) with SYBR-green detection. The primers for the endogenous vATPase (GGTGGAGACTTGTACGGTATTG and TGACAGTACCTTTGGCTTTAGG) and actin (GACCAGCCAAGTCCAAACGA and CCTTTGTGGTAGAGGTCTCAGTT) genes were designed using the PrimerQuest Tool (IDT Integrated DNA Technologies, Inc.). The relative vATPase transcription levels in different RNA samples were normalized with respect to the internal standard actin gene. Triplicate quantitative assays were performed from three biological samples. The relative level of the expression was calculated using the Livak method (Livak and Schmittgen 2001).
Effect on non-target organisms
In order to determine whether the transgenic tomato plants present negative effects on non-target insects that interact with tomato plants, two assays were carried out: the first one with Myzus persicae (Hemiptera: Aphidae), which has the same feeding habit as the whitefly, and the second with Tuta absoluta (Lepidoptera: Gelechiidae).
M. persicae fourth instar nymphs were transferred to tomato plants (50 insects per plant), and aphid numbers were counted after seven days of feeding. Plants were maintained at 25 ± 2°C, with 70–90 % relative humidity, under greenhouse conditions without additional illumination. Six transgenic and non-transgenic plants were used. M. persicae were collected from naturally occurring colonies on weed plants, and kept on tomato plants until used for these experiments.
Tuta absoluta (three L1 to L3 instar caterpillars per leaf) were transferred to five leaves detached from transgenic and non-transgenic tomato plants maintained in two-compartment Petri dishes. After 27 days, the number of emerging adults was recorded, expressed as the percentage of normal adults related to the total number of caterpillars. Insect colonies were obtained from tomato plants cultivated under greenhouse conditions at Embrapa Hortaliças (Brasília, DF).
Statistical analysis
The experiments were carried out in a completely randomized design. Data were analyzed by one-way ANOVA. Percentage data were transformed by arcsine square root before analysis. Levene’s test was applied to check for the homogeneity of variance. Mean comparisons were performed by Dunnett’s test, as implemented in GraphPad Prism 6.0 software. Mortality of B. tabaci nymphs developed on the tomato transgenic line was compared to the control line using a chi-square test (p<0.05), for each nymphal stage. The Shapiro-Wilk normality test was conducted to test for the normality of data and when data was not in a normal distribution, a non-parametric Wilcoxon rank sum test with continuity correction was applied for mean comparisons. The mean number of adults sitting on the plants and eggs laid, in the attraction and preference assays with B. tabaci, was compared between treatments using the t test (p<0.05).
Results
To determine the effectiveness of siRNA to produce tomato plants tolerant to whitefly by silencing vATPase, nine independent transgenic lines were generated with the pC2300ATPase vector, containing the intron-spliced hpRNA interfering cassette (ΔATPase) (Fig. 1a). PCR analyses revealed the presence of both ΔATPase and nptII transgenes in all transgenic lines (Fig. 1b). Progenies were analyzed for the presence of the neomycin phosphotransferase II protein (NPT II) using the immunochromatographic method (Fig. 1c). A strong signal was observed in lines 6.8.4, 6.10.3, 9.8 and 8.9. However, lines 10.20 and 4.12.1 presented a weak signal, while no NPT II band was observed for lines 4.4, 9.2, 10.5 and non-transgenic plants (control) (Fig. 1c).
Except for the line 6.10.3, primary transformants (T0) were cultivated in the greenhouse and presented normal phenology and phenotype (plant height, number of leaves, number of branches and number of flowers and fruits). Line 6.10.3 presented several fruits without seeds and a significantly lower number of seeds per fruit (average of 1.3 seeds/fruit) when compared to the other transgenic lines and control plants (average of 27.9 seeds/fruit). Line 6.10.3 was not used for further studies.
Based on the PCR analyses, the chi-squared test revealed that both ΔATPase and nptII transgenes co-segregated in the T1 generation (self-pollinated), closely fitting the Mendelian ratio of 3:1, as expected for one integration locus (Table 1).
