Abstract
Gene expression in plants is primarily modulated by the promoter region, which is located upstream of the gene coding region. Specific regulatory sequences within the promoter region contribute to the strength, tissue specificity, timing and duration of gene expression. In this study, promoter regions were isolated from 40 different soybean (Glycine max) genes that were grouped into five distinct categories, based on the localization of native gene expression (constitutively expressed genes, root-preferentially expressed genes, seed-preferentially expressed genes), or the associated biological processes of native gene expression (fatty-acid biosynthesis pathway associated genes, and WRKY transcription factor genes). Each of the promoters was placed upstream of the green fluorescent protein coding sequence and promoter functionality was then assessed using transient expression analysis following particle bombardment of lima bean (Phaseolus lunatus) cotyledons, as well as stable expression analysis following Agrobacterium rhizogenes induction of soybean hairy roots. Surprisingly, promoter strength was relatively consistent using the two different validation methods while tissue specific expression was not observed, likely due to a combination of the target tissue, the large amount of DNA that was introduced and minimal epigenetic effects. The differences in gene expression within members of each promoter group indicates that promoters need to be characterized in a transgenic environment, rather than relying on expression predicted using native gene expression profiles. This systematic approach into the characterization of native soybean promoters adds to the knowledge base of soybean gene regulation and the toolbox of promoters for plant biotechnology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The promoter region in plants is located upstream of the transcription start site of a gene and is mostly responsible for the specificity and intensity of gene expression. A variety of sequence motifs that function as cis-regulatory elements flank the core promoter region and contribute to regulation of the gene. Most of these identified regulatory elements measure from 6 to 10 nucleotides long (Odell et al. 1985; Higo et al. 1999), but longer element motifs have also been identified (Hernandez-Garcia and Finer 2016; Korkuć et al. 2014). These regulatory elements recruit transcription factors, which may independently or synergistically modulate gene expression (Hernandez-Garcia and Finer 2014; Potenza et al. 2004). Whether in native plant genes or in transgenes of genetically engineered plants, the promoter region is central in determining the location and condition of gene expression.
The Cauliflower Mosaic Virus 35S (35S) promoter is the most commonly used promoter in plant biotechnology research. It is a strong, constitutive promoter, which was isolated from a plant virus and yields relatively high and constitutive expression in many plants (Odell et al. 1985; Benfey and Chua 1989). The first generations of commercialized transgenic plants with resistance to herbicide and insects, as well as many other experimental transgenic plants utilized transgenes modulated by strong, constitutive promoters (Christensen et al. 1992; Hernandez-Garcia et al. 2009, 2010a; Mann et al. 2011; Wang and Oard 2003; Zhang et al. 1991). However, given the possible metabolic burden and detrimental effects associated with strong, constitutive, non-native promoters on plant development and crop yield (Al-Kaff et al. 1998; McCabe et al. 1999; Park et al. 1996), the modulation of transgenes using native promoters with precisely tailored gene expression or using synthetic promoters, composed of novel combinations of cis-regulatory elements, may be appropriate alternatives (Hernandez-Garcia and Finer 2014; Rushton et al. 2002; Shiel et al. 2014).
The recent availability of the soybean genome has hastened the process of soybean promoter identification (Schmutz et al. 2010). To date, several native soybean promoters with varying expression strengths and spatio-temporal activity have been isolated and characterized through transgene expression (Guo et al. 2011; Hernandez-Garcia et al. 2010a; Li et al. 2012, 2014; Zhang et al. 2015; Zhang and Finer 2015). The present study highlights the characterization of 40 soybean promoters that are grouped into five distinct categories (thematic gene groups) based on the localization of native gene expression (constitutive genes for GmCons, root-preferential genes for GmRoot, seed-preferential genes for GmSeed), and the associated biological processes of native gene expression (genes belonging to the fatty-acid biosynthesis pathway for GmFAB, genes encoding WRKY transcription factors for GmWRKY). In this study, the green fluorescent protein (gfp) gene was placed under regulatory control of each promoter and assayed using two distinct, complementary promoter validation systems. The utility of the two systems, transient expression system using lima bean cotyledons and stable expression system using soybean hairy roots, have been previously described, and are useful for rapid assessment of promoter activity (Hernandez-Garcia et al. 2010a). Several soybean promoters were identified that direct expression levels higher or comparable to the 35S promoter. These promoters hold great promise for new strategies to express transgenes in soybean and other plants.
Materials and methods
Promoter cloning
To develop a toolbox of soybean (Glycine max) promoters, genes were first identified across five diverse gene groups (Table 1). These groups were selected based on either gene expression profile (constitutive, root-specific, seed-specific) or the function of the gene product (fatty acid biosynthesis-associated, WRKY). Genes from the G. max constitutive-expression group (GmCons) were previously identified from a set of soybean genes with relatively consistent transcript levels under diverse conditions (Libault et al. 2008). Genes from the G. max root-specific expression group (GmRoot) were also identified from soybean genes with specific expression in roots (Libault et al. 2010). The G. max seed specific gene group (GmSeed) was identified by back-translating forty amino acids at the N-terminus of previously documented soybean seed-targeted proteins (Kalinski et al. 1989; Natarajan et al. 2007; Perez-Grau and Goldberg 1989; Vodkin and Raikhel 1986) using Phytozome BLAST (http://phytozome.jgi.doe.gov/pz/portal.html). The genes for the G. max fatty acid biosynthesis-associated group (GmFAB) were selected from a prioritized list of genes identified as homologous to genes functioning in fatty acid biosynthesis (Bates et al. 2013) and co-localized to quantitative trait loci for fatty acid composition of the seed (http://www.soybase.org). The G. max WRKY transcription factor genes (GmWRKY) were previously identified, based on the WRKY coding sequence (Rushton et al. 2010, 2012).
