Abstract
A novel automated image collection and analysis system was used to compare two new soybean (Glycine max (L.) Merr.) promoters with the cauliflower mosaic virus 35S (CaMV35S) promoter, which was used as an expression standard. For expression comparisons, various permutations of a soybean polyubiquitin (Gmubi) promoter, a soybean heat shock protein 90-like (GmHSP90L) promoter and the CaMV35S promoter were placed upstream of a green fluorescent protein (gfp) gene. DNA constructs were introduced via particle bombardment into excised cotyledons of germinating lima bean (Phaseolus lunatus L.) seeds, which were arranged in Petri dishes for automated image capture and image analysis. The automated system allowed monitoring and quantification of gfp gene expression in the same piece of tissue over time. The Gmubi promoter, with its intronic region intact, showed the highest expression that was over five times stronger than the CaMV35S promoter. When an intronic region was removed from the Gmubi promoter, GFP expression was reduced, but was still over two times greater than with the CaMV35S promoter. The full-length soybean GmHSP90L promoter was four times stronger than the CaMV35S promoter. Truncation of the GmHSP90L promoter resulted in stepwise decreases in promoter strength, which appear to correspond to removal of regulatory elements. Automated image capture and analysis allowed the rapid and efficient evaluation of these new promoters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Promoter analysis in plants can provide information on both the strength of the promoter and its regulation in different tissues. Promoter analysis studies have been performed with either stably transformed tissues or using transient expression analyses. For stable transformation, the time required for generation of a transgenic plant can be as short as 6 weeks (An et al. 1986), but can often extend beyond 5 months when transformation and plant recovery is slow (Santarém and Finer 1999). Production of stably transformed plants is necessary for a detailed examination of promoter expression; however, quantification of promoter strength and comparative analyses in stably transformed plants can still be difficult due to variation in transgene expression among transgenic clones (Finnegan and McElroy 1994).
Rapid, quantifiable, and reproducible promoter analyses are simplified using transient expression, where gene expression can be observed in as little as 1.5 h post introduction (Ponappa et al. 1999) and gene expression will not be influenced by copy number or site of integration. Transient expression analysis can be performed via direct DNA introduction into protoplasts using electroporation (Christensen et al. 1992) or PEG (Hartmann et al. 1998), or particle bombardment-mediated transformation into intact plant tissues (Rolfe and Tobin 1991). Agroinfiltration of tobacco (Bendahmane et al. 1999; Vaucheret 1994) is also commonly used for rapid analysis of transgene effects. Reporter genes such as luciferase (Ow et al. 1986), β-glucuronidase (Samac et al. 2004; Vain et al. 1996) and chloramphenicol acetyl transferase (Kang et al. 2003) are most commonly utilized for quantification of promoter activity. Unfortunately, visualization of luciferase and β-glucuronidase activity requires the addition of an artificial substrate and quantification of promoter activity using all of these reporter genes requires the extraction of protein from the sample, destroying the sample and eliminating the ability to follow gene expression in the same piece of tissue over time.
The green fluorescent protein (gfp) gene offers tremendous opportunities for promoter analysis in plants since its expression can be followed in the same piece of tissue over extended periods of time (Piston et al. 1999). Although GFP expression has been used in studies to characterize promoter activity (Abebe et al. 2006), reports on the quantification of GFP expression using image analysis (Nagatani et al. 1997), spectofluorometry (Richards et al. 2003) or fluorescence spectroscopy (Stewart et al. 2005) are minimal. Standard methods are needed for the evaluation of promoter strength based on GFP detection. Recently, an automated robotics system was developed for monitoring GFP expression over time in multiple pieces of tissue (Buenrostro-Nava et al. 2005). The robotics system consisted of a two-dimensional robotics platform, a cooled CCD camera, and a dissecting fluorescence microscope, all under computer control. Although the monitoring system was initially used for automated image collection of GFP expression in stably transformed somatic embryos (Buenrostro-Nava et al. 2006) and Agrobacterium (Buenrostro-Nava et al. 2003), it also has utility for rapid quantification of promoter strength using transient expression analyses.