Northern analyses were carried out to detect the ΔATPase siRNA in leaves from phenotypically normal lines of transgenic and non-transgenic tomato. Results revealed the presence of siRNA bands of expected size range for transgenic lines 4.4.1, 6.8.4, 9.2, 10.20, 8.9, 10.5, and 4.12.1. Lines 4.4.1, 6.8.4 and 9.2, which presented strong, intermediate and weak signals, respectively (Fig. 2), were used for the mortality assays. No signal was observed in non-transgenic plants. No direct correlation was observed between nptII gene expression (Fig. 1c) and ΔATPase expression (amounts of siRNA) (Fig. 2).
Line 4.4.1, which presented higher siRNA amounts in the Northern analyses, was chosen for the whitefly feeding experiments. Lines 6.8.4 and 9.2, which presented weaker signals, were also chosen as an attempt to establish the cause-effect relationship between insect tolerance and amounts of siRNA produced by the host. Tomato plants were challenged with newly emerged adult whiteflies and their mortality recorded after five to seven days (Fig. 3a). The results revealed that whiteflies that were fed on the transgenic line 4.4.1 presented statistically significant higher mortality rate (57.1%), when compared to insects fed on lines 6.8.4 (23.5% mortality), 9.2 (27.5% mortality), and non-transgenic control (7.6% mortality) plants (P<0.05) (Fig. 3a). Quantitative real-time PCR was carried out to quantify the transcription level of the endogenous v-ATPase gene in B. tabaci feeding on the transgenic tomato leaves from line 4.4.1. The results showed that vATPase expression was significantly lower (by 50.2%; P<0.05) in insects interacting with the transgenic plants, when compared to the level observed in insects interacting with non-transgenic plants (Fig. 3b). Insect developmental time on tomato line 4.4.1 was similar to that on the non-transgenic control plants, except for the 3rd instar, which was significantly longer on the transgenic plant (P<0.05) (Fig. 3c), suggesting a slight delay in their nymphal stage development. Insect mortality was significantly higher on the transgenic line 4.4.1 than on the control plants only for the 2nd instar (Table 2). In the preference assay, the number of adult insects per plant did not differ between treatments. However, the number of eggs per plant was significantly lower in the transgenic plants (Table 3).
Bioassays using transgenic tomato lines compared to non-transgenic for feeding B. tabaci. a Average mortality of B. tabaci feeding on transgenic tomato lines for a period of 7 days (*p<0.05, n=6). b Relative expression of v-ATPase in insects feeding on transgenic tomato line 4.4.1 for 48 h, determined by quantitative real-time RT-PCR. Data represent means of three replications (*p<0.05, n=3). c Developmental time of B. tabaci nymphal stages on the tomato line 4.4.1, compared to the non-transgenic control line (*p<0.05, n=50–80). Bars represent ± standard errors
Due to the similarities in vATPase gene sequences among insects, two insect species [M. persicae (Hemiptera: Aphidae) and T. absoluta (Lepidoptera: Gelechiidae)], which closely interact with tomato leaves, were chosen for a non-target assay. Results showed no difference in the progeny number of M. persicae and percentage of emerging adults of T. absoluta after feeding on transgenic line 4.4.1 (Fig. 4), as compared to the insects that fed on the non-transgenic plants. The results suggest that the siRNA produced by the expression of the ΔATPase transgene did not significantly impair the expression of endogenous ATPase coding genes in M. persicae and T. absoluta.
