Abstract
The success of plant genetic transformation relies greatly on the strength and specificity of the promoters used to drive genes of interest. In this study, we analyzed gfp gene expression mediated by a polyubiquitin promoter (Gmubi) from soybean (Glycine max) in stably transformed soybean tissues. Strong GFP expression was observed in stably transformed proliferative embryogenic tissues. In whole transgenic plants, GFP expression was observed in root tips, main and lateral roots, cotyledons and plumules in young plants as well as in leaf veins, petioles, flower petals, pollen, pods and developing seeds in mature plants. GFP expression was localized mainly in epidermal cells, leaf mesophyll, procambium and vascular tissues. Introduction of an intron-less version of the Gmubi promoter (Gmupri) displayed almost the same GFP expression pattern albeit at lower intensities. The Gmubi promoter showed high levels of constitutive expression and represents an alternative to viral promoters for driving gene expression in soybean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With the release of genome sequences for different plants, plant promoters are receiving increased attention as one of the primary regulators of gene expression (Potenza et al. 2004). More plant-derived promoters are therefore becoming available for both basic studies on gene expression and use in transgenic plants. Although several constitutive and tissue-specific promoters have been isolated and characterized (Potenza et al. 2004), the highly expressed “constitutive” Cauliflower Mosaic Virus 35S (CaMV35S) promoter remains a commonly utilized promoter for regulation of transgene of interest in many plants. However, native promoters offer a multitude of different regulatory options and possess a wide collection of undiscovered cis-regulatory elements which can be isolated and studied to enhance our understanding of gene expression (Rushton et al. 2002).
As an alternative to the CaMV35S promoter, a soybean (Glycine max) polyubiquitin (Gmubi) promoter was isolated and partially characterized using transient expression in lima bean cotyledonary tissues (Chiera et al. 2007). Since preliminary characterization of this promoter gave such high levels of transient GFP expression, further characterization of this soybean promoter in soybean could be useful to more fully understand its expression and to evaluate its utility for regulation of genes of interest in its native genome.
Although heterologous promoters are often used to regulate transgene expression, factors which regulate promoters may not be present in heterologous systems and gene expression may not be predictable. A study of transgene expression in Gladiolus showed that the rice ActI promoter and maize UbiI promoter exhibited surprisingly low expression levels in that species (Kamo et al. 1995). More recently, a polyubiquitin promoter from Gladiolus was shown to give low levels of transgene expression in freesia, Easter lily, tobacco, rice and rose (Joung and Kamo 2006) but high levels when reintroduced in Gladiolus. Likewise, characterization of the storage protein seed-specific promoters from barley (B-Hor, D-Hor) and wheat (HMW-Glu) revealed failure of these promoters to direct seed-specific gfp expression in transgenic rice, as GFP expression was also observed in leaf and root tissues (Furtado et al. 2008).
Ubiquitin is a highly conserved eukaryotic protein consisting of 76 amino acids. The ubiquitin gene family contains two types of transcription units: polyubiquitin and ubiquitin extension protein genes. Polyubiquitin genes encode a polyprotein comprising several tandem repeats of the ubiquitin-coding unit (Sullivan et al. 2003). The ubiquitin monomers derived from this polyprotein are associated with proteolytic tasks, through tagging proteins for degradation by the proteasome (Sullivan et al. 2003; Dennis and O’Malley 2005). The ubiquitin proteasome system also plays an important role in controlling transcription through the ubiquitylation of histones (Muratani and Tansey 2003). Other functions of ubiquitin include control of translation, DNA repair, regulation of endocytosis and protein trafficking.
The study of plant ubiquitin genes has revealed a large group of constitutive and strongly expressed promoters of plant origin. Polyubiquitin promoters have been isolated and characterized from maize (Christensen and Quail 1996), tobacco (Plesse et al. 2001), Arabidopsis (Callis et al. 1990), sunflower (Binet et al. 1990), potato (Garbarino et al. 1995), tomato (Rollfinke et al. 1998), rice (Wang and Oard 2003; Sivamani and Qu 2006), sugarcane (Wei et al. 2003) and soybean (Chiera et al. 2007). Most of these promoters direct higher levels of gene expression than the CaMV35S promoter in transgenic plants. Since the ubiquitin genes are expressed in almost all plant tissues in their native context, their promoters should drive constitutive gene expression in transgenic plants especially of the same species. Characterization of transgenic plants containing different ubiquitin promoters to date have revealed that these promoters drive gene expression preferentially in young tissues, vascular tissues and pollen grains (Plesse et al. 2001; Rooke et al. 2000). This gene expression pattern is in accordance with the broad range of activities that ubiquitin proteins carry out, especially in metabolically active tissues.
The high gene expression mediated by polyubiquitin promoters is influenced quantitatively by the presence of introns, which are commonly located within the 5′ untranslated region (UTR) of the polyubiquitin genes (Sivamani and Qu 2006; Lu et al. 2008). Introns near the translation initiation codon are important cis-elements responsible for intron-mediated enhancement (IME) of gene expression in plants (Callis et al. 1987; Le Hir et al. 2003). In mammals and plants, it has been observed that IME influences gene expression at transcriptional levels through increased mRNA accumulation, probably by increasing gene transcription (Rose 2004; Lu et al. 2008), at post-transcriptional levels playing an important role in RNA processing and/or export (Samadder et al. 2008), and at translational levels due to an increased association of the mRNA with ribosomes via interactions with the proteins of the exon junction complex (EJC) (Le Hir et al. 2000; Nott et al. 2004). IME is more remarkably observed in monocots than dicots (Vain et al. 1996; Rose and Beliakoff 2000).