Promoter DNA sequences, located upstream of the native start codon, were identified from soybean (G. max cv. ‘Williams 82’) through the Phytozome soybean genome database (http://phytozome.jgi.doe.gov/pz/portal.html). Putative promoter regions were then PCR-amplified using a Failsafe™ PCR kit (Epicentre, Madison, WI) from soybean (G. max cv. ‘Jack’) genomic DNA with specific primers (Online Resource 1), which were designed to contain one restriction site on each end of the promoter amplicon, to allow for directional cloning (HindIII/PstI/SalI site for 5′ primers, NcoI/SphI site for 3′ primers). Restriction sites were selected, based on the presence/absence of sites within the predicted promoter sequence. Isolated promoters ranged from 0.8 to 2.7 kb (Table 1), to generate genomic fragments that did not extend into adjacent predicted genes, while also avoiding long AT-rich sequences. PCR amplification conditions were 5 min at 94 °C, followed by 30 cycles (30 s at 94 °C, 30 s at 54 °C, and 4 min at 68 °C), ending with a 10 °C hold. Promoter amplicons were visualized using gel electrophoresis, and sequenced to confirm the identity of the promoter (Online Resource 2). Promoters were then cloned and analyzed as previously described (Hernandez-Garcia et al. 2010a). In brief, promoter amplicons were purified and concentrated using a DNA Clean and Concentrator™ kit (Zymo Research, Irvine, CA), digested with appropriate restriction enzymes and then inserted into the promoter cloning vector, pFLEV (Finer Laboratory Expression Vector; Hernandez Garcia et al. 2010a; Genbank Accession no. KX156843.1), which was used for transient expression analysis. The pFLEV plasmid contained a promoter cloning region, upstream of smRSGFP coding region (Chiu et al. 1996) and the NOS terminator. The fragment from pFLEV, containing the promoter:gfp:NOS terminator, was isolated following digestion with the appropriate restriction enzymes, and inserted into compatibly-digested pCAMBIA1300 (CAMBIA, Canberra, Australia), which was introduced into Agrobacterium rhizogenes (Strain K599) for production of soybean hairy roots. All promoter:gfp:NOS terminator fragments were directionally inserted into pCAMBIA1300 with the promoter:gfp:NOS terminator fragment always upstream and in the same orientation as the 35S promoter:hpt:NOS terminator (Hernandez-Garcia et al. 2010a).
Transient expression analysis of promoters in lima bean cotyledons
Each soybean promoter as well as the 35S promoter in pFLEV was introduced into lima bean (Phaseolus lunatus cv. ‘Henderson Bush’) cotyledonary tissues via particle bombardment (3 cotyledons per construct) and monitored for GFP expression as previously described (Hernandez-Garcia et al. 2010a, b) with slight modifications. In brief, lima bean seeds were surface sterilized in 5 % (v/v) bleach solution with slow agitation for 20 min, rinsed 10 times with sterile water, and germinated between moistened sterile paper towels in GA7 culture vessels (Magenta, Chicago, IL) at 25 °C with 16/8 h light/dark photoperiod, at 40 µEm−2 s−1 illumination. Four days after germination, cotyledons were aseptically excised from the seedlings, and the adaxial side was bombarded with DNA-coated tungsten particles using a Particle Inflow Gun (Finer et al. 1992). Cotyledons were then placed on OMS culture medium, which contained MS salts (Murashige and Skoog 1962), B5 vitamins (Gamborg et al. 1968), 3 % sucrose and 0.2 % Gelrite™ (Aceto Corporation, Lake Success, NY) at pH 5.7. Between 9 and 12 independently bombarded cotyledons were placed in each 100 × 25 mm Petri dish on 35 ml OMS medium, and the standard lid was replaced with a 5.5 mm polycarbonate lid to minimize condensation (Finer and Finer 2007). Images of cotyledons expressing GFP were captured every hour using a previously described automated image collection system (Buenrostro-Nava et al. 2006; Chiera et al. 2007; Finer 2011), which was composed of a MZFLIII dissecting microscope (Leica, Heerbrugg, Switzerland) with a GFP filter set (Excitation 480 ± 40 nm; emission 510 nm), a Spot-RT CCD digital camera (Diagnostic Instruments, Sterling Heights, MI), and a two-dimensional robotics platform (Arrick Robotics, Hurst, TX), all under computer control.
Images of GFP expression in each bombarded cotyledon were captured at 1600 × 1200 pixels RGB every hour for 100 h. After image collection, each image was resized to 800 × 600 pixels and manually aligned using Adobe ImageReady CS v8.0 (Adobe Systems, San Jose, CA) to correct for slight inaccuracies in sample positioning. Aligned images were cropped to a 400 × 300 pixel area, and the background fluorescence from a 50 × 50 pixel area (not containing GFP-expressing cells) was then subtracted from the red and green channels to produce background-corrected red and green channel images prior to quantification. GFP quantification was calculated using ImageJ v1.44b (Rasband 1997). GFP-expressing cells were separated from the non-expressing background in the red and green channels by adjusting selection threshold levels. Mean grayscale values from the red and green channels were subsequently obtained only from the GFP-expressing cells, and the values were multiplied by the total number of GFP-expressing pixels detected in each channel to obtain GFP expression value for each channel. Total GFP expression at each time point was calculated by the addition of the red and green channel GFP expression values (Chiera et al. 2007). The mean transient GFP expression of the 3 replicates for each promoter construct was reported as the percentage of the peak GFP expression of the 35S promoter.
Stable expression analysis of promoters in soybean hairy roots
For each promoter construct, soybean cotyledons from cv. ‘Williams 82’ were inoculated with A. rhizogenes strain K599 containing the specific promoter sequence and gfp reporter gene in pCAMBIA1300 as previously described (Hernandez-Garcia et al. 2010a). Briefly, soybean cotyledons from seedlings 5 days after germination were excised, and wounded several times on the abaxial side with sterile scalpel dipped in bacterial cultures, which were grown overnight in 2 ml liquid YEP (Yeast Extract Peptone) medium, containing 100 mg l−1 kanamycin. Inoculated cotyledons were then placed abaxial side up on moistened, sterilized P5 Fisherbrand filter paper (Fisher Scientific, Pittsburgh, PA) for 3 days under the same temperature, photoperiod, and illumination as used for the lima bean cotyledons. The soybean cotyledons were then transferred to OMS culture medium containing 400 mg l−1 Timentin (SmithKline Beecham Corp., Philadelphia, PA). After 2–3 weeks, hairy roots were recovered and excised for subculture. Although pCAMBIA1300 contained a plant selectable hygromycin resistance gene, hygromycin selection was not utilized and only GFP expressing roots were selected for subsequent analysis. Seven independent events of the fastest growing, GFP-expressing hairy roots (~2 cm) were transferred to OMS medium containing Timentin to allow the roots to grow for 4 days prior to GFP quantification.
Images of the root tips were manually collected using the same microscope and camera previously utilized for the transient GFP expression analysis. The 1600 × 1200 pixels RGB images were separated into red, green and blue channels, and the background fluorescence (from 100 × 100 pixels area excluding the root) was subtracted from the green channel to produce background-corrected green channel images. Threshold levels were then adjusted to separate the root from the background for quantification of GFP expression. The mean GFP expression was determined by calculating the grayscale mean value of the green channel pixels associated with the root using ImageJ software according to method described in Hernandez-Garcia et al. (2010a). In order to completely eliminate any background root fluorescence, the final GFP grayscale mean intensity value for each promoter construct was calculated by subtracting the average grayscale mean value of 7 roots induced with A. rhizogenes containing no binary vector, from the grayscale mean value of the 7 GFP-expressing hairy roots. One experiment per promoter was conducted for each thematic group for GFP quantification in hairy roots. Mean separation was calculated according to Tukey’s Studentized Range (HSD) test in Proc GLM of SAS 9.4 TS (SAS Institute Inc., Cary, NC).