Soybean (Glycine max (L.) Merr.), a valuable agronomic crop world-wide, has the highest transgenic acreage of any crop. As efforts move forward to produce new and improved transgenic soybean, the need for different types of native soybean promoters will continue to increase. Some soybean promoters have already been identified but these promoters direct expression in a tissue-specific (Chen et al. 1986) or inducible manner (Czarnecka et al. 1989; Liu et al. 1994). A strong, constitutive, native soybean promoter, which could replace the constitutive Cauliflower Mosaic Virus 35S (CaMV35S) was sought. Of the strong constitutive plant promoters that have been used extensively for directing transgene expression, the polyubiquitin promoters have received the most widespread attention (Christensen and Quail 1996). A common feature of polyubiquitin promoters is the presence of a leading intron, which is considered part of the promoter, and can influence transgene expression (Christensen and Quail 1996). Removal of the intron from the promoter region either reduces the strength of the promoter (Plesse et al. 2001) or results in complete loss of promoter activity (Wang and Oard 2003). In addition to a strong constitutive promoter, a developmentally regulated promoter would be useful for comparative studies. Although soybean promoters active during late stages of seed development are available (Chen et al. 1986), EST data from induced soybean somatic embryos (Thibaud-Nissen et al. 2003) now permits the identification of useful early embryo-specific promoters.
In this study, a constitutive G. max polyubiquitin (Gmubi) promoter and an early embryo-specific G. max heat shock protein 90-like (GmHSP90L) promoter were isolated, fused to the gfp coding region, and introduced into cotyledonary tissue of lima bean for rapid evaluation of promoter activity using automated image collection and analysis. The activities of the soybean promoters were compared to that of the constitutive CaMV35S promoter (Odell et al. 1985).
Materials and methods
Plasmid constructs
Eight different promoter constructs were generated for this study (Fig. 1). All promoters were placed upstream of a modified gfp gene (sGFP(S65T); Chiu et al. 1996). Two different forms of a soybean (G. max) polyubiquitin promoter were evaluated, along with five different forms of a soybean heat shock protein 90-like (GmHSP90L) promoter. The CaMV35S promoter was used as a standard.
Maps of promoter constructs used for particle bombardment-mediated DNA introduction into lima bean cotyledons. All the promoters were positioned 5′ to the GFP open reading frame. Gmubi: full length Glycine max ubiquitin promoter with an intronic sequence Gmupri: G. max ubiquitin pre-intronic promoter contained only the nucleotides before the putative intronic sequence. GmHSP90L: G. max heat shock protein 90-like, with promoter truncations leading to promoter sizes of 177, 231, 443, 628, and 830 nucleotides. CaMV35S: the full-length Cauliflower Mosaic Virus 35S promoter. All numbers are in relation to the translational start of the gfp coding sequence
The soybean (G . m ax) polyubiquitin promoter (Gmubi) was identified from GenBank submission D28123 (G. max SUBI-3). Based on similar motifs in this gene’s 5′ region and those found in the maize polyubiquitin 1 promoter (Christensen et al. 1992; Ling et al. 1995), primers were designed to clone the entire 5′ UTR region of the D28123 sequence, adding a 5′ SphI site and a 3′ BamHI site to allow insertion of the promoter in front of the gfp gene. The entire 5′ Gmubi fragment was amplified from soybean genomic DNA using the FailSafe™ PCR system (Epicentre Biotechnologies, Madison, WI, USA). An intron-less version of the Gmubi promoter (Gmupri; G . m ax pre-intronic) was recovered by amplifying the 328 bp segment of Gmubi preceding the intron, adding a 5′ SphI site and a 3′ BamHI site as before. This fragment was amplified directly from the Gmubi promoter fragment and was also inserted in front of the gfp gene.
The GmHSP90L promoter was selected based on early embryogenesis expression data from EST-based microarrays (Thibaud-Nissen et al. 2003). A GenomeWalker™ (Clontech, Palo Alto, CA, USA) library was constructed from soybean genomic DNA according to the manufacturer’s instructions. Nested reverse primers were constructed against the GmHSP90-like EST (AW278784) at the junction between the 5′ UTR and the start of the open reading frame. The first-round primer was complementary to a segment just inside the open reading frame. The secondary primer was designed to be complementary to the 3′ end of the upstream 5′ UTR and the junction of the 5′ UTR with the start codon, but with modifications of the sequences overlapping the start codon to create an NcoI site encompassing the ATG. This allowed transcriptional fusion of the promoter directly to the gfp open reading frame. All the primers used in cloning the promoter regions are available in the supplemental material (S1).
Plant material
All eight promoter constructs were assayed using a newly developed bean cotyledon transient assay system. Cotyledonary tissue from germinating lima bean (Phaseolus lunatus L. cv. “Henderson-Bush”) seeds was targeted for DNA introduction. Lima bean seed source plants were grown in the greenhouse (16/8 h light:dark, 28°C) with supplemental lighting from high pressure sodium lamps. Mature seeds, used in these experiments, were harvested and stored at room temperature for up to 4 weeks prior to use.