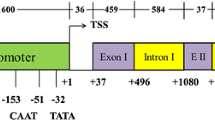
Although many promoters have been identified in other crops, only a few highly expressing promoters have been cloned from and characterized in soybean. In our laboratory, a soybean polyubiquitin promoter (Gmubi) was recently isolated and evaluated using transient expression (Chiera et al. 2007). Tissues, containing the introduced Gmubi::gfp construct and its intron-less version Gmupri::gfp, showed 2–5-fold higher transient GFP expression than those containing a CaMV35S promoter construct (Chiera et al. 2007). Since characterization of novel promoters in stable transgenic plants also requires documentation of spatial and temporal expression, the aim of this study was to characterize stably transformed soybean plants containing the Gmubi promoter and a truncated, intron-less Gmupri version, both fused to the gfp coding region.
Materials and methods
Genetic transformation
Embryogenic tissues of soybean (Glycine max Merr.) cv. Jack were initiated, maintained and transformed according to Finer and McMullen (1991) with some modifications. One week before bombardment, proliferative embryogenic target tissues were subcultured on D20 medium containing 20 mg l−1 2,4-D (Santarém and Finer 1999). Directly prior to bombardment, embryogenic tissues on D20 medium in Petri dishes were uncovered and placed in a hood for 15 min to facilitate partial drying (Vain et al. 1993). Plasmids were delivered using a Particle Inflow Gun (Finer et al. 1992). For gene introductions, the plasmid pHytru containing the selectable hygromycin resistance gene driven by the CaMV35S promoter (Chiera et al. 2004) was co-precipitated onto tungsten particles with a plasmid containing a gfp gene (Chiu et al. 1996) under regulatory control of either the 917 bp Gmubi promoter (Genbank submission U310508), the intron-less 328 bp Gmupri version (Chiera et al. 2007; Wu et al. 2008), or the CaMV35S promoter (Ponappa et al. 1999). After bombardment, tissues were grown in D20 medium for 1 week and then placed on D20 medium containing 30 mg l−1 hygromycin (Calbiochem, La Jolla, CA, USA) to select for transgenic proliferative tissues. Selected tissues were maintained on medium containing 15 mg l−1 hygromycin for a minimum of 3 months and were transferred to embryo development medium lacking the selective agent for plant recovery. Transgenic regenerants were grown in a greenhouse with supplemental lighting for GFP analysis and seed production.
GFP expression analysis
GFP expression was analyzed in transformed embryogenic callus and in organs and tissues from both seedlings and developing plants. Based on consistency and strength of GFP expression, Gmubi events 59, 72 and 135 and Gmupri events 7 and 20 were selected for detailed analysis. For GFP expression analysis with the CaMV35S promoter (Ponappa et al. 1999), event 11 was selected and only seedling tissues were analyzed. For GFP analysis in seedlings, seeds were first germinated in moistened rolls of paper towels. GFP-positive seedlings were selected, transplanted to soil and subsequently grown in a greenhouse. Entire organs and hand-cut sections from roots, hypocotyls, leaves, petioles, flowers, pods and seeds were observed under a MZFLIII stereomicroscope (Leica, Heerbrugg, Switzerland) equipped with a 100 W mercury lamp, a “GFP-2” filter set (Excitation 480 ± 40 nm; Emission 510 nm) and a Nikon Coolpix 990 digital camera. In some cases, images of larger pieces of tissues which could not be viewed in a single field using a dissecting microscope (e.g., germinating seedlings and pods), were assembled using Adobe®Photoshop® from multiple, individual images to form large composite images. Leaf, young stem and flower petal samples were observed using confocal laser scanning microscopy (Leica TCS-SP). Pollen was observed using an inverted epifluorescence Leica DM IRB microscope equipped with a Q Imaging Q19838 digital camera.
Southern blot analysis
Genomic DNA from transgenic plants was extracted according to Murray and Thompson (1980) as modified by Fulton et al. (1995). Southern analysis was performed as described previously (Chiera et al. 2006). Ten micrograms of the extracted DNA from each sample were digested overnight with BsrGI, which cuts pGmubi and pGmupri plasmids at a single site. The positive control was 20 pg of either the pGmubi or the pGmupri plasmid mixed with 10 μg of DNA from a non-transgenic plant, digested with the same enzyme. Restricted DNAs were electrophoretically separated on 0.8% (w/v) agarose gels and then transferred to nylon membranes (Roche Diagnostics GmbH, Indianapolis, IN, USA) as described by Sambrook et al. (1989). The probe was a 717 bp amplicon of the gfp coding sequence generated via PCR from pGmubi using the primers 5′ATGGTGAGCAAGGGCGAGGAGCTG3′ and 5′TTACTTGTACAGCTCGTCCATG3′. The purified probe was labeled with [α-32P]-dCTP (Perkin-Elmer, Boston, MA, USA) using the Prime-It® II Random Labeling Kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s instructions and two PCR cycles, 95°C for 5 min and 37°C for 10 min. For hybridization, the labeled probe was hybridized to the membranes and incubated overnight at 60°C using a hybridization incubator (FisherBiotech, Pittsburgh, PA, USA). The hybridized membranes were exposed to a phosphor screen holder for 24 h. Autoradiographs were visualized with a Storm 860 PhosphorImager™ System (Molecular Dynamics, Sunnyvale, CA, USA).