Bioinformatic analyses
RNAseq expression profiles of soybean cv. ‘Williams 82’ young leaf, flower, 1 cm pod, pod shell 10 days after flowering, pod shell 14 days after flowering, seed 10 days after flowering, seed 14 days after flowering, seed 21 days after flowering, seed 25 days after flowering, seed 28 days after flowering, seed 35 days after flowering, seed 42 days after flowering, root, and nodule were collected in normalized reads/kilobase/million of raw data for each gene based on their respective gene ID from Soybase “SoySeq” RNA-Seq online database (http://www.soybase.org; Severin et al. 2010). In addition, RNAseq expression profile of cv. ‘Williams 82’ hairy roots was also collected. For the hairy root data, total RNA was extracted from 51 to 66 days old (from inoculation date) hairy root tissue using the Macherey–Nagel Plant kit (Macherey–Nagel, Bethlehem, PA). Libraries were prepared using Illumina’s TruSeq Stranded mRNA Sample Preparation kit and sequenced using the Illumina HiSeq2500 System (Illumina, San Diego, CA) for paired-end reads at 100 cycles. Sequencing data were processed using FastQC 0.10.1 (Andrews 2010), Scythe-adapter-trimming (Buffalo 2014), and sickle-quality-base-trimming (Joshi and Fass 2011). Alignments to the G. max Wm82.a2.v1 reference genome from the Phytozome database (http://www.phytozome.org) were performed using TopHat2-PE (Kim et al. 2013). Read counts were generated using HTSeq (Anders et al. 2015) and gene expression counts were normalized using an adjusted FPKM method as applied to single-end reads (Severin et al. 2010). Sequencing data were processed using the iPlant platform (http://www.iplantcollaborative.org; Goff et al. 2011).
For identification of putative cis-regulatory elements, each promoter sequence was also assessed for the presence of previously identified plant promoter element motifs using the PLACE online database (http://www.dna.affrc.go.jp/PLACE; Higo et al. 1999). Motifs were gathered from all promoters and sorted according to their presence within all 5 thematic groups and each thematic group.
Results
Transient expression analyses following particle bombardment (Figs. 1, 2, 3, 4, 5) allowed us to rapidly determine promoter sequence functionality, measure the relative strength of the promoter and gather some information on the temporal kinetics of expression. In parallel to the transient expression analysis, stable expression in soybean hairy roots (Figs. 1, 2, 3, 4, 5) provided a complementary tool for gfp transgene expression analysis for all of the promoters in each thematic group in stably transformed tissue. RNAseq profiles (Table 2) provided yet additional information on expression of the native gene, in its native context. Finally, a detailed list of predicted motifs within each promoter (Online Resource 3–7) showed common predicted cis-regulatory element motifs among the promoter groups.
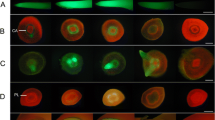
Transient expression (a) and hairy root expression (b) of GmCons promoters. Transient GFP expression is shown as the percentage of peak GFP expression of the 35S promoter. Internal legends are shown from highest to lowest expression, with 35S represented by a solid black line. Hairy root expression values are displayed as mean ± SD. Columns followed by the same letter are not significantly different at p < 0.05 using Tukey’s Studentized Range (HSD) test. Representative images for peak transient expression and hairy roots are displayed below respective graphs. In the CaMV35S image, bar = 500 microns
Transient expression (a) and hairy root expression (b) of GmRoot promoters. Transient GFP expression is shown as the percentage of peak GFP expression of the 35S promoter. Internal legends are shown from highest to lowest expression, with 35S represented by a solid black line. Hairy root expression values are displayed as mean ± SD. Columns followed by the same letter are not significantly different at p < 0.05 using Tukey’s Studentized Range (HSD) test. Representative images for peak transient expression and hairy roots are displayed below respective graphs. In the CaMV35S image, bar = 500 microns
Transient expression (a) and hairy root expression (b) of GmSeed promoters. Transient GFP expression is shown as the percentage of peak GFP expression of the 35S promoter. Internal legends are shown from highest to lowest expression, with 35S represented by a solid black line. Hairy root expression values are displayed as mean ± SD. Columns followed by the same letter are not significantly different at p < 0.05 using Tukey’s Studentized Range (HSD) test. Representative images for peak transient expression and hairy roots are displayed below respective graphs. In the CaMV35S image, bar = 500 microns
Transient expression (a, b) and hairy root expression (c) of GmFAB promoters. Transient GFP expression is shown as the percentage of peak GFP expression of the 35S promoter. Internal legends are shown from highest to lowest expression, with 35S represented by a solid black line. Hairy root expression values are displayed as mean ± SD. Columns followed by the same letter are not significantly different at p < 0.05 using Tukey’s Studentized Range (HSD) test. Representative images for peak transient expression and hairy roots are displayed below respective graphs. In the CaMV35S image, bar = 500 microns
Transient expression (a, b) and hairy root expression (c) of GmWRKY promoters. Transient GFP expression is shown as the percentage of peak GFP expression of the 35S promoter. Internal legends are shown from highest to lowest expression, with 35S represented by a solid black line. Hairy root expression values are displayed as mean ± SD. Columns followed by the same letter are not significantly different at p < 0.05 using Tukey’s Studentized Range (HSD) test. Representative images for peak transient expression and hairy roots are displayed below each respective graphs. In the CaMV35S image, bar = 500 microns
GmCons promoters
Among the 3 promoters isolated from the constitutively expressed genes [GmCons4, GmCons6 and GmCons10 (naming convention derived from Libault et al. 2008)], GmCons10 displayed the highest transient and stable expression levels (Fig. 1). In contrast, GmCons4 and GmCons6 promoters displayed very low levels of detectable transient expression in bombarded lima bean cotyledonary tissue, and showed low levels of expression in stably-transformed hairy roots. RNAseq expression data of the genes modulated by GmCons4, GmCons6 and GmCons10 indicate that all three genes showed constitutive but moderate transcript levels at all of the different developmental stages and parts of soybean plant (Table 2). Thirty-one common putative cis-regulatory elements were identified among the three GmCons promoters (Online Resource 3), aside from the common motifs identified among all of the 40 analyzed promoters.
GmRoot promoters
Among the 7 GmRoot promoters analyzed (Fig. 2), GmRoot7 had the highest peak transient expression, at more than 3.5× of the peak expression of the 35S promoter. Interestingly, the expression from the GmRoot7 promoter in stably transformed hairy roots was statistically lower than GmRoot3, which was the highest expressing GmRoot promoter. GmRoot6 and GmRoot3 displayed similar transient expression profiles as the 35S promoter (Fig. 2a). The expression of GmRoot2 was the lowest of the 7 GmRoot promoters, based on both transient and stable expression. RNAseq data for the GmRoot promoters confirmed that all 7 promoters were largely root-specific (Table 2) and GmRoot3 showed the highest reads. Surprisingly, the GmRoot7 promoter showed the highest transient expression level and the second highest expression in stably transformed roots. RNAseq data showed the lowest transcript level for the GmRoot7 gene in the root, but the highest transcript level in the GmRoot group in hairy roots. A total of 18 putative cis-regulatory elements were also shared among the GmRoot promoters (Online Resource 4), aside from the common motifs identified among all of the 40 analyzed promoters.
