Abstract
Indoleamine 2,3-dioxygenase-1 (IDO-1) is a heme-containing enzyme that initiates the kynurenine pathway by catalyzing the first step in l-tryptophan catabolism. IDO-1 has been shown to play an important role in immunosuppressive mechanisms and tumor evasion, making it an attractive target for therapeutic intervention, a computer-aided drug design (CADD) approach was applied to a set of 34 of 4,5-Disubstituted 1,2,3-triazole derivatives that had been evaluated for their IC50 values against indoleamine 2,3-dioxygenase 1 (IDO1) enzyme. By employing atom-based 3-QSAR pharmacophores and field-based 3D-QSARs, the study identified specific chemical groups and structural locations that could be modified to enhance the compounds’ activity against IDO1 in order to discover more effective inhibitors. The study utilized atom-based 3-QSAR pharmacophores and field-based 3D-QSARs to model the 4,5-Disubstituted 1,2,3-triazole derivatives against IDO1, leading to the identification of specific chemical groups and structural regions that could be altered to enhance the compounds’ activity. The triazole derivatives were subjected to molecular docking, yielding docking scores of − 7.12 kcal/mol for compound 25, and − 7.1, − 7.4, − 7.4, − 8.1, and − 8.3 kcal/mol for the five newly designed molecules T01-T05, respectively. Moreover, an assessment of the free energy was conducted to determine the energetic factors that contribute to the stability of the compounds within the enzyme’s binding site.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently, cancer research sees immunotherapy as a major and very promising advance [1, 2]. Recent successes in clinical trials against a wide range of cancers, with immune checkpoint inhibitors such as anti-PD-1, anti-CTLA4, and anti-PD-L1 monoclonal antibodies, have shown therapeutic results remarkable in many cases [3, 4]. Because of this, boosting the body’s immune system is considered one of the safest approaches to fighting cancer and many other diseases [5]. Excess catabolism of the essential amino acid L-tryptophan (L-Trp) allows cancer cells to escape the normal immune attack. This leads to the disproportionate production of kynurenine and other metabolites that suppress T cell functions [6]. Blocking this immunosuppressive mechanism is considered a promising approach to treat cancer, neurological disorders, autoimmune diseases and other diseases which are immune mediated.
IDO1 is a heme-containing dioxygenase that catalyzes the initial and rate-limiting step of the kynurenine pathway by oxidatively cleaving tryptophan to N-formylkynurenine. It is responsible for over 90% of tryptophan metabolism in humans. Under certain conditions such as pregnancy, chronic infections, organ transplantation, and tumors, IDO expression can be significantly increased by interferon (IFN) or other cytokines. Local depletion of tryptophan and accumulation of toxic catabolites, such as kynurenines, create a tolerogenic microenvironment that leads to local immunosuppression characterized by T cell anergy and regulatory T cell activation. This type of microenvironment is involved in a wide range of human diseases, including infections, autoimmune diseases, and cancer tumors [7, 8].
Of note, upregulation of IDO1 expression has been commonly observed in several types of cancers, such as colorectal cancer, non-small cell lung carcinoma, ovary, melanoma, and hepatocarcinoma, and is associated with a poor prognosis [9, 10]. Therefore, IDO1 and TDO are considered as potential targets for cancer immunotherapy. The discovery of inhibitors for IDO1 has garnered a significant amount of interest from both academic and industrial communities in recent years. Various potent IDO1 inhibitors have been reported [11,12,13,14,15]. Some of these inhibitors have been examined in clinical trials, such as NLG 919, Epacadostat, and PF-0684003 (Fig. 1). However, the failure of the IDO1 inhibitor Epacadostat in a phase III clinical trial (ECHO-301) for the treatment of metastatic melanoma has significantly dampened the enthusiasm for research and development of drugs targeting IDO1 [13, 16].