Results
Transgenic event recovery
The Gmubi promoter gave rise to high levels of GFP expression in embryogenic transgenic tissues in nine of eleven events (Fig. 1a). However, the pre-intronic Gmupri promoter showed only low to moderate GFP expression in embryogenic callus (Fig. 1b). All T0 Gmubi and Gmupri plants produced seeds and had a phenotype similar to the soybean cv. Jack, which was the cultivar used for transformation.
GFP expression in different events of transgenic embryogenic soybean tissues growing on D20 medium. a Embryogenic tissues transformed with the soybean polyubiquitin promoter Gmubi or b the intron-less Gmupri promoter. NT is non-transformed embryogenic tissue from soybean cv. Jack. The numbers represent individual transgenic events for each construct
GFP expression in seeds and seedlings
A detailed analysis of GFP expression was conducted primarily in imbibed seeds and various tissues from transgenic seedlings carrying either the CaMV35S, Gmubi or Gmupri promoter, focusing on the highly expressing Gmubi events 59, 72 and 135 and on the Gmupri events 7 and 20, and CaMV35S event 11, which expressed GFP at more moderate levels.
GFP expression was evident in newly emerged root tips and cotyledons of germinating transgenic seeds, 2 days after seed imbibition. Five-day-old progeny seedlings from transgenic events containing the Gmubi promoter exhibited strong GFP expression in root tips, main and lateral roots, hypocotyls, cotyledons and plumules (Fig. 2a). Tissues of seedlings containing the Gmupri promoter showed GFP expression in root tips, main and lateral roots and cotyledons, but at somewhat lower levels (Fig. 2b).
Composite image collected from 5-day-old transgenic soybean seedlings showing GFP expression driven by two promoters. a Seedling containing the Gmubi promoter event 72, b the intron-less Gmupri promoter event 20, c non-transformed seedling from soybean cv. Jack. Images were collected using a dissecting microscope, equipped for GFP detection
In root tissues of seedlings containing either the Gmubi, Gmupri or CaMV35S construct, GFP expression was highest in the young root, primarily in the elongation zone behind the root tip (Fig. 3). Once again, expression driven by the Gmupri promoter followed a pattern similar to Gmubi but at lower levels. To further characterize tissue-specific expression in roots and other seedling tissues, hypocotyls and root sections from 4-day-old seedlings were hand-cut for observation of GFP expression using a dissecting microscope (Fig. 3). Cross-sections of roots with lateral root primordia showed high GFP expression in epidermal cells and procambial tissues. The lateral root primordia were evident in both Gmubi- and Gmupri-containing events because of their striking GFP expression. Cross-sections of seedling hypocotyl tissues containing Gmubi also showed GFP in the epidermis and parenchyma cells of the cortex and procambium. GFP expression in hypocotyl tissues of seedlings containing Gmupri was quite low. Plumules of 4-day old seedlings revealed high levels of GFP expression with the Gmubi promoter, while expression was also present, albeit at reduced levels, in plumules of seedlings containing the Gmupri construct. Transgenic tissues from CaMV35-containing seedlings showed lower GFP expression than those containing either the Gmubi or Gmupri promoters (Fig. 3).
GFP expression in soybean transgenic seedlings carrying the Gmubi (event 72), Gmupri (event 20) and CaMV35S (event 11) promoters. Root tip images were collected from 3-day-old seedlings. Hypocotyl and root images are from hand-cut cross-sections of 4-day-old seedlings. Plumules were observed from 4-day-old seedlings. Non-transformed tissues and organs are from non-transformed seedlings of soybean cv. Jack. Images were collected using a dissecting microscope, equipped for GFP detection
GFP analysis in plants
The expression of GFP directed by the Gmubi or Gmupri promoter was primarily studied using a dissecting microscope in entire leaf blades and in tissues from transverse sections of median veins of leaves and cross-sections of petioles and stems. In young leaves of plants containing the Gmubi promoter, GFP expression was observed in the median and lateral veins on both adaxial and abaxial leaf surfaces (Fig. 4a). GFP expression was not detected in Gmupri-containing leaves and in non-transgenic control leaves. Hand-cut transverse sections of leaf tissues containing the median leaf vein (Fig. 4a) from plants transformed with Gmubi confirmed GFP expression in veinal tissues such as pith, parenchyma and phloem. In Gmubi-containing plants, GFP was also detected at high levels in petiole cross-sections, with GFP expression in the epidermis, pith, phloem and xylem (Fig. 4a). No GFP expression was observed in petiole tissues from plants containing the Gmupri promoter (Fig. 4a). In cross sections of stems with little secondary growth from plants containing the Gmubi promoter, GFP was strongly expressed in epidermal cells, secondary phloem and pith cells (Fig. 4b). No GFP expression was seen in cross-sections of stems containing the Gmupri promoter, which were similar to cross sections from non-transformed control plants (Fig. 4b).