Non-target assays using transgenic tomato plants to feed Myzus persicae (Hemiptera: Aphidae) or Tuta absoluta (Lepidoptera: Gelechiidae). a Progeny average of 50 M. persicae adults per replicate (plants of line 4.4.1). b Percentage of emerging adults of T. absoluta on detached leaves of transgenic line 4.4.1 (*p < 0.05, n=5)
Discussion
Whiteflies are among the most important insect pests that impair the production of vegetable crops worldwide, especially in tropical regions, where their control is even more challenging. In high-incidence regions, their control is essentially based on the use of insecticides, with partial efficacy, negative environmental effects, reduction of abundance and diversity of natural enemies, and selection of pesticide-resistant whitefly populations. Thus, the best way to manage this pest in the field is to develop plant genetic resistance or tolerance. The concept of using RNA interference (RNAi) constructs to silence key genes in insect pests promotes the development of novel approaches toward plant protection in a sustainable way (Yan et al. 2020).
Several target genes have been proposed for engineering insect resistance in plants using plant-mediated RNAi strategies. vATPase coding genes are among the most promising targets for gene silencing, which has been applied to some model plants and commercial crops (Yan et al. 2020; Liu et al. 2020; Ibrahim et al. 2017). In the present study, we showed that the expression of siRNA corresponding to sequences of a vATPase gene from B. tabaci conferred tolerance to this insect pest in genetically modified tomato. In addition, a significant reduction in endogenous gene expression was observed, revealed by RT-qPCR analysis, associated with 57% mortality in adult insects. Previously, we had expressed vATPase siRNA in lettuce plants and observed whitefly adult mortality between 83.8 and 98.1% within 5 days of feeding on transgenic plants (Ibrahim et al. 2017). Although the percentage of insect mortality was lower, our present results with this tomato line suggest a reasonable level of tolerance to the target insect pest, which might represent a promising tool to be used along with other management strategies. Baum et al. (2007) demonstrated that vATPaseA dsRNA supplied in an artificial diet, and expressed in transgenic maize, triggered RNA interference in Diabrotica virgifera, resulting in larval stunting and mortality. Thakur et al. (2014) reported a 34 to 85% mortality among whiteflies feeding on leaf discs from transgenic tobacco expressing siRNA from a v-ATPase. Our results showed higher mortality of 2nd instar nymphs and a slight delay in the development of 3rd instar nymphs feeding on the tomato line 4.4.1, suggesting that the development of 2nd and 3rd instars was impaired by the transgenic line. Additionally, although the attraction of insects was not significantly different between treatments, the number of eggs laid by the insects on the transgenic plants was significantly lower, corroborating the results obtained with the lettuce transgenic plants carrying the same RNAi construct as we used in the present work (Ibrahim et al. 2017). In phloem feeders, host plant selection starts with olfactory cues, meaning that the insects assess the suitability of a host plant mainly by the plant volatiles (Powell et al. 2006). Our data shows that the number of adults settling on the transgenic plants did not differ from the conventional control plants, suggesting that the volatiles produced by both plants might be similar. However, other works have shown that even when the attraction of adults does not change among different treatments, a reduction in the oviposition might occur (Jiao et al. 2012; Li et al. 2017), suggesting that attraction of adults and preference for oviposition are not necessarily correlated. The host plant selection for oviposition by insects might be affected by differences in nutritional and defensive chemistry (Bernays and Chapman, 1994), such as the amount of wax on the leaves, presence of trichomes and glucosinolate, salicylic acid accumulation or the contents of nitrogen, carbohydrate, and phenolic compounds or chlorophyl, for example (Oriani and Vendramim, 2010; Jiao et al. 2012; Moshitzky and Morin, 2014; Ueda et al. 2019; Jafarbeigi et al. 2020; Abbasi et al. 2020). Moreover, treating plants with chemicals, such as essential oils, amino acids or insecticides, for example, can potentially reduce the preference of whiteflies for oviposition (Prabhaker et al. 1999; Hammad et al. 2001; Baldin et al. 2007; Yang et al. 2010), suggesting that oviposition might be reduced when the females perceive a potential hazard to the progeny (Moshitzky and Morin, 2014), or in other words, oviposition preference correlates with host suitability for their offspring performance (preference–performance hypothesis) (Jaenike, 1978). Therefore, several factors might lead to a reduction in insect oviposition, and so we cannot rule out the potential effect of silencing a gene involved in many cellular processes, as the vATPase. Further research will be conducted in the future to investigate this hypothesis.