In recent years, the field of drug design has greatly benefited from the use of three-dimensional quantitative structure–activity relationship (3D-QSAR) as a computational tool [17,18,19,20]. Specifically, techniques such as atom-based 3D-QSAR and field-based 3D-QSAR have been instrumental in identifying essential structural characteristics and guiding the modification of compounds to enhance their effectiveness, particularly in the development of Indoleamine 2,3-Dioxygenase 1 enzyme inhibitors [21, 22]. Several other computational approaches have also been utilized for this purpose. A theoretical investigation into the area of drug design for IDO1 inhibitors was carried out by Lu Zhang and colleagues. Firstly, they developed and verified a QSAR pharmacophore model that utilized the ferrous bonding group (FBG). This model was then utilized for virtual screening. Subsequently, molecular docking simulations and similarity analysis were performed to identify the bioactive conformations of the inhibitors [23]. Sanskar Jain et al. performed a classification QSAR analysis via Monte Carlo optimization by identifying structural fingerprints to design inhibitors against the enzyme IDO1 [24]. Yeheng Zhou and colleagues employed several methods including Lipinski’s Rule of Fives, Veber’s rules filter, molecular anchoring, HipHop Pharmacophores, 3D-quantitative structure activity relation (3D-QSAR) studies, and Pan-assay interference compounds (PAINS) filter. Pharmacophores and 3D-QSAR models were constructed based on the results obtained from these approaches. Docking and QSAR models were used in conjunction to identify critical components for developing novel inhibitors with superior IDO1 inhibitory properties.
In the present work, the analysis of atom-based 3D QSAR and field-based 3D-QSAR were performed on IDO1 inhibitors to determine structural guidelines for the computational design of novel inhibitors with higher potency. Molecular docking is applied to understand the mechanism of interaction between a studied ligand and its protein target and to predict the best binding conformation [25]. To further ensure the orientation of the ligands inside the active sites of IDO1, we performed the docking of the designed compounds and the more active compound 25 [26, 27]. In addition, the MM/PBSA calculation provided free energy binding of the compound studies to assess the energetic component involved in the active site of IDO1 [28].
Methods and computational materials
Data set and ligand preparation
Derivatives of 4,5-Disubstituted 1,2,3-triazole have been synthesized and their anticancer biological activities have been evaluated by Subhankar Panda et al. [29]. In this study, a set of 34 compounds was selected to develop a pharmacophore model and discovery basic features of chemicals structure for the inhibitory activity effect. For a linear layout, experimental inhibitory IC50 values of 34 compounds in the dataset were converted to pIC50 values (pIC50 = log(IC50 (10–6))) [30]. The values of inhibitory activities of the compounds studied varied from 4.64 to 7.123 (pIC50). The molecules were drawn in 2D and then converted to 3D using Maestro software package (Schrödinger Release, 2021). The energy minimization of the structure of the compounds was carried out by applying the LigPrep module (Schrödinger Release: LigPrep, 2021) of the Maestro software using OPLS_2005 (optimized potentials for liquid simulations) with an implicit distance-dependent dielectric solvation treatment [31, 32]. The Epik (Schrödinger Release: Epik, 2021), which is based on the more accurate Hammett and Taft methodologies was applied to generate the ionization/tautomeric states [33].
Determination of pharmacophore
In the present work, the PHASE module was used to generate a pharmacophore model [34]. A database of Fig. 1 was aligned using a common scaffold alignment method, then conformers were created using the macropattern search method with a maximum of 1000 per structure, then minimized by OPLS-2005 the force field using a minimization of 100 steps [35, 36]. PHASE provides six built-in pharmacophore characteristics such as hydrogen bond acceptor (A), hydrogen bond donor (D), hydrophobic group (H), ring aromaticity (R), positively ionizable groups (P), and negatively ionizable (N) [37]. The number of features in the hypothesis ranged from 4 to 6, which generated 10 different pharmacophore hypotheses. Based on activity, the compounds were classified as active if they had a pIC50 value greater than 5.7 (13 compounds) and inactive if they had a pIC50 value less than 5.2 (9 compounds). Compounds falling in between were referred to as intermediates. The process involved searching for pharmacophores in 13 active compounds, using a final box size of 1 Å and a minimum intersite distance of 2 Å. Subsequently, ten pharmacophore hypotheses were generated and evaluated based on survival scores, site scores, vector scores, volume scores, selectivity, and phase hyposcores, with the rankings reflecting the outcome of this evaluation.
In the context of the pharmacophore hypothesis, the different scores can be explained as follows:
Survival score: The survival score assesses the ability of a compound to survive or maintain its biological activity under a specific set of conditions. It is used to assess the stability and robustness of compounds in a given biological environment.
Site score: Site score evaluates a compound’s ability to specifically bind to an active site or target region on a biological molecule. It measures the affinity and bond strength between the compound and the desired site of interaction. A high site score suggests a better ability of the compound to interact with the target site, which may be beneficial for the desired pharmacological activity.