GFP expression in leaves and stems of T1 soybean plants containing the Gmubi (event 72) or Gmupri promoter (event 20). a Images of different leaf structures and b stem cross-sections showing GFP expression. Images were collected from both adaxial and abaxial surface of young leaves from 1-month-old developing plants. The median vein images were taken from transverse sections of leaves, while petiole and stem images were taken from hand-cut cross-sections. Non-transformed control corresponds to soybean cv. Jack. Images were collected using a dissecting microscope, equipped for GFP detection. Pi pith, Ep epidermis, Sp secondary phloem
For more detailed analysis of GFP expression in transgenic soybean tissues, confocal microscopy was employed. Stem longitudinal-sections from young plants containing the Gmubi promoter showed high GFP expression localized in the epidermis and cells forming the vascular tissues (Fig. 5a). Although stem sections of plants transformed with the Gmupri promoter showed the same expression pattern, GFP expression was at more moderate levels. In leaf blades from plants carrying the Gmubi promoter, GFP was detected in the epidermis including stomata, specifically in the nucleus of epidermal and guard cells (Fig. 5b, top panel). The Gmupri promoter showed the same GFP expression pattern as Gmubi in the leaf blade although at lower levels. In leaf hand-cut cross-sections of leaves containing Gmubi, GFP was strong in epidermal cells located on both leaf surfaces. The cells forming the spongy and palisade mesophylls from the same leaf sections also showed high GFP expression (Fig. 5b, bottom panel). Cross-sections of leaves carrying Gmupri also displayed GFP expression in epidermal and mesophyll cells but at low intensity (Fig. 5b, bottom panel).
GFP expression in different tissues from T1 transgenic soybean plants containing the gfp gene under regulatory control of either the Gmubi (event 72) or Gmupri promoter (event 20). a, b and d Confocal microscopy analysis of young stems, leaves and flower petals, respectively. a Longitudinal-sections of young stems showing GFP in epidermis and vascular tissues. b Top panel leaf epidermis with high GFP expression in epidermal cells, guard cells, nucleus and cytoplasm. b Bottom panel leaf cross-sections showing GFP in both palisade and spongy mesophylls. c Epifluorescence microscopy of pollen grains with and without (arrow) GFP expression. d Flower petal tissues (top panel brightfield; bottom panel fluorescence) showing green fluorescence from GFP expression located in nucleus and cytoplasm. Non-transformed controls were soybean cv. Jack. The bars are equivalent to 50 μm
The expression of GFP was also studied in floral structures from transgenic plants. GFP was localized in pollen grains from flowers transformed with the Gmubi promoter and analyzed with an epifluorescence microscope (Fig. 5c). In flowers from Gmubi-containing T1 heterozygous plants, the GFP-positive pollen grains were seen along with non-fluorescent pollen grains, similar to those from non-transformed control plants (Fig. 5c). Confocal microscopy of high GFP-expressing flower petals from plants transformed with the Gmubi promoter showed that GFP was expressed at high levels in cells of flower petals, especially in the nucleus and the non-vacuole portion of the cytoplasm (Fig. 5d). On the other hand, no fluorescence was observed in flower petals from non-transformed plants (Fig. 5d). High GFP expression was also detected in flower organs from plants carrying the Gmubi promoter using a dissecting fluorescence microscope (Fig. 6a). Flowers from plants containing the Gmupri promoter showed only low GFP expression, which seemed to be restricted to the pollen grains (Fig. 6a).
GFP expression in flowers and pods from T1 transgenic soybean plants containing the gfp gene under regulatory control of the Gmubi or Gmupri promoter. a Mature flowers. b Longitudinal-sections of immature T2 seeds (top panel) and a later developmental stage (bottom panel). c GFP expression in pods from two different events containing the Gmubi promoter, event 59 (left panel) and event 72 (right panel). Non-transformed controls were soybean cv. Jack. Emb embryo, En endosperm, Hi hilum, Co cotyledon, Hy hypocotyl of developing embryo, Ra radicle, Ow ovary wall
GFP expression under regulatory control of the Gmubi or Gmupri promoter was also analyzed in pods and immature seeds at two different developmental stages (Fig. 6b, c). Pods with seeds at late cotyledonary stage from plants containing either the Gmubi or the Gmupri promoter showed GFP expression in the endosperm and the region surrounding the hilum (Fig. 6b, first panel), while seeds at later developmental stages showed GFP expression in cotyledons and along the embryo axis (Fig. 6b, second panel). Developing seeds containing the Gmupri promoter showed moderate GFP only in the embryo axis but not in cotyledons (Fig. 6b, second panel). High levels of GFP expression was also observed in the inner layer of the ovary wall of pods from the Gmubi event 59 (Fig. 6c, left). In pods from T1 heterozygous Gmubi-containing plants, both GFP-positive and GFP-negative developing seeds were seen in the same pod (Fig. 6c).