GmSeed promoters
All 9 GmSeed promoters analyzed (Fig. 3) showed peak transient expression below that of 35S promoter. GmSeed10 showed the highest transient expression, at about 80 % of the peak expression of the 35S promoter. GmSeed12, GmSeed6, GmSeed8, and GmSeed3 displayed an intermediate level of transient expression, while GmSeed11, GmSeed2, and GmSeed5 gave the lowest detectable transient expression. GmSeed7 did not display detectable transient expression in all three replicates. Peak expression of GmSeed10, GmSeed12, GmSeed6, GmSeed8, and GmSeed3 seemed to lag behind approximately 10 h relative to the peak expression of 35S promoter. In stably transformed hairy roots, all 9 GmSeed promoters actively regulated GFP expression, with mean GFP intensities ranging from 12.41 for GmSeed7 to 42.13 for GmSeed8. The soybean RNAseq profile of GmSeed genes showed that all GmSeed promoters, except for GmSeed8, were transcriptionally active during seed development (Table 2). A total of 19 putative cis-regulatory elements were also shared among the GmSeed promoters (Online Resource 5), aside from the common motifs identified among all of the 40 analyzed promoters.
GmFAB promoters
Among the 9 GmFAB promoters analyzed (Fig. 4), GmFAB17 had the highest transient and stable expression levels, at approximately 1.5× peak transient expression of the 35S promoter, and 30.60 mean GFP intensity in stably transformed hairy roots. GmFAB11 had the next highest transient expression, with expression levels comparable to those obtained with the 35S promoter. In stably-transformed hairy roots, expression of GmFAB11 was not statistically different from GmFAB5, GmFAB1, GmFAB2, GmFAB8, GmFAB3, GmFAB9 and GmFAB10, all of which had peak transient expression below 30 % of the levels obtained with the 35S promoter. Among the lowest expressing GmFAB promoters, GmFAB3 had the lowest peak transient expression, at <5 % relative to 35S promoter, as well as the lowest stable expression, at 9.26 mean GFP intensity. RNAseq data also indicated that the GmFAB3 gene was the least transcriptionally active in comparison to the other GmFAB genes (Table 2). All transient expression profiles for the GmFAB group were similar with the exception of GmFAB11, which showed later peak expression and a delayed decline in expression (Fig. 4). RNAseq analysis of GmFAB promoters as a group did not show preferred expression in a specific soybean tissue or growth stage with the exception of GmFAB17, which modulated higher gene expression in soybean seeds, specifically in seeds 35 days after flowering (Table 2). A total of 14 shared putative cis-regulatory elements were identified among the 9 GmFAB promoters (Online Resource 6), aside from the common motifs identified among all of the 40 analyzed promoters.
GmWRKY promoters
Among the 12 GmWRKY promoters, GmWRKY17 had the highest transient expression, with peak expression around 3× higher than the 35S promoter (Fig. 5). GmWRKY13 and GmWRKY27 showed transient expression levels that were similar to that of 35S promoter while most of the GmWRKY promoters had relatively low, but detectable transient expression levels relative to the 35S promoter: GmWRKY43, GmWRKY67, GmWRKY82, GmWRKY79, GmWRKY85 and GmWRKY80, GmWRKY162 all showed low transient expression while transient expression using the GmWRKY21 and GmWRKY54 promoters was not detected. Expression in stably transformed hairy roots containing the 12 GmWRKY promoters followed the same general trend that was observed with the transient expression with the exception of GmWRKY21 and GmWRKY67, which showed high expression in stably-transformed roots and low transient expression (Fig. 5). RNAseq analysis of GmWRKY genes as a group did not show any pattern of localization in a specific soybean tissue or growth stage (Table 2), with GmWRKY21 showing the lowest level of transcript (undetected) across different conditions. A total of 8 putative cis-regulatory elements were also shared among the GmWRKY promoters (Online Resource 7), aside from the common motifs identified among all of the 40 analyzed promoters.
Discussion
Among all 40 promoters characterized through transient expression analysis, differences in the timing and intensity of peak expression, along with the decline of GFP expression were commonly observed. The specific gfp gene used in this research (Chiu et al. 1996) was selected because the translated GFP protein displays good fluorescence and is relatively unstable. Other versions of fluorescent proteins can display higher fluorescence and stability (Stewart 2001), but may not be suitable for promoter analysis studies, as the protein may be detectable long after the promoter becomes inactive. For the specific version of the gfp gene used in this study, detectable and quantifiable GFP fluorescence was mostly indicative of promoter activity (Hernandez-Garcia et al. 2010a).
The use of GFP also allowed semi-continuous monitoring of gene expression in the same tissue over time, which provided expression profiles, reflected in the shape of the graphs showing promoter activity (Figs. 1, 2, 3, 4, 5). Assays for activity of other marker genes such as luciferase and β-glucuronidase are unfortunately destructive and therefore only allow single time point expression determinations. In this research, time-lapse animations of transient GFP expression (Online Resource 8) were also evaluated to confirm transient expression profiles. Finally, concerns of background fluorescence and chlorophyll interference with GFP detection (Finer 2011) were avoided by monitoring expression in achlorophyllous lima bean cotyledons and soybean hairy roots. The use of hardware and software corrections using appropriate filter sets and background gray value subtraction respectively, led to a reduction or elimination of all background fluorescence, and problems of interference with GFP detection were therefore minimized (Hernandez-Garcia et al. 2010a).
Although soybean promoters were isolated and studied in this research, lima bean and not soybean cotyledonary tissues were used as a target tissue for transient expression. Unfortunately, soybean cotyledonary tissues were not suitable for transient expression and image analysis as the GFP protein diffuses so rapidly from targeted soybean cells following particle bombardment (Hernandez-Garcia et al. 2010a), leading to a rapid decline in GFP detection and loss of definition of expression in the targeted cells. In addition, the adaxial surface of the lima bean cotyledons remained flat for the 100-h duration of the automated image capture, which allowed the tissue to remain in the same focal plane during the automated process of image capture.