For germination, seeds were sterilized with a 10% bleach solution for 20 min and rinsed five times with sterile deionized water. Sterilized seeds were placed in Magenta GA7 containers between layers of a folded white paper towel that was saturated with 25 ml of sterile water. After 4 days (40 μEm−2 s−1; 16/8 h light:dark, 25°C,), the light-green cotyledons were excised from the germinating seedlings and placed in Petri dishes containing OMS medium (pH 5.7), which consisted of MS salts (Murashige and Skoog 1962), B5 vitamins (Gamborg et al. 1968), 3% sucrose and 0.2% Gelrite™ (Aceto Corporation; Lake Success, NY, USA).
DNA introduction
Cotyledons were placed, adaxial surface up, on top of an inverted modified baffle, which was comprised of a 500 μm nylon screen melted to the bottom of a 400 ml polypropylene beaker (Finer et al. 1992) with triangular slots cut in the bottom to allow expelled helium gas to flow around the sample and minimize tissue displacement. Each DNA construct was precipitated onto tungsten particles and introduced into the target tissue using the Particle Inflow Gun as described previously (Finer et al. 1992). Following the introduction of each DNA construct, cotyledons were immediately placed adaxial side up in Petri dishes containing OMS medium. Gmubi, Gmupri, and CaMV35S promoter constructs were bombarded in two independent experiments of three replicates each. The GmHSP90L constructs were bombarded in three independent experiments of three replicates each.
Image collection and analysis
Once GFP expression was detected (1–3 h), the Petri dishes were placed on the robotics platform of the automated image collection system (Buenrostro-Nava et al. 2006) which consisted of a MZFLIII dissecting microscope (Leica, Heerbrugg, Switzerland) equipped with a GFP2 filter set (Ex. 480 ± 40 nm; Em. 510 nm LP), a Spot-RT CCD digital camera (Diagnostic Instruments Inc., Sterling Heights, MI, USA), and a two-dimensional robotics platform (Arrick Robotics Inc., Hurst, TX, USA), all under computer control. Images (1,600 × 1,200 pixels; 256 gray levels for each of the three channels) of each cotyledon were collected every hour for at least 95 h. Following image collection, GFP expression was quantified using the software package ImageJ (Rasband 1997–2006). Each series of images was opened, resized to 800 × 600 pixels, and aligned using the TurboReg plugin (Thévenaz et al. 1998). After alignment, an area comprising 400 × 300 pixels containing the highest number of expressing cells was cropped from the series of images and used for quantification of GFP.
Each series of images was separated into red, green, and blue channels. Due to background autofluorescence, the contribution of the background to the overall GFP intensity was first subtracted from the entire image. A 20 × 20 pixel area was selected in the background of the red and green channels (from an area not containing GFP-expressing cells) for determination of background gray value (typically 30–40). The background gray value of the red and green channels was subtracted from every pixel in the respective channel to yield background-corrected images which were used for all expression determinations.
Mean grayscale values in the red and green channels were determined by first segmenting the expressing pixels from the background by adjusting the threshold levels. The mean grayscale values were calculated using only the segmented (expressing) pixels. The “Total Expression” value was calculated by multiplying a mean grayscale value per pixel from the red and green channels by the total number of GFP-expressing pixels in the respective channel and then adding these two values. The green channel was automatically segmented using an entropy threshold algorithm (Sahoo et al. 1988) and the number of GFP-expressing foci was counted.
Results
Target tissue
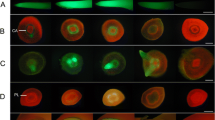
Lima bean cotyledonary tissues showed even and consistent distribution of GFP-expressing foci over much of the adaxial surface of the cotyledons (Fig. 2). Expression was localized to single targeted cells although very high levels of expression resulted in a low level of reflected fluorescence through adjacent cells. Occasionally, discrete areas of the cotyledon did not show GFP expression, although they were clearly targeted. These patches were either slow to turn green (observed under brightfield conditions) or did not turn green over the course of the experiment, suggesting patches of non-viable or low viability tissues. Similar areas were observed in non-bombarded cotyledons.
Analysis of Gmubi, Gmupri, and CaMV35S promoters
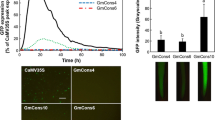
Although major differences in promoter strengths were easily visualized (Fig. 2), quantification of the level of GFP expression provided additional information that was not easily evaluated by the eye alone. Total Expression values were fivefold greater with the Gmubi promoter compared to the CaMV35S promoter, whereas the Focus Number was only 40% greater. Although the expression levels of GFP driven by the Gmupri promoter appeared to be similar to the expression levels of CaMV35S-regulated GFP based on visual analysis, a twofold greater Total Expression value for Gmupri was measured using image analysis. The Focus Numbers obtained for the Gmupri and CaMV35S promoters were not significantly different.