Southern blot analysis
Southern blot hybridizations were performed to observe hybridization patterns and to determine the copy number of the transgene in soybean T1 plants and progeny. Southern hybridization analysis showed a range in copy number for the introduced transgene and different hybridization patterns for each event. High copy number events for Gmupri were events 5, 24 and 42 (Fig. 7a). For Gmubi, high copy number transgenic events included 46, 68 and 69 (Fig. 7b). The remaining events, which made up the majority of the transgenic events, contained from 1 to 4 hybridization bands. A hybridization band, which was a little less than 4 kb, was observed in almost all lanes (Fig. 7a, b).
Southern blot analysis of different soybean transgenic events containing the Gmubi or Gmupri promoter. Genomic DNA was digested with BsrGI and hybridized with a 717 bp probe, which was PCR-amplified from the gfp ORF. DNA hybridization patterns from seedlings containing the intron-less Gmupri promoter (a) or the Gmubi promoter (b). c DNA hybridization analysis of plants from two transgenic events containing the Gmubi promoter through two or three generations. Jack − is non-transformed soybean cv. Jack DNA used as a negative control, Jack + lane contains DNA from non-transformed cv. Jack plus 20 pg pGmubi or pGmupri, representing a single transgene copy in the soybean genome. Numbers represent the different transgenic events. Asterisks show the events selected for detailed GFP analysis
The presence of the gfp gene was evaluated and detected in two or three generations of plants from Gmubi events 135 and 72, respectively. The same hybridization pattern was observed in all three generations of event 72 and two generations of events 135 (Fig. 7c). These same two distinct hybridization patterns were also observed with DNA from proliferating embryogenic D20 tissues (data not shown).
Discussion
The most widely used promoter for directing constitutive high levels of expression in plants is the CaMV35S promoter (Odell et al. 1985). In grasses, the maize polyubiquitin promoter (Christensen et al. 1992) appears to be the most commonly used promoter when constitutive expression is desired. To expand the availability of promoters for driving constitutive gene expression, we initially isolated and performed preliminary characterization of a G lycine m ax polyubiquitin promoter (Gmubi; Chiera et al. 2007) using transient expression. In this present study, a more extensive characterization of the Gmubi promoter was performed using a gfp reporter gene in soybean, the source of the promoter. Knowledge of expected expression patterns of a promoter is critical, when a promoter is being considered for regulation of genes of interest, for both basic research and crop improvement (Rooke et al. 2000).
GFP expression overview
In this work, the use of the Gmubi promoter led to constitutive, high levels of GFP expression in many of the tissues analyzed. High expression levels were initially observed in proliferative transgenic tissues (Fig. 1), and later in regenerated whole plants and progeny (Figs. 2, 3, 4, 5, 6). A comparison of GFP expression driven by the CaMV35S, Gmubi and Gmupri promoters in seedling tissues (Fig. 3) showed that the Gmubi promoter was much stronger than the CaMV35S promoter while Gmupri-driven GFP expression was only moderately higher. Using a quantitative lima bean cotyledon transient expression assay (Chiera et al. 2007), the Gmubi and Gmupri promoters were shown to be 5- and 2-times stronger than the CaMV35S promoter, respectively. The quantitative expression data from bombardment of lima bean cotyledons with the Gmubi, Gmupri and CaMV35S promoters (Chiera et al. 2007) appear to correlate well with the intensity of GFP expression observed with seedling tissues in this study (Fig. 3). These findings suggest that our transient expression analysis might be useful as a general and early indicator of promoter strength in stably transformed tissues. GFP expression in transgenic plants containing the CaMV35S promoter was not easily observed (Ponappa et al. 1999), and these events were therefore not further evaluated at the whole plant level in this study. However, the Gmubi promoter was clearly of sufficient strength to allow easy GFP visualization (Fig. 4) using the same version of the gfp gene and the same detection system.
For analysis of GFP evaluation in the present study, it should be stressed that three different GFP detection microscopies were utilized. GFP expression profiles were generally consistent regardless of the detection method but the images that were collected allowed resolution at different levels. When using the dissecting fluorescent microscope (Figs. 1, 2, 3, 4, 6), resolution (both optical and GFP detection) was the lowest, but observations at low magnification were necessary for the most rapid and thorough analysis of expression in many different tissues and events. Due to the optics and mechanics of the fluorescent dissecting microscope, GFP detection was enhanced at higher magnifications. At high magnification, the excitation light became more focused and the area under direct observation was smaller, allowing higher resolution for GFP detection. Therefore, high magnification image collection yields brighter green fluorescence when viewed by eye and when using standard camera exposures. When performing direct comparisons between the Gmubi and Gmupri promoters in the same tissues, images were always collected using the same magnification. To generate Fig. 2, composite images were assembled from multiple images, collected at low magnification from each germinating seedling. Each of the individual images in the composite was collected under the same conditions, validating the comparison between promoters. Using confocal (Fig. 5a, b, d) and epifluorescence (Fig. 5c) microscopies, resolution of GFP expression was additionally enhanced while the viewing field was reduced.