Vector score: Vector score assesses the structural similarity between a compound and a reference vector or template representing desired pharmacophoric characteristics. It is used to measure how closely the compound matches the structural characteristics sought for the specific biological activity. A high end vector score indicates a better match between the compound and the pharmacophore template, which may suggest a greater likelihood of desired biological activity.
These different scores provide crucial information to assess the potential of a compound under the pharmacophore hypothesis. They allow quantification of stability, interaction with active sites, and conformity to desired structural features, which helps researchers select the most promising compounds for further experimental studies or for potential drug development.
Generation of field-based 3D-QSAR model
To create a 3D QSAR model that accurately predicts the activity, it is important to properly align the optimized conformers of dataset structure in a fixed network [38]. In this particular study, the molecules were aligned using different structure alignment tools in Maestro 12.9, including shape-based flexible alignment, common scaffold alignment, maximum common substructure, and intelligence as shown in Fig. 2. The grid spacing was set at 0.1 Å and extended 3.0 Å beyond the train set limits. The force field values were set to ignore anything less than 2.0 Å, with steric and electrostatic truncated force field preset to 30.0 kcal mol − 1. Variables with a standard deviation (SD) of < 0.01 were suppressed, and 1 ligand was excluded from cross-validation. The model was constructed using 5 features: steric, electrostatic, H-bond acceptor, bond donor H, and hydrophobic field.
3D-QSAR modeling by partial least square (PLS) analysis
Atom-based and field-based 3D-QSAR models were built using the PHASE module of Schrödinger’s software. These models were developed to correlate the structural characteristics of a set of compounds and anticancer biological activity. Partial least squares (PLS) analysis was used to develop 3D-QSAR models, taking pIC50 values as dependent variables and Gaussian intensities as descriptors (independent variables) [39]. Some statistical parameters such as standard deviation (SD), uncross-validated correlation coefficient (R2), cross-correlation validation was determined based on the predictions generated by a (LOO) (R2CV), R2 scramble, Fischer’s test (F-test), variance ratio (p-value), root mean square error (RMSE), Pearson-r values, correlation coefficient of external validation (Q2), and Pearson-r were estimated to assess the reliability of the established models. To assess the predictive ability of the developed QSAR models.
In the atom-based, field-based 3D-QSAR model, the dataset was randomly split into a train set containing 26 (76%) compounds and the remaining 8 compounds were reserved for test set (24%). PLS factors of 1 to 5 have been selected for the generation of atom-based and field-based QSAR models. A total of 10 models are built into the atomic and field-based QSAR models. Field-based models were represented as steric, electrostatic, hydrophobic, hydrogen fields description of hydrogen bond donor and acceptor [40, 41].
Docking study
The crystal structure of IDO1 was obtained from the Protein Data Bank (PDB) using code 4PK5 at a resolution of 2.79 Å [42, 43]. The A-chain of the structure underwent preparation, which included adding hydrogens, determining ionization states at pH 7.0, and optimizing/minimizing the structure with the Protein Preparation Wizard tool in Maestro 10.1 (Schrödinger Inc.) [44]. Prime 3.9 was used to rebuild the unresolved loop 361–379. The iron ion in the structure was defined as FeIII based on experimental evidence that imidazole-based molecules preferentially bind to the oxidized form of the enzyme. The ligands were prepared with LigPrep 3.3, generating all possible ionization states at pH 7.2. Docking studies were conducted using Glide 6.6 in standard precision (XP) mode, and the top ten bonding poses for each molecule were recorded as output (Schrödinger Release: Glide, 2020). Grids were defined for each crystal, with the center located at the center of mass of the co-crystallized ligand. The inner grid box was set to 20 × 20 × 20 angstroms, and all residual rotating groups within the outer box were considered. All other docking parameters were set to their default values.
Binding free energy calculation
The docked complexes were subjected to prediction of binding free energies using the generalized born surface method of molecular mechanics (MM-GBSA) [45]. This approach is considered as an interesting tool for the correct classification of inhibitors. The local optimization feature of Prime (v4.3) was used for structure minimization of anchored ligand-enzyme complexes [46]. Simulations were performed using partial loadings of input ligand without applying stress on the flexible residues. Binding free energies were calculated with the OPLS3 force field using the continuous solvent MM-GBSA and the solvation model VSGB 2.0 [47, 48].