This study employed two expression analysis systems for rapid screening of promoter functional activity, and each system carried unique advantages and limitations. For the most part, the transient lima bean cotyledon expression analysis data and the stable soybean hairy roots expression data were relatively consistent and comparable, with respect to GFP intensities among the promoters in thematic groups. General consistencies between transient expression using lima bean cotyledons and stable expression in soybean hairy roots using either groups of native soybean promoters (Hernandez-Garcia et al. 2010a; Zhang et al. 2015) or variants of individual soybean promoters (De La Torre and Finer 2015; Hernandez-Garcia and Finer 2016) have been reported previously. Although most of these previous studies utilized constitutive promoters (Zhang et al. 2015; De La Torre and Finer 2015) where tissue specificity and inducibility are not major influences on promoter activity, these same tools and targets were used for the wound inducible GmERF3 promoter (Hernandez-Garcia and Finer 2016). Some inconsistencies between measured promoter strengths among these two expression analysis systems were observed in the present study, which may not be unusual considering that root specific promoters were evaluated in bombarded lima bean cotyledons (Fig. 2) and seed specific promoters were evaluated in stably transformed soybean hairy roots (Fig. 3). With transient expression using the gene gun, large amounts of DNA are introduced into plant cells and the observed rapid expression likely results from extrachromosomal activity of the introduced genes (Hernandez-Garcia and Finer 2014). As an extrachromosomal entity, gene expression may not be impacted by epigenetic effects, including chromosome structure or chromatin-based regulation, allowing promoter strength to be directly assessed. Soybean hairy roots present another model for evaluation of promoter-mediated gene expression, but expression in this stably transformed tissue was likely impacted by epigenetic effects and by copy number of the introduced reporter gene (Hernandez-Garcia et al. 2010a). Although hairy roots are hormone autotrophic and show the altered hairy root phenotype, they can be induced on stem tissue and used to support plant growth (Veena and Taylor 2007). Due to this altered phenotype, epigenetic effects, and copy number of the introduced DNA, hairy roots may not display the same tissue specificity and inducibility exhibited by native promoters in their native context. Soybean hairy roots were generated from cv. ‘Williams 82’, due to the availability of the annotated genome and RNAseq data (http://www.soybase.org) of this cultivar. Although GFP expression levels in hairy roots containing the 35S promoter was not directly assessed in the current study, grayscale mean expression levels of approximately 50 were consistently obtained in previous studies using the 35S promoter with the same hairy root expression system (Hernandez-Garcia et al. 2010a; Zhang et al. 2015).
The RNAseq data (Table 2) highlighted in this study captured the mRNA transcript levels of the native soybean genes in each gene group, in various soybean tissues and stages of development. However, transcript levels do not necessarily correlate well with protein expression, since post-transcriptional regulation also contributes to protein expression (Maier et al. 2009). Accordingly, the RNAseq data in this study for each promoter serve as an indicator of promoter activity of genes in their native context as a regulator of transcription, but it is not a conclusive evidence of the specificity of protein expression. In addition, for use of these promoters in a transgenic context, gene expression mediated by these promoters should be specifically studied by observing transgenic expression, either through transient or stable expression analyses. Although these soybean promoters should ideally be studied in transgenic soybean plants, soybean transformation remains consistent but inefficient and the time needed for production of transgenic soybean plants is typically 4–9 months (Finer and Larkin 2008). The rapid validation tools utilized here provided some useful information on promoter strength, although tissue specificity could not be extensively studied with transient expression in lima bean cotyledons and stable expression in hairy roots.
Some interesting differences between transient expression and stable expression results were observed in the present study, of which GmWRKY21 was the most dramatic (Fig. 5). In the case of GmWRKY21, no transient GFP expression in lima bean cotyledons was detected, while expression in stably transformed hairy root expression was among the highest in the GmWRKY thematic group. It is possible that the ~1.4 kb size of this promoter (Table 1) was small and led to elimination of essential native promoter elements, but this was a standard size used for most of the cloned soybean promoters. Because the transient expression system is sensitive to gene silencing of highly expressed genes (Dhillon et al. 2009), silencing of the native promoter may have led to rapid silencing of the transgenic promoter, which may have been hypersensitive to silencing. WRKY genes are transcription factors, which usually express at low levels in most tissues (Rushton et al. 2012). Finally, the GmWRKY21 gene showed the lowest expressing transcript in all tissues of all the WRKY genes (Table 2), suggesting that some component of the hairy root expression system led to this unexpected high expression.
GmCons promoters
The three GmCons promoters characterized in this study displayed transient expression that was below that of the 35S promoter (Fig. 1). Given that multiple studies using the same hairy root expression system consistently gave the 35S promoter a grayscale mean GFP expression level of approximately 50 (Hernandez-Garcia et al. 2010a; Zhang et al. 2015), GFP expression levels from the hairy root data in this study was compared to previous 35S promoter expression data. While the GmCons10 promoter displayed a mean transient GFP expression below the 35S promoter, expression of GMCons10 was slightly higher than the 35S promoter in soybean hairy roots (Fig. 1). Expression of the constitutive promoters was consistent among the group as GmCons10 displayed the highest transgene expression using both validation tools, while GmCons4 and GmCons6 were lower. Other constitutively expressing native soybean promoters have been identified, which regulate transgene expression at much higher levels than the 35S promoter (Hernandez-Garcia et al. 2010a; Zhang et al. 2015). Because the selection conditions for the genes for the constitutive promoters reported here was based on consistent RNA-seq expression data in all tissues (Libault et al. 2008), constitutive but moderately expressing genes were identified (Table 2). The GmCons promoters identified in this study may be useful alternatives for constitutive transgene expression in soybean, especially for targeted expression at levels lower than the 35S, Gmubi (Hernandez-Garcia et al. 2009) or GmScream (Zhang et al. 2015) promoters.
GmRoot promoters
The highest transiently expressing GmRoot promoter was GmRoot7, which had more than 3× higher transient expression than the 35S promoter, more than 2× higher than the 35S promoter in stably transformed hairy roots. Interestingly, RNAseq data for GmRoot7 indicated that the promoter was indeed root-specific, but this gene showed the lowest transcript level of all GmRoot genes in non-transformed tissue (Table 2). Interestingly the GmRoot7 transcript was the highest of all GmRoot genes in soybean hairy roots (Table 2). High expression of this GmRoot7 promoter during transient expression and in hairy roots suggests that this root-specific promoter may be inducible in bombarded cotyledons and hairy roots. The annotation of GmRoot7 as a germin-like protein gene (Table 1) further suggests that the promoter for this gene may be either stress or pathogen inducible (Lu et al. 2010). As stated previously, high transient expression in lima bean cotyledons for many of the root specific promoters was unexpected but consistent, and may have resulted from extrachromosomal expression of large amounts of the introduced expression vector. Transient expression results seem to be more reflective of promoter strength rather than tissue specificity. The GmRoot3 promoter showed transient expression levels similar to that of the 35S promoter, but hairy root expression was approximately 4× higher than the 35S promoter. RNAseq analysis (Table 2) showed that GmRoot3 and GmRoot6 were the highest expressing root specific genes and the promoters from these genes also showed high expression using both validation tools (Fig. 2). Other root-associated soybean promoters (GmEXPB2 and GmPAP21) are both root-associated and induced by low phosphorus levels (Guo et al. 2011; Li et al. 2012, 2014), while induction of the root-associated GmPRP2 promoter was not reported (Chen et al. 2014). GmEXPB2, GmPAP21 and GmPRP2 were not the same as any of the GmRoot promoters identified in this study. Taken together, promoters from this thematic group may provide alternatives for directed transgene expression in roots, which may be useful for developing soybean varieties with increased resistance to root pathogens, pests and some abiotic stresses.