In soybean embryogenic cultures and whole plants, GFP detection is often impaired by red autofluorescence in chlorophyll-containing tissues. In achlorophyllous tissues of soybean (roots, pollen, leaf epidermal cells, flower petals), visualization of GFP was more straightforward as interference from chlorophyll was not an issue (Figs. 3, 5c, d, 6a). Chlorophyll may interfere with GFP fluorescence by either competing with GFP for excitation light or masking GFP fluorescence from an overabundance of red fluorescence (Zhou et al. 2005). To eliminate chlorophyll and allow more efficient detection of GFP in embryogenic soybean cultures, we have previously included the herbicide isoxaflutole in the D20 culture medium (Wu et al. 2008). Isoxaflutole caused tissue bleaching while not affecting growth of in vitro cultures, permitting detection of even low levels of GFP (Wu et al. 2008). GFP expression was low in tissues containing GFP under regulatory control of Gmupri promoter in this study (Fig. 1b) and inclusion of isoxaflutole in the culture medium in a previous study led to reduced chlorophyll levels and easier observations of GFP in Gmupri-containing transgenic events (Wu et al. 2008). Although the beneficial effects of isoxaflutole on GFP visualization was not further explored in this study, partial bleaching and some improvement of GFP detection in leaf tissues could be obtained with sublethal applications of this herbicide to leaves of whole plants (Finer unpublished data).
GFP expression patterns
The expression patterns observed in soybean tissues containing either Gmubi or Gmupri were similar but the intensity of GFP expression was always much lower with the intron-less Gmupri promoter. In general, introns appear to affect the quantitative level of expression rather than qualitative aspects of gene expression such as tissue specificity or induction (Le Hir et al. 2003). Intron effects on the level of transgene expression have been well-documented using both transient expression (Callis et al. 1987; Vain et al. 1996) and stable transformation (Callis et al. 1987; Wang and Oard 2003; Genschik et al. 1994). With other polyubiquitin promoters from both monocots and dicots (Garbarino et al. 1995; Joung and Kamo 2006; Sivamani and Qu 2006), inclusion of the intron in the 5′ UTR of the promoter region led to enhancements of reporter gene expression of up to 10-fold. Deletion of introns from polyubiquitin promoters of rice (Wang and Oard 2003) and tobacco (Genschik et al. 1994) also led to a considerable reduction in gene expression levels. Using transient expression to evaluate Gmubi and Gmupri, Chiera et al. (2007) reported a 2-fold increase in GFP expression levels with the intron-containing Gmubi promoter, compared to the intron-less Gmupri. Although a quantitative evaluation of GFP expression levels was not undertaken in the present study, the Gmubi promoter clearly gave higher levels of gene expression compared to Gmupri (Figs. 1, 2, 3, 4, 5, 6).
Gmubi-driven GFP expression was observed in many different tissues of the transgenic soybean, with very high levels of expression in vascular tissues, epidermal cells, stomata and young roots (Figs. 3, 4, 5). This expression pattern has also been observed with polyubiquitin promoters from other plants (Cornejo et al. 1993; Rooke et al. 2000; Plesse et al. 2001). Ubiquitin promoters typically show high activity in young, actively growing tissues and organs such as vascular tissue, epidermis and root tip. Interestingly, the CaMV35S promoter also drives high levels of gene expression in these same tissues (Battraw and Hall 1990), suggesting that “constitutive” promoters show heightened activity in young, actively growing tissues. Induction of the Gmubi promoter in transgenic soybean, in response to various stress phenomena, was not observed in this study (data not shown). Ubiquitin promoters from other plant species, regulating various transgene, were inducible by heat stress as well as wounding (Takimoto et al. 1994; Garbarino et al. 1995; Nagatani et al. 1997). Either the GFP detection method utilized in this research was not sufficiently sensitive to discriminate increases in expression levels or the specific ubiquitin promoter that was used in this work (Chiera et al. 2007) is not wound- or heat-inducible. Considering the relatively small size of the Gmubi promoter (917 bp; including 591 bp intron), additional upstream sequences may contain additional regulatory elements which would confer induction to various factors. Promoters from other soybean polyubiquitin genes (Xia et al. 1994) may show other patterns of gene expression.
Most of the detailed analysis of promoter activity reported here utilized three Gmubi events and two Gmupri events. Additional events were generated but not characterized (Fig. 1). These other transgenic events did not display GFP that could be accurately assessed. The variation in transgene expression among events is not unusual even when transgenic plants are generated using the same conditions and DNA constructs (Butaye et al. 2005). The DNA regions flanking the transgene can influence gene expression (De Bolle et al. 2003). Southern hybridization analysis revealed that the five events, which were studied in detail in this work, contained 1–3 copies of the introduced constructs (Fig. 7). Other events, which were not selected for more extensive analysis, contained from 1 to approximately 20 copies of the introduced DNA. Copy number may also influence expression of the transgene.
Among those events studied in detail, the GFP expression pattern driven by the Gmubi and Gmupri promoters was fairly consistent among all soybean events and organs/tissues analyzed. The use of transgenic soybean for evaluation of soybean promoters (Buenrostro-Nava et al. 2006) is noteworthy as many studies of soybean promoters do not utilize soybean to evaluate promoter activity (Li et al. 1994; Strömvik et al. 2004; Waclawovsky et al. 2006). The high levels of promoter activity observed in the young endosperm of the immature seed (Fig. 6b) and the ovary wall (Fig. 6c) of the pod, using soybean, may not be as apparent or possibly not even be visible if tobacco or Arabidopsis were utilized because of the small size or absence of the corresponding structures.