Absorption, distribution, metabolism, and excretion predictions
The QikProp module (QikProp, v. 6.6, Schrödinger, LLC) has been applied to predict ADME properties, to assess the pharmacocentical behavior of compounds [49, 50]. QikProp is used to predict pharmacologically relevant characteristics of organic compounds. The QikProp also provides limits to contrast the properties of a certain compound with those of 95% of known drugs [51].
Results and discussion
Scoring for best pharmacophore hypothesis
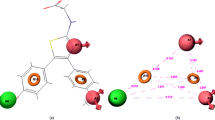
In this study, ten pharmacophore hypotheses were generated and ranked based on their scores. The selection of the best hypothesis was determined by considering survival score, site score, and vector score, as shown in Table 1. The most optimal six-feature pharmacophore hypothesis, AAADHR_1, was chosen for further investigation. The distances between different sites of this selected model can be observed in Fig. 3A.
Representation of cross-site AAADHR_1 pharmacophore model (A) distances in Å unit. Hydrogen bond acceptor A: Pink sphere with arrow; Aromatic ring (R): yellow open circle; Hydrogen bond donor D: blue sphere with arrow; B alignment of active compounds to the generated pharmacophore template and C alignment of inactive compounds to the generated pharmacophore template
Using the AAADHR_1 model as a reference, both the training set and test set were aligned. Additionally, a statistically significant atom-based 3D-QSAR model was developed through partial least squares (PLS) regression, utilizing the 19 aligned active molecules from the training set. The model’s statistical significance and predictivity increased as the number of PLS factors increased, up to a maximum of five factors (see Table 2). Figure 3B, C depicts the alignment of active and inactive ligands with the generated AAADHR_1.
Atom-based 3D-QSAR models
In the current study, both atom-based and field-based models were developed using a training set consisting of 26 compounds, with an additional 8 compounds used for testing. The models were developed with a PLS factor set to 5. To assess the statistical stability of the models, internal validation methods were employed.
Table 3 presents the statistical results for the developed models. The atom-based models performed statistically better with a PLS factor of 5. The coefficient of determination for the training set (R2) was found to be 0.8881. The cross-validated R2 value obtained through the LOO method (R2CV) was 0.5248. The Q2 value for predicted activities (Q2 for atom-based) was 0.8333.
Additional statistical parameters were evaluated to assess the performance of the models. The standard deviation (SD) was determined to be 0.28, while the root mean square error (RMSE) was calculated to be 0.71. The variance ratio significance level (P) was reported as 7.45 × 10−9, and the variance ratio (F) was found to be 31.7. These results indicate that the developed 3D-QSAR models are effective and suitable for selecting the best models in this study.
The contribution percentages of different fields to the developed models are presented in Table 4. In the best model, utilizing a PLS factor of 5, the hydrophobic property emerged as the predominant factor, contributing 63.2% to the overall model. The Electron-withdrawing and Negative ionic properties accounted for 17.8% and 13.4%, respectively. On the other hand, the H-bond donor property made minimal contributions to the established model, representing approximately 5.7% of the overall contribution.
The correlation between experimental and predicted activity was visualized for both the training and test sets in Fig. 4. Overall, the developed model exhibited less dispersion between the experimental and predicted values.
Further to verify the trustworthiness of this structure-based pharmacophore receiver operating characteristics (ROC), area under accumulation curve (AUAC) as well as the enrichment factor (EF) are calculated to evaluate its ability to detect active and inactive set of compounds with known biological activity [52, 53]. The ROC value represents the sensitivity (proportion of true positives) and a function of specificity (proportion of false positives). The value of the area under accumulation curve (AUAC) provides an objective measure of the overall performance of a classifier [54]. An AUAC value equal to 1 (or 100%) indicates that the active and inactive compounds are perfectly discriminated. The enrichment factor describes the number of active compounds found using a specific pharmacophore model as opposed to the number hypothetically found if compounds were screened randomly. In pharmacophore hypothesis AAADHR_1 when tested on active set of compounds showed promising results with Enrichment factor 80.80%, ROC 0.91 and AUAC 0.52 (Area under accumulation curve) (Fig. 5), it confirmed that pharmacophore hypothesis AAADHR_1 is good model to predict active molecule.