GmSeed promoters
All GmSeed promoters displayed transient and stable expression levels that were below that of the 35S promoter. RNAseq data (Table 2) confirmed that these promoters were distinctly seed-specific, with the exception of GmSeed8 (undetected transcript) and GmSeed7 (also expressed in nodules). Similar to the observed transient expression of root-specific promoters in lima bean cotyledons (Fig. 2), these seed specific promoters showed stable expression in soybean hairy roots (Fig. 3). It is likely that these transgenic promoters were more active or displayed less specificity due to their altered copy number or lack of epigenetic regulation, relative to the native promoter. The soyAP1 gene promoter also has high expression in seed (Zhao et al. 2012) but this promoter does not share the same promoter identity as the GmSeed promoters in this study. The GmSeed10 promoter, which drives the expression of the soybean lectin gene, has been previously compared to the 35S promoter through expression in soybean developing somatic embryos using automated image capture (Buenrostro-Nava et al. 2006). The soybean lectin protein was one of the first isolated and characterized seed-specific protein in soybean (Vodkin and Raikhel 1986), and its seed-specific expression has been attributed to the promoter sequence (Cho et al. 1995). Levels of transient expression in lima bean cotyledons and stable expression in hairy roots data presented in this present study are comparable to the GFP expression data reported previously (Buenrostro-Nava et al. 2006), as the level of peak GFP expression driven by GmSeed10 (lectin) promoter was similar to that of the 35S promoter. Taken together, these seed-specific soybean promoters could provide alternatives for regulating transgene expression in seeds, such as for improvement in oil, protein and nutritional content.
GmFAB promoters
Transient and stable expression profiles of the 9 GmFAB promoters showed that these promoters mostly had lower levels of expression compared to the 35S promoter, with the exception of the transient expression of GmFAB17 and GmFAB11 (Fig. 4). These same two promoters displayed unusual expression profiles, where GmFAB17 showed a delayed peak expression while GmFAB11 gave more sustained expression, characterized by a slower decline in transient GFP expression, compared to all other promoters. The profile showing delayed peak transient expression may have resulted from stress induction of the promoter (Hernandez-Garcia and Finer 2016) and the sustained expression (and slower decline) of GFP may indicate less sensitivity to promoter-mediated gene silencing (Dhillon et al. 2009). The variation in transient expression profiles, as well as the diversity in transcript levels from the RNAseq data (Table 2) indicate that the GmFAB group of promoters function across a variety of soybean developmental stages and tissues. As soybean fatty acid biosynthesis can be light induced (Willms et al. 1999), we detected an IBOXCORE motif for light regulation in all of the GmFAB promoter sequences. We did not validate light inducibility for any of the GmFAB promoters, although GmFAB17 and GmFAB11 have 7 and 10 copies of the IBOXCORE motif respectively (Online Resource 6).
GmWRKY promoters
A wide variation in patterns and intensities of transient and stable expression of the GmWRKY promoters was observed (Fig. 5), along with broad differences in the RNAseq data (Table 2). WRKY transcription factors are integral components for an array of plant biotic and abiotic stress responses, as well as seed dormancy and germination (Rushton et al. 2010, 2012). These transcription factors function by direct binding to response elements of other gene promoters (Rushton et al. 1996), as well as interactions with a variety of protein partners, among which are MAP kinases, MAP kinase kinases, histone deacetylases, resistance proteins, 14-3-3 proteins, calmodulin, and other WRKY factors (Rushton et al. 2010). For many of the GmWRKY promoters, transient and stable expression levels were significantly lower than that obtained with the 35S promoter. In addition to the interesting expression results demonstrated with GmWRKY21 reported earlier in this discussion (undetected transient expression and high stable expression), the GmWRKY17 promoter is also of particular interest, as it showed high transient and stable expression levels (higher than that of 35S promoter), but did not show high transcript levels in any of the soybean tissues analyzed via RNAseq (Table 2). This may simply indicate that more tissues and inductive conditions need to be analyzed for the detection of high gene expression using RNAseq. Recent studies of the GmWRKY17 and GmWRKY67 promoters in stably transformed hairy roots showed 12.7-fold and 4.8-fold increases in expression following drought induction respectively (Tripathi et al. 2016). Additionally, Tripathi et al. (2016) observed that the GmWRKY17 promoter responded to ABA and both promoters were responsive to cold. As the GmWRKY17 and GmWRKY67 promoters are strongly induced by various abiotic stresses, they may have shown low transcript levels in the RNAseq analysis as tissues used in those studies were not stressed. High expression of the GmWRKY17 promoter in the present study (Fig. 5) may reflect stress induction following bombardment, which can cause some damage to the target tissue. The GmWRKY17 promoter, which has previously shown little background expression and high inducibility by abiotic stress (Tripathi et al. 2016), may prove extremely useful for driving transgenes for improving abiotic stress responses.
Element identification
A large number of shared motifs that represent putative cis-regulatory elements were identified within each promoter family (Online Resources 3–7). Databases for cis-acting regulatory elements, such as PLACE (http://www.dna.affrc.go.jp/PLACE; Higo et al. 1999) have been developed as prediction tools to identify significant motifs within the promoter region based on previously identified motifs. Although these databases are useful in identifying numerous potential elements, confirmation through element functional validation is necessary. With the large number of putative elements identified using PLACE in this study of 40 different soybean promoters, it becomes challenging to assign functionality to all of the putative promoter elements. In addition, the modular assembly model for plant promoters, based on a model developed for the 35S promoter (Odell et al. 1985), may need to be revised, as regulatory elements that are much larger than ~6 nucleotides have been identified (Hernandez Garcia and Finer 2014, 2016). Because so many putative elements are typically identified using cis-regulatory element databases, most of the identified motifs are likely biologically inaccurate. Assessing each promoter, promoter family, and promoter element individually through coupling motif-prediction databases with mutation analysis and validation of quantifiable transgene expression will yield promoter characterization that is more biologically accurate (De La Torre and Finer 2015; Hernandez-Garcia et al. 2009; Liu et al. 2014; Rushton et al. 2002; Zhang et al. 2015). Taken together, this study provides starting data for a relatively large number of soybean promoters, grouped into thematic families. In addition, we have identified several native soybean promoters, which may be of interest for further cis-regulatory element analysis and for the development of synthetic promoters with improved gene expression specificity. These promoters add to the repertoire of native soybean promoters, which may be utilized for production of transgenics for basic research and crop improvement.