We report here a detailed characterization of expression patterns using a soybean polyubiquitin promoter in transgenic soybean. This is the first report of an extensive characterization of a soybean polyubiquitin promoter in transgenic plants. The Gmubi promoter directed high levels of gene expression in young rapidly growing tissues, while lower levels of expression could be observed in most parts of the transgenic soybean plants. When promoters are evaluated for regulation of transgene, it would be best if the evaluation were performed in the eventual target plant species. For this work, soybean was targeted, making these results more meaningful and relevant for both understanding native promoter regulation and use for production of commercial transgenics in soybean.
References
Battraw MJ, Hall TC (1990) Histochemical analysis of CaMV 35S promoter-β-glucuronidase gene expression in transgenic rice plants. Plant Mol Biol 15:527–538
Binet MN, Lepetit M, Weil JH, Tessier LH (1990) Analysis of a sunflower polyubiquitin promoter by transient expression. Plant Sci 79:87–94
Buenrostro-Nava MT, Ling PP, Finer JJ (2006) Comparative analysis of 35S and lectin promoters in transgenic soybean tissue using and automated image acquisition system and image analysis. Plant Cell Rep 25:920–926
Butaye K, Cammue B, Delauré S, De Bolle M (2005) Approaches to minimize variation of transgene expression in plants. Mol Breed 16:79–91
Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Genes Dev 1:1183–1200
Callis J, Raasch JA, Vierstra RD (1990) Ubiquitin extension proteins of Arabidopsis thaliana––structure, localization, and expression of their promoters in transgenic tobacco. J Biol Chem 265:12486–12493
Chiera JM, Finer JJ, Grabau EA (2004) Ectopic expression of a soybean phytase in developing seeds of Glycine max to improve phosphorus availability. Plant Mol Biol 56:895–904
Chiera JM, Streeter JG, Finer JJ (2006) Ononitol and pinitol production in transgenic soybean containing the inositol methyl transferase gene from Mesembryanthemum crystallinum. Plant Sci 171:647–654
Chiera JM, Bouchard RA, Dorsey SL, Park EH, Buenrostro-Nava MT, Ling PP, Finer JJ (2007) Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26:1501–1509
Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Christensen AH, Quail PH (1996) Ubiquitin promoter-based vectors for high levels of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Christensen AH, Sharrock RA, Quail PH (1992) Sequence analysis and transcriptional regulation by heat shock of polyubiquitin transcripts from maize. Plant Mol Biol 18:675–689
Cornejo M-J, Luth D, Blankenship KM, Anderson OD, Blechl AE (1993) Activity of a maize ubiquitin promoter in transgenic rice. Plant Mol Biol 23:567–581
De Bolle M, Butaye K, Coucke W, Goderis I, Wouters P, van Boxel N, Broekaert W, Cammue B (2003) Analysis of the influence of promoter elements and a matrix attachment region on the inter-individual variation of transgene expression in populations of Arabidopsis thaliana. Plant Sci 165:169–179
Dennis AP, O’Malley BW (2005) Rush hour at the promoter: How the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol 93:139–151
Finer JJ, McMullen MD (1991) Transformation of soybean via particle bombardment of embryogenic suspension culture tissue. In Vitro Cell Dev Biol Plant 27:175–182
Finer JJ, Vain P, Jones MW, McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:232–238
Fulton TM, Chunzoongse J, Tanksley SD (1995) Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol Biol Rep 13:207–209
Furtado A, Henry RJ, Takaiwa F (2008) Comparison of promoters in transgenic rice. Plant Biotech J 6:679–693
Garbarino JE, Oosumi T, Belknap WR (1995) Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiol 109:1371–1378
Genschik P, Marbach J, Uze M, Feuerman M, Plesse B, Fleck J (1994) Structure and promoter activity of a stress and developmentally regulated polyubiquitin-encoding gene of Nicotiana tabacum. Gene 148:195–202
Joung YH, Kamo K (2006) Expression of a polyubiquitin promoter isolated from Gladiolus. Plant Cell Rep 25:1081–1088
Kamo K, Blowers A, Smith F, Van Eck J, Lawson R (1995) Stable transformation of Gladiolus using suspension cells and callus. J Am Soc Hortic Sci 120:347–352
Le Hir H, Moore MJ, Maquat LE (2000) Pre-mRNA splicing alters mRNP composition: evidence for stable association of proteins at exon-exon junctions. Genes Dev 14:1098–1108
Le Hir H, Nott A, Moore MJ (2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28:215–220
Li Y, Liu ZB, Shi X, Hagen G, Guilfoyle TJ (1994) An auxin-inducible element in soybean SAUR promoters. Plant Physiol 106:37–43
Lu J, Sivamani E, Azhakanandam K, Samadder P, Li X, Qu R (2008) Gene expression enhancement mediated by the 5′ UTR intron of the rice rubi3 gene varied remarkably among tissues in transgenic rice plants. Mol Genet Genomics 279:563–572
Muratani M, Tansey WP (2003) How the ubiquitin-proteasome system controls transcription. Nat Rev Mol Cell Biol 4:192–201
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4325