Luo et al. (2017) have shown that transgenic tomato plants engineered to express a dsRNA 370-bp fragment from the green fluorescent protein gene (dsGFP) was partially degraded when ingested by B. tabaci. In addition, they demonstrated that most of the dsGFP degradation was suppressed when it was orally co-administrated with dsRNA targeting insect dsRNases. Although the co-suppression of insect RNases genes could prevent the degradation of dsRNA for target genes, high levels of silencing could be achieved by higher expression levels of the intron-hairpin constructs (Luan et al. 2013, Thakur et al. 2014, Raza et al. 2016, Malik et al. 2016, Ibrahim et al. 2017 and Xia et al. 2021).
Since vATPase sequences share up to 83% nucleotide-sequence identities among insect pest and beneficial insect genomes, negative effects on the life cycle of non-target insect species may occur (Lundgren and Duan 2013). It may not be a problem in pest management (Yan et al. 2020) but it could affect the commercial approval of genetically modified crops. We tested transgenic tomatoes against two non-target insects, M. persicae and T. absoluta, which closely interact with tomato leaves. Despite similarities in the vATPase gene sequences of 83% and 85% for M. persicae and T. absoluta, respectively, we observed no negative effect on insect reproduction, survival and development. However, although the draft genome assembly of T. absoluta (Tabuloc et al. 2019) is available, only a few incomplete ATPase sequences are currently available in the NCBI GenBank. In silico comparison with M. persicae using the si-Fi Software (Lück et al. 2019) revealed just a few putative sequences presenting potential similarity for an unexpected gene silencing effect on this non-target insect species. Nevertheless, biosafety studies should be further carried out under field conditions to determine the effect of this tomato transgenic line on a greater number of insect species.
Although a correlation between siRNA amounts found in tomato leaves and tolerance has been established, no correlation was observed between the expression of the nptII selective gene and the ΔATPase transgenes. Similarly, in the transgenic common bean engineered to express dsRNA targeting the rep gene from Bean golden mosaic virus, no correlation was found between the amount of rep siRNA and the ahas gene expression, used for selection of transformed plants (Aragão et al. 2013).
RNAi efficiency is dependent on the siRNA (or dsRNA) dose (Vogel et al. 2019). In this study, the double 35 S CaMV promoter was used to control ΔATPase. Better promoters might be identified to promote higher levels of transcription and consequently greater amounts of dsRNA into tomato leaves.
Collectively, the results presented here demonstrated the generation of whitefly-tolerant tomato plants mediated by the RNAi strategy. A correlation between the amount of siRNA vATPase and the insect tolerance was observed for the transgenic line 4.4.1. RT-qPCR analysis established the cause-effect relationship between silencing of the endogenous BtvATPase gene and adult mortality. Results presented here may form the foundation for the generation of elite tomato varieties resistant to whitefly, a devastating insect pest for horticultural crops.
Data availability
The data generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- 35SCaMV :
-
35 S promoter from the cauliflower mosaic virus.
- AChE :
-
Acetylcholinesterase.
- AMV :
-
Alfalfa mosaic virus.
- Atms-i3 :
-
Intron 3 of the malate synthase gene from Arabidopsis thaliana.
- ATP :
-
Adenosine triphosphate.
- B5 :
-
Basal salt mixture containing micro and macro elements with vitamins as described by Gamborg et al. (1968)
- BAP :
-
6-Benzylaminopurine.
- cDNA :
-
Complementary DNA.
- dsRNA :
-
Double-stranded RNA.
- EcR :
-
Ecdysone receptor.
- EHA105 :
-
Elizabeth Hood Agrobacterium 105.
- hpRNA :
-
Hairpin RNA.
- IAA :
-
Indole-3-acetic acid.