References
Al-Kaff NS, Covey SN, Kreike MM, Page AM, Pinder R, Dale PJ (1998) Transcriptional and posttranscriptional plant gene silencing in response to a pathogen. Science 279:2113–2115
Anders S, Pyl PT, Huber W (2015) HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinform. http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 29 June 2016
Bates PD, Stymne S, Ohlrogge J (2013) Biochemical pathways in seed oil synthesis. Curr Opin Plant Biol 16:358–364
Benfey PN, Chua NH (1989) The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J 8:2195–2202
Buenrostro-Nava MT, Ling PP, Finer JJ (2006) Comparative analysis of 35S and lectin promoters in transgenic soybean tissue using an automated image acquisition system and image analysis. Plant Cell Rep 25:920–926
Buffalo V (2014) Scythe—a Bayesian adapter trimmer (v. 0.981). GitHub. https://www.github.com/vsbuffalo/scythe. Accessed 29 June 2016
Chen L, Jiang B, Wu C, Sun S, Hou W, Han T (2014) GmPRP2 promoter drives root-preferential expression in transgenic Arabidopsis and soybean hairy roots. BMC Plant Biol 14:245
Chiera JM, Bouchard RA, Dorsey SL, Park E, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26:1501–1509
Chiu W-l, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Cho MJ, Widholm JM, Vodkin LO (1995) Cassettes for seed-specific expression tested in transformed embryogenic cultures of soybean. Plant Mol Biol Rep 13:255–269
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18:675–689
De La Torre CM, Finer JJ (2015) The intron and 5’ distal region of the soybean Gmubi promoter contribute to very high levels of gene expression in transiently and stably transformed tissues. Plant Cell Rep 34:111–120
Dhillon T, Chiera JM, Finer JJ (2009) Quantitative evaluation of six different viral suppressors of silencing using image analysis of transient GFP expression. Plant Cell Rep 28:639–647
Finer JJ (2011) Visualizing transgene expression. In: Stewart CN, Touraev A, Citovsky V, Tzfira T (eds) Plant transformation technologies. Wiley, Hoboken, pp 109–122
Finer JE, Finer JJ (2007) A simple method for reducing moisture condensation on Petri dish lids. Plant Cell Tiss Organ Cult 91:299–304
Finer JJ, Larkin KM (2008) Genetic transformation of soybean using particle bombardment and SAAT approaches. In: Kirti PB (ed) Handbook of new technologies for genetic improvement of legumes. CRC Press, Boca Raton, pp 103–125
Finer JJ, Vain P, Jones MW, McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:323–328
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures for soybean root cells. Exp Cell Res 50:151–158
Goff SA, Vaughn M, McKay S et al (2011) The iPlant collaborative: cyberinfrastructure for plant biology. Front Plant Sci. doi:10.3389/fpls.2011.00034
Guo W, Zhao J, LiX Qin L, Yan X, Liao H (2011) A soybean β-expansin gene GmEXPB2 intrinsically involved in root system architecture responses to abiotic stresses. Plant J 66(3):541–552
Hernandez-Garcia CM, Finer JJ (2014) Identification and validation of promoters and cis-acting regulatory elements. Plant Sci 217–218:109–119
Hernandez-Garcia CM, Finer JJ (2016) A novel cis-acting element in the GmERF3 promoter contributes to inducible gene expression in soybean and tobacco after wounding. Plant Cell Rep 35:303–316
Hernandez-Garcia CM, Martinelli AP, Bouchard RA, Finer JJ (2009) A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28:837–849
Hernandez-Garcia CM, Bouchard RA, Rushton PJ, Jones ML, Chen X, Timko MP, Finer JJ (2010a) High level transgenic expression of soybean (Glycine max) GmERF and Gmubi gene promoters isolated by a novel promoter analysis pipeline. BMC Plant Biol 10:237
Hernandez-Garcia CM, Chiera JM, Finer JJ (2010b) Robotics and dynamic image analysis for studies of gene expression in plant tissues. J Vis Exp. http://www.jove.com/video/1733/robotics-dynamic-image-analysis-for-studies-gene-expression-plant. Accessed 29 June 2016
Higo K, Ugawa Y, Iwamoto M, Korenaga T (1999) Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Res 27:297–300
Joshi NA, Fass JN (2011) Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (Version 1.0). https://github.com/najoshi/sickle. Accessed 29 June 2016
Kalinski A, Weisman JM, Mathews BF, Herman M (1989) Molecular cloning of a protein associated with soybean seed oil bodies that is similar to thiol proteases of the papain family. J Biol Chem 265:13843–13848
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions, and gene fusions. Genome Biol. doi:10.1186/gb-2013-14-4-r36
Korkuć P, Schippers JHM, Walther D (2014) Characterization and identification of cis-regulatory elements in Arabidopsis based on single-nucleotide polymorphism information. Plant Physiol 164:181–200
Li CC, Gui SH, Yang T, Liao H, Wang XR (2012) Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann Bot 109:275–285
Li C, Zhang H, Wang X, Liao H (2014) A comparison study of Agrobacterium-mediated transformation methods for root-specific promoter analysis in soybean. Plant Cell Rep 33:1921–1932
Libault M, Thibivilliers DD, Bilgin DD, Radwan O, Benitez M, Clough SJ, Stacey G (2008) Identification of four soybean reference genes for gene expression normalization. Plant Genome 1:44–54
Libault M, Farmer A, Trupti J, Takahashi K, Langley RJ, Franklin LD, He J, Dong X, May G, Stacey J (2010) An integrated transcriptome atlas of the crop model Glycine max and its use in comparative analysis in plants. Plant J 63:86–99
Liu W, Mazarei M, Peng Y, Fethe MH, Rudis MR, Lin J, Millwood RJ, Arelli PR, Stewart CN Jr (2014) Computational discovery of soybean promoter cis-regulatory elements for the construction of soybean cyst nematode-inducible synthetic promoters. Plant Biotechnol J 12:1015–1026
Lu M, Han Y-P, Gao J-G, Wang X-J, Li W-B (2010) Identification and analysis of the germin-like gene family in soybean. BMG Genom 11:1–15
Maier T, Güell M, Serrano L (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583:3966–3973
Mann DG, King ZR, Liu W, Joyce BL, Percifield RJ, Hawkins JS, LaFayette PR, Artelt BJ, Burris JN, Mazarei M, Bennetzen JL, Parrott WA, Stewart CN Jr (2011) Switchgrass (Panicum virgatum L.) polyubiquitin gene (PvUbi1 and PvUbi2) promoters for use in plant transformation. BMC Biotechnol 11:74
McCabe MS, Schepers F, van der Arend A, Mohapatra U, de Laat AMM, Power JB, Davey MR (1999) Increased stable inheritance of herbicide resistance in transgenic lettuce carrying a petE promoter-bar gene compared with a CaMV35S-bar gene. Theor Appl Genet 99:587–592