With semi-continuous tracking of GFP expression, it was possible to determine precisely the peak expression times for all of the different promoter constructs. The times for maximum expression of the CaMV35S, Gmubi and Gmupri promoters occurred at hours 22, 27 and 29, respectively (Fig. 3). At those peak expression points, the Gmubi and the intron-less Gmupri promoters had fivefold (1.58 × 106 ± 0.35 × 106; P = 0.01) and over twofold (0.76 × 106 ± 0.09 × 106; P = 0.002) greater Total Expression compared to the CaMV35S (0.30 × 106 ± 0.04 × 106) promoter. Gmubi and Gmupri promoters had an average of 105 ± 21 (P = 0.1) and 92 ± 24 (P = 0.3) foci expressing during their peak expression levels, respectively; whereas the CaMV35S promoter averaged 61 ± 7 foci during its peak expression (Fig. 3). Although Total Expression values for Gmubi continually increased to the peak expression at 27 h (Fig. 3), the Focus Number for Gmubi reached a plateau at hour 15 and did not increase with increasing expression levels. An increase in expression paralleled the increase in focus number for all of the other constructs.
Expression profiles of Gmubi, Gmupri, and CaMV35S promoters were calculated from replicated (n = 6) series of images for each promoter. The Total Expression calculation is described in the “Materials and methods”. Focus Number was the number of foci expressing GFP in the same area used for Total Expression determinations
Analysis of GmHSP90L promoters
Analysis of the five different lengths of the GmHSP90L promoter showed that the largest promoter (GmHSP90L-830) displayed both the highest Total Expression value (1.15 × 106 ± 0.27 × 106) and Focus Number (141 ± 29; Fig. 4). Truncation of the GmHSP90L promoter region to either 628 or 443 nucleotides resulted in roughly 50% reductions in both the Total Expression value and Focus Number. These two truncated promoters appeared to act similarly with regard to intensity and timing of GFP expression. A further reduction of the GmHSP90L promoter to either 231 or 177 nucleotides resulted in a further decline in Total Expression and the Focus Number (Fig. 4).
Expression profiles of GmHSP90L promoter constructs: 177, 231, 443, 628, and 830. Quantification as described in Fig. 3 (n = 9)
Discussion
Lima bean cotyledonary transient assay
Lima bean cotyledons, which were used as the target tissue in this study, have physiological and morphological characteristics that make them ideal for monitoring transient expression of GFP using the automated image collection system. The adaxial surface of the lima bean cotyledon is flat, which minimizes the variation in focal planes, allowing the collection of sharp, focused images. In addition, the tissue contains relatively large cells which yield distinct, GFP-expressing foci following successful DNA introduction. Lastly, this tissue contains minimal amounts of chlorophyll which may interfere with GFP fluorescence (Billinton and Knight 2001). Evaluation of embryogenic cultures of soybeans (Finer 1988) and soybean seedling cotyledons (prepared similarly to the lima bean cotyledons) did not give consistent or useful results (data not shown). The small unresponsive areas in the lima bean cotyledons that were occasionally observed did not result from bombardment damage, as they were also observed (as light areas) in cotyledons that were never bombarded. These areas were avoided in image analysis and may represent parts of the cotyledon that were late to resume active metabolism following seed imbibition. Lima bean cotyledons provide the most robust and consistent transient GFP expression results of any system that we have evaluated. To our knowledge, this is the first report of the use of lima bean cotyledonary tissue for evaluating plant promoters or transient GFP expression.
Automated image capture
The ability to collect and analyze images from the same pieces of tissue over time adds another dimension to promoter characterization. This system allows tracking of GFP expression dynamics in single pieces of tissue, which is impossible with other reporter genes. Data was generated on both promoter strength and the timing of maximum GFP expression. As a recommendation for general observation of transient GFP expression, 24 h post-introduction provided relatively consistent results with this target tissue.
In general, the promoters which generated high Focus Numbers also yielded the highest Total Expression values. Focus Number, or “spot count”, has been the classical method for reporting successful DNA introduction via particle bombardment (Klein et al. 1987). As DNA introduction via particle bombardment is a physical process, the same number of cells should be penetrated by the particles for each bombardment. The Focus Numbers reported here suggest that there is a threshold for the detection of GFP expression. At the lower levels of GFP expression, a threshold appears to be required before a GFP focus can be visualized. Using our detection methods, many of the cells that were penetrated by particles carrying the CaMV35S-controlled gfp construct obviously did not express GFP at detectable levels. For the Gmubi and Gmupri comparisons, the same maximum numbers of cells were visualized using each promoter, with the increase in expression levels resulting from higher expression levels per cell.