Field-based 3D-QSAR models
The 3D QSAR models were developed using PLS analysis, incorporating five descriptors: steric, electrostatic, hydrophobic, H-bond Donor, and H-bond Acceptor properties. The most effective model, based on the Gaussian field, was achieved with a PLS factor of 5. The statistical parameters obtained are summarized in Table 5.
The R2cv cross-validation coefficient for the model is 0.603, indicating a moderate level of predictive ability. The standard deviation of the regression (SD value) is 0.3895, which represents the average distance between the predicted and actual values. The Fischer ratio F is 12.5, indicating a statistically significant model. The stability of the model is determined to be 0.668. In the best established model, the contributions from steric and electrostatic fields are 21.6% and 21.8%, respectively (Table 6). The hydrophobic field makes the highest contribution of 38.4%, followed by H-bond Acceptor with 17.7%, and H-bond Donor with a minimal contribution of 0.4%. These results suggest that the hydrophobic properties of the protein–ligand interaction fields play a more significant role compared to other fields.
Overall, the findings highlight the importance of hydrophobic interactions in the protein–ligand binding process, supported by the higher contributions observed in the hydrophobic field compared to the other fields.The plot of the correlation between the experimental biological activity and that predicted from the train and test sets is presented in Fig. 6.
3D-QSAR visualization
Atom-based 3D-QSAR visualization
The biological activity studied was correlated with different structural properties and described in the form of contour maps. The atom-based 3D-QSAR model for Indoleamine 2,3-dioxygenase-1 (IDO-1) inhibition was applied to the most active compound 25 (pIC50 = 7.161) to visualize the most important favorable and unfavorable regions for H-bond donor, hydrophobic groups, negative ionic, and Electron-withdrawing (Fig. 7A–D). The contour visualization technique aids in identifying preferred functional groups and highlighting specific positions within a compound that contribute to its activity.
Visual representation of atom-based 3D-QSAR models. A H-bond donor—blue color indicates a positive coefficient or increase in activity and red cubes indicate a negative coefficient or decrease inactivity. B Hydrophobic—yellow shade indicates a positive coefficient, and cyan e cubes indicate a negative coefficient. C Negative ionic—light red cubes indicate a positive coefficient, and orange cubes indicate a negative coefficient. D Electron-withdrawing—purple cubes indicate a positive coefficient, and green cubes indicate a negative coefficient
For hydrogen bond donor attributes, favorable regions contributing to ligand interactions with the target enzyme are represented by blue cubes, while red cubes indicate unfavorable features affecting activity. As shown in Fig. 7A, red cubes appeared around the NH of the triazole moiety suggesting that the hydrogen bond donating groups at this position are unfavorable for inhibitory antiproliferative activity. Further, it is apparent from the appearance of blue cubes around the oxygen of the amide group indicating that the hydrogen bond donor group at this position is favorable to activity.
The hydrophobic character is an important factor which has an impact on the activity (38.4%). Yellow and cyan cubes indicate favorable and unfavorable characteristics contributing to biological activity. As shown in Fig. 7B. The appearance of yellow cubes at the C2 and C5 positions of the phenyl ring, around the triazole group and around the NH2 group of the amide indicated the preference of the hydrophobic group at these positions, while the appearance of cyan cubes around the phenyl substituted chlorine atoms, and around the oxygen atom of the amide group indicating the preference of the hydrophilic group at these positions. Generally, the yellow cubes are more, reflecting the hydrophobic substitution can increase the antiproliferative inhibition activity.
The negative ion function map is shown in Fig. 7C in which the only orange color cubes have been appeared indicating the positive coefficients on the activity. The NH2 group of amide group promotes the substitution of the negative ion group, and it can increase the activity. Regarding the electron attractor volume occlusion maps, as shown in Fig. 7D, the purple and green cubes indicate respectively the positive and negative coefficients on the activity. The red cubes are localized more in the vicinity of the nitrogen atoms of the triazole group while the amide group is covered by both types of the cubes but more by the purple cubes, which indicates the preference of the substitution of electron withdrawing groups, which can increase inhibitory activity.
Gaussian-based 3D-QSAR model visualization
The developed model based on the PLS five-component Gaussian field was applied to visualize the contour map visualization of the steric field, electrostatic field, hydrophobic field of the hydrogen bond acceptor, hydrogen bond donor, and as the represented in Fig. 8. Always compound 25 was taken to locate structural features generated by 3D field-based QSAR analysis.