Murashige T, Skoog F (1962) A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plant 15:473–497
Natarajan S, Xu C, Bae H, Bailey BA (2007) Proteomic and genomic characterization of Kunitz trypsin inhibitors in wild and cultivated soybean genotypes. Plant Physiol 164:756–763
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Park Y, Papp I, Moscone EA, Iglesias VA, Vaucheret H, Matzke AJM, Matzke MA (1996) Gene silencing mediated by promoter homology occurs at the level of transcription and results in meiotically heritable alterations in methylation and gene activity. Plant J 9:183–194
Perez-Grau L, Goldberg RB (1989) Soybean seed protein genes are regulated spatially during embryogenesis. Plant Cell 1:1095–1109
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Rasband W (1997) ImageJ, US National Institutes of Health, Bethesda. http://rsbweb.nih.gov/ij/
Rushton PJ, Torrea JT, Parniske M, Wenert P, Hahlbrock K, Somssich IE (1996) Interaction of elicitor-induced DNA-binding proteins with elicitor response elements in the promoters of parsley PR1 genes. EMBO J 15:5690–5700
Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14:749–762
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Rushton DL, Tripathi P, Rabara RC, Lin J, Ringler P, Boken AK, Langum TJ, Smidt L, Boomsma DD, Emme NJ, Chen X, Finer JJ, Shen QJ, Rushton PJ (2012) WRKY transcription factors: key components in abscisic acid signaling. Plant Biotechnol J 10:2–11
Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu S, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du J, Tian Z, Zhu L, Gill N, Joshi T, Libault M, Sethuraman A, Zhang X, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA (2010) Genome sequence of the palaeopolyploid soybean. Nature 463:178–183
Severin AJ, Woody JL, Bolon Y, Joseph B, Diers BW, Farmer AD, Muehlauer GJ, Nelso RT, Grant D, Specht JE, Graham MA, Cannon SB, May GD, Vance CP, Shoemaker RC (2010) RNA-Seq atlas of Glycine max: a guide to the soybean transcriptome. BMC Plant Biol 10:1471–2229
Shiel DL, Tao Y, Albert H, Dong Y, Heckert M, Madrigal A, Lincoln-Cabatu B, Lu J, Fenwick T, Bermudez E, Sandoval M, Hom C, Green JM, Hale T, Pagano P, Clark J, Udranszky IA, Rizzo N, Bourett T, Howard RJ, Johnson DH, Vogt M, Akinsola G, Castle LA (2014) Broad 4-hydroxyphenylpyruvate dioxygenase inhibitor herbicide tolerance in soybean with an optimized enzyme and expression cassette. Plant Physiol 166:1162–1176
Stewart CN (2001) The utility of green fluorescent protein in transgenic plants. Plant Cell Rep 20:376–382
Tripathi P, Rabara RC, Reese RN, Miller MA, Rohila JS, Subramanian S, Shen QJ, Morandi D, Bücking H, Shulaev V, Rushton PJ (2016) A toolbox of genes, proteins, metabolites and promoters for improving drought tolerance in soybean includes the metabolite coumestrol and stomatal development genes. BMC Genom 17:102
Veena V, Taylor C (2007) Agrobacterium rhizogenes: recent developments and promising applications. In Vitro Cell Dev Biol Plant 43:383–403
Vodkin LO, Raikhel NV (1986) Soybean lectin and related proteins in seeds and roots of Le+ and Le− soybean varieties. Plant Physiol 81:558–565
Wang J, Oard J (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22:129–134
Willms JR, Salon C, Lay DB (1999) Evidence for light-stimulated fatty acid synthesis in soybean fruit. Plant Physiol 120:1117–1127
Zhang Z, Finer JJ (2015) Soybean actin, heat shock protein, and ribosomal protein promoters direct tissue-specific transgene expression in transgenic soybean. In Vitro Cell Dev Biol Plant 51:9–18
Zhang W, McElroy D, Wu R (1991) Analysis of Rice Act1 5’ region activity in transgenic rice plants. Plant Cell 3:1155–1165
Zhang N, McHale LK, Finer JJ (2015) Isolation and characterization of “GmScream” promoters that regulate the highest expressing soybean (Glycine max Merr.) genes. Plant Sci 241:189–198
Zhao Y, Shao S, Li X, Zhai Y, Zhang Q, Qian D, Wang Q (2012) Isolation and activity analysis of a seed-abundant soyAP1 gene promoter from soybean. Plant Mol Biol Rep 30:1400–1407
Acknowledgments
The authors are grateful for the work of Dr. Robert A. Bouchard in identifying the promoters belonging to GmSeed promoter family, as well as for carrying out a number of the experiments. The authors also thank Ning Zhang for valuable technical inputs. Salaries and research support for this work were provided by the United Soybean Board, and State and Federal Funds appropriated to the Ohio Agricultural Research and Development Center, The Ohio State University. Mention of trademark or proprietary products does not constitute a guarantee or warranty of the product by OSU/OARDC and also does not imply approval to the exclusion of other products that may also be suitable.
Author contributions
J.J.F. and P.J.R. conceived and designed the research. A.G. gathered data and performed the majority of the analyses. L.K.M. and A.H.G. analyzed RNAseq data. J.J.F., P.J.R., and L.K.M. defined the promoter groups. A.G., P.J.R., L.K.M. and J.J.F performed data interpretation. A.G., A.H.G., P.J.R., L.K.M. and J.J.F. wrote the manuscript. All authors read and approved the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Primers used for promoter amplification (XLSX 12 kb)
Online Resource 2
Promoter sequences in FASTA format (PDF 157 kb)
Online Resource 3
Occurrence of putative cis-regulatory elements identified using PLACE in GmCons promoters (XLSX 54 kb)
Online Resource 4
Occurrence of putative cis-regulatory elements identified using PLACE in GmRoot promoters (XLSX 111 kb)
Online Resource 5
Occurrence of putative cis-regulatory elements identified using PLACE in GmSeed promoters 5 (XLSX 133 kb)
Online Resource 6
Occurrence of putative cis-regulatory elements identified using PLACE in GmFAB promoters (XLSX 141 kb)
Online Resource 7
Occurrence of putative cis-regulatory elements identified using PLACE in GmWRKY promoters (XLSX 168 kb)
Online Resource 8
Animations showing transient GFP expression in lima bean cotyledonary tissue over 100 hours for all promoters (ZIP 523853 kb)
Rights and permissions
About this article
Cite this article
Gunadi, A., Rushton, P.J., McHale, L.K. et al. Characterization of 40 soybean (Glycine max) promoters, isolated from across 5 thematic gene groups. Plant Cell Tiss Organ Cult 127, 145–160 (2016). https://doi.org/10.1007/s11240-016-1038-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-016-1038-x