- IBA :
-
Indole-3-butyric acid.
- MEAM1 :
-
Middle East Asia Minor – 1 = Biotype B.
- miRNA :
-
MicroRNA.
- mtCOI :
-
Mitochondrial cytochrome oxidase I.
- NPTII :
-
Neomycin Phosphotransferase II.
- PVP :
-
Polyvinylpyrrolidone.
- RNAi :
-
RNA interference.
- siRNA :
-
Small interfering RNAs.
- vATPase :
-
Vacuolar ATPase.
References
Abbasi A, Sufyan M, Arif MJ, Sahi TS (2020) Effect of silicon on oviposition preference and biology of Bemisia tabaci (Gennadius) (Homoptera: Aleyrodidae) feeding on Gossypium hirsutum (Linnaeus). Int J Pest Manag. https://doi.org/10.1080/09670874.2020.1802084
Andrade CM, Tinoco MLP, Rieth AF, Maia FCO, Aragão FJL (2016) Host-induced gene silencing in the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Pathol 65(4):626–632. https://doi.org/10.1111/ppa.12447
Aragão FJL, Nogueira EOPL, Tinoco MLP, Faria JC (2013) Molecular characterization of the first commercial transgenic common bean immune to the Bean golden mosaic virus. J Biotechnol 166(1–2):42–50. https://doi.org/10.1016/j.jbiotec.2013.04.009
Baldin ELL, Vendramim JD, Lourenção AL (2007) Interaction between resistant tomato genotypes and plant extracts on Bemisia tabaci (Genn.) biotype B. Sci. agric. (Piracicaba. Braz) 64(5):476–481. https://doi.org/10.1590/S0103-90162007000500004
Baum JA, Bogaert T, Clinton W, Heck GR, Feldmann P, Ilagan O, Johnson S, Plaetinck G, Munyikwa T, Pleau M, Vaughn T, Roberts J (2007) Control of coleopteran insect pests through RNA interference. Nat Biotechnol 25:1322–1326. https://doi.org/10.1038/nbt1359
Bernays EA, Chapman RE (eds) (1994) Chemicals in Plants. In: Host-Plant Selection by Phytophagous Insects. Springer US, Boston, pp 14–60
Beyenbach KW, Wieczorek H (2006) The V-type H+ ATPase: molecular structure and function, physiological roles and regulation. J Exp Biol 209:577–589. https://doi.org/10.1242/jeb.02014
Bonfim K, Faria JC, Nogueira EOPL, Mendes EA, Aragão FJL (2007) RNAi-mediated resistance to Bean golden mosaic virus in genetically engineered common bean (Phaseolus vulgaris). Mol Plant-Microbe Interact 20(6):717–726. https://doi.org/10.1094/MPMI-20-6-0717
Byrne DN, Bellows Junior TS (1991) Whitefly biology. Annu Rev Entomol 36:431–457. https://doi.org/10.1146/annurev.en.36.010191.002243
Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW (1974) Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. Proc Natl Acad Sci USA 71(9):3672–3676. https://doi.org/10.1073/pnas.71.9.3672
Datla RS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G (1991) A bifuntional fusion between beta-glucuronidase and neomycin phosphotransferase: a broad-spectrum marker enzyme for plants. Gene 101(2):239–246. https://doi.org/10.1016/0378-1119(91)90417-a
Dorais M, Ehret DL, Papadopoulos AP (2008) Tomato (Solanum lycopersicum) health components: from the seed to the consumer. Phytochem Rev 7:231–250. https://doi.org/10.1007/s11101-007-9085-x
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bull 19:11–15
Gordon KHJ, Waterhouse PM (2007) RNAi for insect-proof plants. Nat Biotechnol 25(11):1231–1232. https://doi.org/10.1038/nbt1107-1231
Greathead AH( (1986) Host plants. In: Cock) MJW (ed) Bemisia tabaci. A Literature Survey on the Cotton Whitefly with an Annotated Bibliography. C.A.B International Institute of Biological Control, Silwood Park, pp 17–26