Gmubi and Gmupri promoters
Although ubiquitin promoters have been isolated and characterized from maize (Christensen et al. 1992), tobacco (Plesse et al. 2001), Arabidopsis (Callis et al. 1987), potato (Garbarino et al. 1995), tomato (Rollfinke et al. 1998) and rice (Wang and Oard 2003), this is the first report describing a ubiquitin promoter from soybean. All of these ubiquitin promoters show similar characteristics, including the presence of a leading intron which can contribute to increased promoter strength. Removal of the intron from the rice promoter resulted in a complete loss of promoter activity (Wang and Oard 2003) while removal of the intron from the tobacco promoter resulted in a reduction in promoter strength and an alteration in tissue-specific expression (Plesse et al. 2001). The maize ubiquitin promoter with its intron has been used extensively over the years to obtain high levels of transgene expression in cereals (Callis et al. 1987; Fennell and Hauptman 1992; Christensen and Quail 1996). In the present report, expression of the gfp gene controlled by the intron-containing Gmubi promoter was much higher than with the pre-intronic Gmupri and the standard CaMV35S promoters (Figs. 2, 3). However, even without the intronic region, the Gmupri promoter was still substantially stronger than the CaMV35S promoter. Although many different forms of the CaMV35S promoter are available for plant transformation research, the version used here (Chiu et al. 1996) was the 423 nucleotide version and did not contain additional enhancer elements or regions of promoter duplication. Use of an even smaller CaMV35S promoter (−343 from the transcriptional start; Odell et al. 1985) did not give reduced transcript levels compared to the full-length promoter, showing that the 423 nucleotide CaMV35S promoter used in the present report is comparable to the full-length promoter. Given the high levels of GFP expression obtained with the Gmubi and Gmupri promoters used here, the addition of enhancer elements or duplication of promoter regions could potentially lead to even higher levels of gene expression.
In addition to the ubiquitin intron, other introns from maize (act3, adh1-S, bz-1, sh1; Callis et al. 1987; Luehrsen and Walbot 1991; Vasil et al. 1989), rice (act1; McElroy et al. 1990) and oat (phyA3; Bruce and Quail 1990) have been used to enhance transgene expression in cereals. In dicot species, the effects of including an intronic region on promoter activity have been less definitive, resulting in no or minimal gene expression enhancements (Paszkowski et al. 1992; Plessse et al. 2001; Tanaka et al. 1990; Vancanneyt et al. 1990). Recently, however, a five- to sixfold enhancement of constitutive expression using a prolifin intron was reported in Arabidopsis (Jeong et al. 2006). Similarly, Chung et al. (2006) obtained a twofold increase in transient expression using the Arabidopsis EF1α leading intron in Agroinfiltrated Nicotiana benthalmiana leaves. Internal truncations of this 5′UTR intron suggested that the absolute size of the intron along with at least three internal elements contributed to this twofold enhancement. Introns may enhance transgene expression through increased mRNA translational activity acquired from intron splicing and processing (Bourdon et al. 2001; Matsumoto et al. 1998). A large enhancement of GFP expression was observed in the present study when the intronic region from Gmubi was included in the promoter. The soybean intronic region may also contain one or more enhancer elements, as has been reported in the ubiquitin promoter regions of rice (Wang and Oard 2003), maize (Christensen and Quail 1996) and tobacco (Genschik et al. 1994). Clearly, further work will be required to untangle the specifics of the soybean ubiquitin intron-enhancement of transgene expression reported here, as well as general intron enhancement effects.
GmHSP90L promoters
Based on the Total Expression values and Focus Numbers, the GmHSP90L promoter constructs segregated into three distinct groups; the high expressing 830, the more moderately expressing 628 and 443 and the low expressing 231 and 177 (Fig. 4). Apparently, the regions that were eliminated between the groups contained important regulatory regions that contributed to promoter activity. This promoter complexity was not unexpected, since regulation of plant Hsp90 genes can be intricate. For example, two maize Hsp90 genes exhibited distinct patterns of expression and heat shock induction in various tissues (Marrs et al. 1993), as did a pair of Arabidopsis cytosolic Hsp90 genes studied during embryo development (Prasinos et al. 2005).
The Total Expression value calculated for the GmHSP90L-830 construct (Fig. 4) was comparable to the value obtained with the Gmubi promoter (Fig. 3). The largest soybean promoters, therefore, showed the highest activities in this study. The Gmupri and the GmHSP90L-628 and GmHSP90L-443 promoters presented similar expression profiles while the smallest GmHSP90L promoters 177 and 231 showed expression levels that were comparable to the CaMV35S promoter, which had the lowest activity of all the promoters. Although evaluation of these promoter constructs gave fairly consistent results, transient gene expression may be influenced by the nature of the target tissue and the means of DNA introduction. For example, exposure of plants to a variety of stresses, including wounding, caused changes in transcript levels for both Hsp90 in tobacco (Rizhsky et al. 2002) and for hsf genes that regulate all hsp genes in Arabidopsis (Miller and Mittler 2006). In addition, stress-induction of the Ubi.U4 gene has been reported in tobacco (Plesse et al. 2001). Particle bombardment may contribute to stress induction, resulting in an increase in gene expression. Bombardment of tissues, stably transformed with GmHSP90L-gfp and Gmubi-gfp constructs with “blank” particles, should help to address questions of wound induction of these promoters.