Nagatani N, Takumi S, Tomiyama M, Shimada T, Tamiya E (1997) Semi-real time imaging of the expression of a maize polyubiquitin promoter-GFP gene in transgenic rice. Plant Sci 124:49–56
Nott A, Le Hir H, Moore MJ (2004) Splicing enhances translation in mammalian cells: an additional function of the exon junction complex. Genes Dev 18:210–222
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Plesse B, Criqui MC, Durr A, Parmentier Y, Fleck J, Genschik P (2001) Effects of the polyubiquitin gene Ubi.u4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol 45:655–667
Ponappa T, Brzozowski AE, Finer JJ (1999) Transient expression and stable transformation of soybean using the jellyfish green fluorescent protein (GFP). Plant Cell Rep 19:6–12
Potenza C, Aleman L, Sengupta-Gopalan C (2004) Targeting transgene expression in research, agricultural, and environmental applications: promoters used in plant transformation. In Vitro Cell Dev Biol Plant 40:1–22
Rollfinke IK, Silber MV, Pfitzner UM (1998) Characterization and expression of a heptaubiquitin gene from tomato. Gene 211:267–276
Rooke L, Byrne D, Salgueiro S (2000) Marker gene expression driven by the maize ubiquitin promoter in transgenic wheat. Ann App Biol 136:167–172
Rose AB (2004) The effect of intron location on intron-mediated enhancement of gene expression in Arabidopsis. Plant J 40:744–751
Rose AB, Beliakoff JA (2000) Intron-mediated enhancement of gene expression independent of unique intron sequences and splicing. Plant Physiol 22:535–542
Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE (2002) Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14:749–762
Samadder P, Sivamani E, Lu J, Li X, Qu R (2008) Transcriptional and post-transcriptional enhancement of gene expression by the 5′ UTR intron of rice rubi3 gene in transgenic rice cells. Mol Genet Genomics 279:429–439
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Santarém ER, Finer JJ (1999) Transformation of soybean (Glycine max (L.) Merrill) using proliferative embryogenic tissue maintained on semi-solid medium. In Vitro Cell Dev Biol Plant 35:451–455
Sivamani E, Qu R (2006) Expression enhancement of a rice polyubiquitin gene promoter. Plant Mol Biol 60:225–239
Strömvik MV, Sundararaman VP, Vodkin LO (2004) A novel promoter from soybean that is active in a complex developmental pattern with and without its proximal 650 base pairs. Plant Mol Biol 41:217–231
Sullivan JA, Shirasu K, Deng XW (2003) The diverse roles of ubiquitin and the 26S proteasome in the life of plants. Nat Rev Genet 4:948–958
Takimoto I, Christensen AH, Quail PH, Uchimiya H, Toki S (1994) Non-systemic expression of a stress-responsive maize polyubiquitin gene (Ubi-1) in transgenic rice plants. Plant Mol Biol 26:1007–1012
Vain P, McMullen MD, Finer JJ (1993) Osmotic treatment enhances particle bombardment-mediated transient and stable transformation of maize. Plant Cell Rep 12:84–88
Vain P, Finer KR, Engler DE, Pratt RC, Finer JJ (1996) Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep 15:489–494
Waclawovsky AJ, Freitas RL, Rocha CS, Contim LAS, Fontes EPB (2006) Combinational regulation modules of GmSBP2 promoter: A distal cis-regulatory domain confines the SBP2 promoter activity to the vascular tissue in vegetative organs. Biochem Biophys Acta 1759:89–98
Wang J, Oard JH (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22:129–134
Wei HR, Wang ML, More PH, Albert HH (2003) Comparative expression analysis of two sugarcane polyubiquitin promoters and flanking sequences in transgenic plants. J Plant Physiol 160:1241–1251
Wu C, Chiera JM, Ling PP, Finer JJ (2008) Isoxaflutole treatment leads to reversible tissue bleaching and allows for more effective detection of GFP in transgenic soybean tissues. In Vitro Cell Dev Biol Plant 6:540–547
Xia B-S, Waterhouse RN, Watanabe Y, Kajiwara H, Komatsu S, Hirano H (1994) Nucleotide sequence of a soybean (Glycine max L. Merr.) ubiquitin gene. Plant Physiol 104:805–806
Zhou X, Carranco R, Vitha S, Hall TC (2005) The dark side of green fluorescent protein. New Phytol 168:313–322
Acknowledgments
We would like to thank Ms. Cheri Nemes for generation of the transgenic events and plants and Dr. Tea Meulia (MCIC/OARDC/OSU) for the technical assistance during the confocal microscopy analysis. Salaries and research support were provided by the United Soybean Board, and by State and Federal funds appropriated to The Ohio State University/Ohio Agricultural Research and Development Center. This research was partially supported through fellowships from CAPES, Brazil, to APM and from CONACYT, Mexico, to CMHG. Mention of trademark or proprietary products does not constitute a guarantee or warranty of the product by OSU/OARDC and also does not imply approval to the exclusion of other products that may also be suitable. Journal Article No HCS 08–22.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Jordan.
Rights and permissions
About this article
Cite this article
Hernandez-Garcia, C.M., Martinelli, A.P., Bouchard, R.A. et al. A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep 28, 837–849 (2009). https://doi.org/10.1007/s00299-009-0681-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-009-0681-7