Hammad EMA-F, Zournajian H, Talhouk S (2001) Efficacy of extracts of Melia azedarach L. callus, leaves and fruits against adults of the sweetpotato whitefly Bemisia tabaci (Hom. Aleyrodidae) J Appl Entomol 125(8):483–488. https://doi.org/10.1046/j.1439-0418.2001.00577.x
Head GP, Carroll MW, Evans SP, Rule DM, Willse AR, Clark TL, Storer NP, Flannagan RD, Samuel LW, Meinke LJ (2017) Evaluation of SmartStax and SmartStax PRO maize against western corn rootworm and northern corn rootworm: efficacy and resistance management. Pest Manage Sci 73(9):1883–1899. https://doi.org/10.1002/ps.4554
Ibrahim AB, Monteiro TR, Cabral GB, Aragão FJL (2017) RNAi-mediated resistance to whitefly (Bemisia tabaci) in genetically engineered lettuce (Lactuca sativa). Transgenic Res 26(5):613–624. https://doi.org/10.1007/s11248-017-0035-0
Jaenike J (1978) On optimal oviposition behavior in phytophagous insects. Theor Popul Biol 14(3):350–356. https://doi.org/10.1016/0040-5809(78)90012-6
Jafarbeigi F, Samih MA, Alaei H, Shirani H (2020) Induced tomato resistance against bemisia tabaci triggered by salicylic acid, β-aminobutyric acid, and trichoderma. Neotrop Entomol 49(3):456–467. https://doi.org/10.1007/s13744-020-00771-0
Jiao X, Xie W, Wang S, Wu Q, Zhou L, Pan H, Liu B, Zhang Y (2012) Host preference and nymph performance of B and Q putative species of Bemisia tabaci on three host plants. J Pest Sci 85:423–430. https://doi.org/10.1007/s10340-012-0441-2
Jones DR (2003) Plant viruses transmitted by whiteflies. Eur J Plant Pathol 109(3):195–219. https://doi.org/10.1023/A:1022846630513
Li J, Rao L, Xie H, Schreiner M, Chen L, Liu Y (2017) Morphology and glucosinolate profiles of chimeric Brassica and the responses of Bemisia tabaci in host selection, oviposition and development. J Integr Agric 16(9):2009–2018. https://doi.org/10.1016/S2095-3119(16)61617-9
Liu S, Jaouannet M, Dempsey D’MA, Imani J, Coustau C, Kogel K-H (2020) RNA-based technologies for insect control in plant production. Biotechnol Adv 39:107463. https://doi.org/10.1016/j.biotechadv.2019.107463
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Luan J-B, Ghanim M, Liu S-S, Czosnek H (2013) Silencing the ecdysone synthesis and signaling pathway genes disrupts nymphal development in the whitefly. Insect Biochem Mol Biol 43(8):740–746. https://doi.org/10.1016/j.ibmb.2013.05.012
Lück S, Kreszies T, Strickert M, Schweizer P, Kuhlmann M, Douchkov D (2019) siRNA-Finder (si-Fi) Software for RNAi-Target Design and Off-Target Prediction. Front Plant Sci 10:1023. https://doi.org/10.3389/fpls.2019.01023
Lundgren JG, Duan JJ (2013) RNAi-based insecticidal crops: potential effects on nontarget species. Bioscience 63(8):657–665. https://doi.org/10.1525/bio.2013.63.8.8
Luo Y, Chen Q, Luan J, Chung SH, Eck JV, Turgeon R, Douglas AE (2017) Towards an understanding of the molecular basis of effective RNAi against a global insect pest, the whitefly Bemisia tabaci. Insect Biochem Mol Biol 88:21e29. https://doi.org/10.1016/j.ibmb.2017.07.005
Malik HJ, Raza A, Amin I, Scheffler JA, Scheffler BE, Brown JK, Mansoor S (2016) RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci Rep 6(1):38469. https://doi.org/10.1038/srep38469