Although peak GFP expression times were somewhat different for the various constructs, all of the GFP expression curves generated in this study showed very similar profiles (Figs. 3, 4). An initial rapid increase in expression was followed by a slow decline. Although the early fate of the introduced DNA may be of paramount importance for the recovery of transgenics, very little is known about the early events following DNA introduction. The decline in transient expression could result from degradation of unincorporated DNAs or silencing of either unincorporated or integrated DNAs through RNAi (Baulcombe 2004). The decline could also result from cell death, as the expressing cells have walls that have been compromised from particle introduction. Lastly, a small number of foci did not fade but suddenly stopped expressing between one time point to the next (1 h). Disappearing foci were easily visualized through generation of time-lapse animations of the assembled image series (S2).
The combination of digital imaging and image analysis, along with the use of GFP for gene expression tracking constitutes a powerful tool to evaluate promoters and the dynamics of gene expression. The automated image analysis system permits the continual monitoring and rapid quantification of gene expression over time while bombardment of lima bean cotyledons provides a standard transient expression assay system with reduced variation for use in comparative expression studies. Although a transient assay may not precisely reflect expression in stably transformed tissues, this transient assay system quantitatively evaluates a large number of independent events. Use of the automated system can quickly provide valuable information on the characteristics of different promoters and factors which can influence gene expression, prior to the generation of stably transformed plants.
Abbreviations
- CaMV35S:
-
Cauliflower mosaic virus 35S
- gfp:
-
Green fluorescent protein
- Gmubi:
-
Glycine max ubiquitin
- GmHSP90L:
-
Glycine max Heat shock protein 90-like
References
Abebe T, Skadsen R, Patel M, Kaeppler H (2006) The Lem2 gene promoter of barley directs cell- and development-specific expression of gfp in transgenic plants. Plant Biotech J 4:35–44
An G, Watson BD, Chiang CC (1986) Transformation of tobacco, tomato, potato and Arabidopsis thaliana using a binary Ti vector system. Plant Physiol 81:301–305
Baulcombe D (2004) RNA Silencing in plants. Nature 431:356–363
Bendahmane A, Kanyuka K, Baulcombe DC (1999) The Rx gene from potato controls separate virus resistance and cell death responses. Plant Cell 11:781–791
Billinton N, Knight AW (2001) Seeing the wood through the trees: a review of techniques for distinguishing green fluorescent protein from endogenous autofluorescence. Anal Biochem 291:175–197
Bourdon V, Harvey A, Lonsdale DM (2001) Introns and their positions affect the translational activity of mRNA in plant cells. EMBO Rep 2:394–398
Bruce WB, Quail PH (1990) cis-Acting elements involved in photoregulation of an oat phytochrome promoter in rice. Plant Cell 2:1081–1089
Buenrostro-Nava MT, Ling PP, Finer JJ (2003) Development of an automated image collection system for generating time-lapse animations of plant tissue growth and green fluorescent protein gene expression. In: Vasil I (ed) Plant biotechnology 2002 and beyond, Kluwer, Dordrecht, pp 293–295
Buenrostro-Nava MT, Ling PP, Finer JJ (2005) Development of an automated image acquisition system for monitoring gene expression and tissue growth. Trans Am Soc Agric Eng 48:841–847
Buenrostro-Nava MT, Ling PP, Finer JJ (2006) Comparative analysis of 35S and Lectin promoters in transgenic soybean tissue using an automated image acquisition system and image analysis. Plant Cell Rep 25:920–926
Callis J, Fromm M, Walbot V (1987) Introns increase gene expression in cultured maize cells. Gene Dev 1:1183–1200
Chen ZL, Schuler MA, Beachy RN (1986) Functional analysis of regulatory elements in a plant embryo-specific gene. PNAS USA 83:8560–8564
Chiu W-L, Niwa Y, Zeng W, Hirano T, Kobayashi H, Sheen J (1996) Engineered GFP as a vital reporter in plants. Curr Biol 6:325–330
Christensen AH, Quail PH (1996) Ubiquitin promoter based vectors for high level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res 5:213–218