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY (2007) Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol 25(11):1307–1313. https://doi.org/10.1038/nbt1352
Moshitzky P, Morin S (2014) Bemisia tabaci females from the Mediterranean (Q) species detect and avoid laying eggs in the presence of pyriproxyfen, a juvenile hormone analogue. Pest Manag Sci 70(10):1468–1476. https://doi.org/10.1002/ps.3698
Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S (2011) Emerging virus diseases transmitted by whiteflies. Annu Rev Phytopathol 49(15):219–248. https://doi.org/10.1146/annurev-phyto-072910-095235
Obbard DJ, Gordon KHJ, Buck AH, Jiggins FM (2009) The evolution of RNAi as a defence against viruses and transposable elements. Phil Trans R Soc Lond B Biol Sci 364(1513):99–115. https://doi.org/10.1098/rstb.2008.0168
Oliveira MRV, Henneberry TJ, Anderson P (2001) History, current status, and collaborative research projects for Bemisia tabaci. Crop Prot 20(9):709–723. https://doi.org/10.1016/S0261-2194(01)00108-9
Oriani MA, de G, Vendramim JD, (2010) Influence of trichomes on attractiveness and ovipositional preference of Bemisia tabaci (Genn) B biotype (Hemiptera: Aleyrodidae) on tomato genotypes. Neotrop Entomol 39(6):1002–1007. https://doi.org/10.1590/S1519-566-2010000600024
Perring TM, Stansly PA, Liu TX, Smith HA, Andreason SA (2018) Whiteflies: Biology, Ecology, and Management. In: Wakil W, Brust GE, Perring TM Sustainable Management of Arthropod Pests of Tomato. Academic Press - Elsevier Science 372: 73-110 https://doi.org/10.1016/B978-0-12-802441-6.00004-8.
Powell G, Tosh CR, Hardie J (2006) Host plant selection by aphids : Behavioral, evolutionary, and applied perspectives. Annu Rev Entomol 51:309–330. https://doi.org/10.1146/annurev.ento.51.110104.151107
Prabhaker N, Toscano NC, Henneberry TJ (1999) Comparison of neem, urea, and amitraz as oviposition suppressants and larvicides against Bemisia argentifolii (Homoptera: Aleyrodidae). J Econ Entomol 92(1):40–46. https://doi.org/10.1093/jee/92.1.40
Raza A, Malik HJ, Shafiq M, Amin I, Scheffler JA, Scheffler BE, Mansoor S (2016) RNA Interference based Approach to Down Regulate Osmoregulators of Whitefly (Bemisia tabaci): Potential Technology for the Control of Whitefly. PLoS One 11(4):e0153883. https://doi.org/10.1371/journal.pone.0153883
Shah MMR, Liu TX (2013) Feeding Experience of Bemisia tabaci (Hemiptera: Aleyrodidae) Affects Their Performance on Different Host Plants. PLoS One 8(10):e77368. https://doi.org/10.1371/journal.pone.0077368
Sun HJ, Uchii S, Watanabe S, Ezura H (2006) A highly efficient transformation protocol for Micro-Tom, a model cultivar for tomato functional genomics. Plant Cell Physiol 47(3):426–431. https://doi.org/10.1093/pcp/pci251
Swevers L, Smagghe G (2012) Use of RNAi for control of insect crop pests. In: Smagghe G, Diaz I (eds) Arthropod-plant interactions: novel insights and approaches for IPM, Progress in Biological Control 14, Netherlands, pp 177–197. https://doi.org/10.1007/978-94-007-3873-77
Tabuloc CA, Lewald KM, Conner WR, Lee Y, Lee EK, Cain AB, Godfrey KE, Arnó J, Agustí N, Perini CR, Guedes JC, Zalom FG, Chiu JC (2019) Sequencing of Tuta absoluta genome to develop SNP genotyping assays for species identification. J Pest Sci 92:1397–1407. https://doi.org/10.1007/s10340-019-01116-6