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18:675–689
Chung BYW, Simons C, Firth AE, Brown CM, Hellens RP (2006) Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics 7:120
Czarnecka E, Key JL, Gurley WB (1989) Regulatory domains of the Gmhsp17.5-E heat shock promoter of soybean. Mol Cell Biol 9:3457–3463
Fennell A, Hauptmann R (1992) Electroporation and PEG delivery of DNA into maize microspores. Plant Cell Rep 11:567–570
Finer JJ (1988) Apical proliferation of embryogenic tissue of soybean [Glycine max (L.) Merrill]. Plant Cell Rep 7:238–241
Finer JJ, Vain P, Jones MW, McMullen MD (1992) Development of the particle inflow gun for DNA delivery to plant cells. Plant Cell Rep 11:232–238
Finnegan J, McElroy D (1994) Transgene inactivation: plants fight back! Biotechnology 12:883–888
Gamborg OL, Miller RA, Ojima K (1968) Nutrient requirements of suspension cultures of soybean root cells. Exp Cell Res 50:150–158
Garbarino JE, Oosumi T, Belknap WR (1995) Isolation of a polyubiquitin promoter and its expression in transgenic potato plants. Plant Physiol 109:1371–1378
Genschik P, Marbach J, Uze M, Feuerman M, Plesse B, Fleck J (1994) Structure and promoter activity of a stress and developmentally regulated polyubiquitin-encoding gene of Nicotiana tabacum. Gene 148:195–202
Hartmann U, Valentine WJ, Christie JM, Hays J, Jenkins GI, Weisshaar B (1998) Identification of UV/blue light-response elements in the Arabidopsis thaliana chalcone synthase promoter using a homologous protoplast transient expression system. Plant Mol Biol 36:741–754
Jeong Y-M, Mun J-H, Lee I, Woo JC, Hong CB, Kim S-G (2006) Distinct roles of the first introns on the expression of Arabidopsis prolifin gene family members. Plant Physiol 140:196–209
Kang TJ, Kwon TH, Kim TG, Loc NH, Yang MS (2003) Comparing constitutive promoters using CAT activity in transgenic tobacco plants. Mol Cell 16:117–122
Klein TM, Wolf ED, Wu R, Sanford JC (1987) High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 327:70–73
Ling L, Maillet DS, Frappier JRH, Walden DB, Atkinson BG (1995) Characterization, chromosomal mapping, and expression of different polyubiquitin genes in tissues from control and heat-shocked maize seedlings. Biochem Cell Biol 73:19–30
Liu ZB, Ulmasov T, Shi X, Hagen G, Guilfoyle (1994) Soybean GH3 promoter contains multiple auxin-inducible elements. Plant Cell 6:645–657
Luehrsen KR, Walbot V (1991) Intron enhancement of gene expression and the splicing efficiency of introns in maize cells. Mol Gen Genet 225:81–93
Marrs KA, Casey ES, Capitant SA, Bouchard RA, Dietrich PS, Mettler IJ, Sinibaldi RM (1993) Characterization of two maize HSP90 heat shock protein genes: expression during heat shock, embryogenesis, and pollen development. Dev Genet 14:27–41
Matsumoto K, Montzka-Wasserman K, Wolffe AP (1998) Nuclear history of a pre-mRNA determines the translational activity of cytoplasmic mRNA. EMBO J 17:2107–2121
McElroy D, Zhang W, Cao J, Wu R (1990) Isolation of an efficient actin promoter for use in rice transformation. Plant Cell 2:163–171
Miller G, Mittler R (2006) Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann Bot 98:279–288
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nagatani N, Takuni S, Tomiyama M, Shimada T, Tamiya E (1997) Semi-real time imaging of the expression of a maize polyubiquitin promoter-GFP gene in transgenic rice. Plant Sci 124:49–56
Odell JT, Nagy F, Chua NH (1985) Identification of DNA sequences required for activity of the cauliflower mosaic virus 35S promoter. Nature 313:810–812
Ow DW, Wood KV, Deluca M, de Wet JR, Helinski DR, Howell SH (1986) Transient and stable expression of the firefly luciferase gene in plant cells and transgenic plants. Science 234:856–859
Paszkowski J, Peterhans A, Bilang R, Filipowicz W (1992) Expression in transgenic tobacco of the bacterial neomycin phosphotransferase gene modified by intron insertions of various sizes. Plant Mol Biol 19:825–836
Piston DW, Patterson GH, Knobel SM (1999) Quantitative imaging of the green fluorescent protein (GFP). Method Cell Biol 58:31–48
Plesse B, Criqui M-C, Durr A, Parmentier Y, Fleck J, Genschik P (2001) Effects of the polyubiquitin gene Ubi.U4 leader intron and first ubiquitin monomer on reporter gene expression in Nicotiana tabacum. Plant Mol Biol 45:655–667
Ponappa T, Brzozowski AE, Finer JJ (1999) Transient expression and stable transformation of soybean using the jellyfish green fluorescent protein. Plant Cell Rep 19:6–12
Prasinos C, Krampis K, Samakovli D, Hatzopoulos P (2005) Tight regulation of expression of two Arabidopsis cytosolic Hsp90 genes during embryo development. J Exp Bot 56:633–644
Rasband WS (1997–2006) ImageJ, U. S. National Institutes of Health, Bethesda, Maryland, USA. http://www.rsb.info.nih.gov/ij/
Richards HA, Halfhill MD, Millwood RJ, Stewart CN (2003) Quantitative GFP fluorescence as an indicator of recombinant protein synthesis in transgenic plants. Plant Cell Rep 22:117–121
Rizhsky L, Liang H, Mittler R (2002) The combined effect of drought stress and heat shock on gene expression in tobacco. Plant Physiol 130:1143–1151
Rolfe SA, Tobin EM (1991) Deletion analysis of a phytochrome-regulated monocot rbcS promoter in a transient assay system. PNAS USA 88:2683–2686
Rollfinke IK, Silber MV, Pfitzner UM (1998) Characterization and expression of a heptaubiquitin gene from tomato. Gene 211:267–276
Sahoo PK, Soltani S, Wong AKC (1988) A survey of thresholding techniques. Comput Vision Graph 41:233–260
Samac DA, Tesfaye M, Dornbusch M, Saruul P, Temple SJ (2004) A comparison of constitutive promoters for expression of transgenes in alfalfa (Medicago sativa). Transgenic Res 13:349–361
Santarém ER, Finer JJ (1999) Transformation of soybean (Glycine max (L.) Merrill) using proliferative embryogenic tissue maintained on semi-solid medium. In Vitro Cell Dev Biol Plant 35:451–455
Stewart CN, Millwood RJ, Halfhill MD, Ayalew M, Cardoza V, Kooshki M, Capelle GA, Kyle KR, Piaseki D, McCrum G, DiBenedetto J (2005) Laser-induced fluorescent imaging and spectroscopy of GFP transgenic plants. J Fluoresc 15:697–705
Tanaka A, Mita S, Ohta S, Kyozuka J, Shimamoto K, Nakamura K (1990) Enhancement of foreign gene expression by a dicot intron in rice but not in tobacco is correlated with an increased level of mRNA and an efficient splicing of the intron. Nucleic Acids Res 18:6767–6770
Thévenaz P, Ruttimann UE, Unser M (1998) A pyramid approach to subpixel registration based on intensity. IEEE Trans Image Process 7:27–41
Thibaud-Nissen F, Shealy RT, Khanna A, Vodkin LO (2003) Clustering of microarray data reveals transcript patterns associated with somatic embryogenesis in soybean. Plant Physiol 132:118–136
Vain P, Finer KR, Engler DE, Pratt RC, Finer JJ (1996) Intron-mediated enhancement of gene expression in maize (Zea mays L.) and bluegrass (Poa pratensis L.). Plant Cell Rep 15:489–494
Vancanneyt G, Schmidt R, O’Connor-Sanchez A, Willmitzer L, Rocha-Sosa M (1990) Construction of an intron-containing marker gene: splicing of the intron in transgenic plants and its use in monitoring early events in Agrobacterium-mediated plant transformation. Mol Gen Genet 220:245–250
Vasil V, Clancy M, Ferl RJ, Vasil IK, Hannah LC (1989) Increased gene expression by the first intron of maize Shrunken-1 locus in grass species. Plant Physiol 91:1575–1579
Vaucheret H (1994) Promoter-dependent trans-inactivation in transgenic tobacco plants; kinetic aspects of gene silencing and gene reactivation. C R Acad Sci 317:310–323
Wang J, Oard JH (2003) Rice ubiquitin promoters: deletion analysis and potential usefulness in plant transformation systems. Plant Cell Rep 22:129–134
Acknowledgments
Salaries and research support were provided by the United Soybean Board, the Yeungnam University research grants in 2006 and by State and Federal funds appropriated to The Ohio State University/Ohio Agricultural Research and Development Center. Mention of trademark or proprietary products does not constitute a guarantee or warranty of the product by OSU/OARDC and also does not imply approval to the exclusion of other products that may also be suitable. Journal Article No HCS-07-07.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by D.A. Somers.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Chiera, J.M., Bouchard, R.A., Dorsey, S.L. et al. Isolation of two highly active soybean (Glycine max (L.) Merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep 26, 1501–1509 (2007). https://doi.org/10.1007/s00299-007-0359-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-007-0359-y