Thakur N, Upadhyay SK, Verma PC, Chandrashekar K, Tuli R, Singh PK (2014) Enhanced whitefly resistance in transgenic tobacco plants expressing double stranded RNA of v-ATPase A gene. PLoS One 9(3):e87235. https://doi.org/10.1371/journal.pone.0087235
Tinoco MLP, Dias BBA, Dall’Astta RC, Pamphile JA, Aragão FJL (2010) In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol 8(27):27. https://doi.org/10.1186/1741-7007-8-27
Ueda H, Kugimiya S, Tabata J, Kitamoto H, Mitsuhara I (2019) Accumulation of salicylic acid in tomato plant under biological stress affects oviposition preference of Bemisia tabaci. J Plant Interact 14(1):73–78. https://doi.org/10.1080/17429145.2018.1550220
Upadhyay SK, Chandrashekar K, Thakur N, Verma PC, Borgio JF, Singh PK, Tuli R (2011) RNA interference for the control of whiteflies (Bemisia tabaci) by oral route. J Biosci 36(1):153–161. https://doi.org/10.1007/s12038-011-9009-1
Vogel E, Santos D, Mingels L, Verdonckt T-W, Broeck JV (2019) RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front Physiol 9:1912. https://doi.org/10.3389/fphys.2018.01912
Xia J, Guo Z, Yang Z, Han H, Wang S, Xu H, Yang F, Wu Q, Xie W, Zhou X, Dermauw W, Turlings TCJ, Zhang Y (2021) Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184:1–13. https://doi.org/10.1016/j.cell.2021.02.014
Yan S, Ren B, Zeng B, Shen J (2020) Improving RNAi efficiency for pest control in crop species. Biotechniques 68(5):283–290. https://doi.org/10.2144/btn-2019-0171
Yang N-W, Li A-L, Wan F-H, Liu W-X, Johnson D (2010) Effects of plant essential oils on immature and adult sweetpotato whitefly, Bemisia tabaci biotype B. Crop Prot 29(10):1200–1207. https://doi.org/10.1016/j.cropro.2010.05.006
Yoo BC, Kragler F, Varkonyi Gasic E, Haywood V, Archer Evans S, Lee YM, Lough TJ, Lucas WJ (2004) A systemic small RNA signaling system in plants. Plant Cell 16:1979–2000 https://doi.org/10.1105/tpc.104.023614
Acknowledgements
We would like to thank Dr. Miguel Michereff Filho (Embrapa Hortaliças, Brasília, DF, Brazil) for providing the T. absoluta and Dr. Josias C. Faria (Embrapa Arroz e Feijão, Santo Antônio de Goiás, GO, Brazil) for providing the whitefly colony. This research was partially supported by the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq, Brazil, Grant Number 429161/2018-2).
Funding
Funding was provided by Empresa Brasileira de Pesquisa Agropecuária, Conselho Nacional de Desenvolvimento Científico e Tecnológico and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior.
Author information
Authors and Affiliations
Contributions
Investigation [CSRP, WRR, MCM, ALF, PVP, GBC]; Methodology [CSRP, WRR, MCM, FJLA]; Formal analysis [CSRP, PVP, FJLA]; Writing- Reviewing & Editing [CSRP, PVP, GBC, FJLA]; Conceptualization [FJLA]; Funding acquisition [FJLA], Supervision [FJLA]. All of the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no declarations of interest.
Additional information
Communicated by Sergio J. Ochatt.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Pizetta, C.S.R., Ribeiro, W.R., Ferreira, A.L. et al. RNA interference-mediated tolerance to whitefly (Bemisia tabaci) in genetically engineered tomato. Plant Cell Tiss Organ Cult 148, 281–291 (2022). https://doi.org/10.1007/s11240-021-02185-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02185-1