In the Gaussian steric contour (Fig. 8A), the green color indicates regions favorable for steric interactions. We can observe two green contours around the nitrogen and oxygen atoms of the amide group, indicating that substitution with steric groups at this position can increase the activity.
The electrostatic contour map is represented by red and blue colors, indicating favorable and unfavorable regions for the studied activity. As depicted in Fig. 8B, three blue contour maps around the benzene ring, the amide group, and the chlorine atom in the Meta position represent electropositive properties that are favorable to the activity. Conversely, a primary red contour covers the chlorine atom, suggesting that electronegative substitutions at this position are unfavorable for the activity.
The hydrophobic contour maps, represented by the yellow color, suggest favorable regions for activity, such as the phenyl group substituted by the two chlorine atoms and around the oxygen atom of the amide group (Fig. 8C). On the other hand, the hydrophilic contour maps, represented by a larger gray area around the NH2 amide group but slightly further away, suggest regions that are disadvantaged for activity, although with a low impact.
In Fig. 8D, the contour maps associated with the hydrogen bond acceptor field, represented by the red color, are observed around the NH2 amide group and the oxygen atom, indicating regions favorable for activity. Similarly, the hydrogen bond donor field is depicted, showing purple-colored maps around the NH3 amide group, which are favored for activity (Fig. 8E).
Molecular design of novel chemical compounds
Based on the generated contour maps from the models, five inhibitors (T01-T05) were designed based on compound 25, which demonstrated high bioactivity. The 3D structures of these inhibitors were sketched using Schrodinger and prepared using the same LigPrep protocol. The bioactivities of all inhibitors were predicted using both the atom-based 3D QSAR models and field-based 3D QSAR models created previously, and the results are summarized in Table 7.
The triazole and phenyl groups were retained in Fig. 9, and further modifications were made based on the contour maps of the developed models. Bulky groups were added to the amide function, and hydrophilic groups were eliminated. Electronegative groups, such as chlorine or trifluoromethyl, were also introduced.
Docking studies
The examination of the binding site was carried out using the SiteMap tool of the Schrödinger software (Schrödinger Release: SiteMap, 2021) [55]. SiteMap found five sites which were expressed in terms of site score, size, volume, amino acid exposure, enclosure, contact, hydrophobicity, hydrophilicity, and donor/acceptor ratio. Generally, sites with a site score of 1 or greater can be considered an ideal site for ligand binding. The best level of potential receptor binding sites was identified using SiteMap as shown in Table 8 and Fig. 10.
The best site had a score of 1.281 site score, 1.363 Dscore, 126.224 volume score, 4.3880 hydrophobic score, 0.334 hydrophilic score, and 1.489 hydrogen bond donor/acceptor score. The active site residues were identified as Gly-251 | Glu-254 | Asp-255 | Pro- 256 | Glu-258 | Phe-259 | Ala-260 | Gln-280 | Gln-281 | Thr-282 | Ala-283 | Glu-311 | Ser-315 | Thr-395 | Glu-396 | Lys 397 | Leu-400 | Glu-402 | Gly-403, respectively. Thus, active site 3 was most likely to perform the docking process to know the intermolecular interaction of the ligand with the targeted receptor by applying the Glide module.
Thus, in our work, a docking score was used to assess the stability of the simulated study complexes. The results of the molecular docking analysis are presented in Table 9 and Fig. 11. The docking affinity obtained for the inhibitor 25 was − 7.7. The newly designed compounds T01-T05 exhibited docking score of − 8.3, − 8.1, − 7.4, − 7.4, and − 7.1, respectively. These results indicate that the proposed molecules not only showed high predicted activity but were also more stable than the 25/4PK5 complex.
In addition, among the inhibitors studied, the docking results of ligand 25 revealed that the oxygen atom of the amide group acted as a hydrogen bond donor. This interaction led to the formation of a hydrogen bond with Ala-264. The presence of this hydrogen bond suggests a potential stabilizing effect and indicates favorable binding of ligand 25 to the target receptor. These findings provide important insights into the specific molecular interactions and binding modes of the ligand, contributing to our understanding of its activity and potential as a therapeutic agent.
Regarding the designed compounds, for the T01 ligands, the sulfonamide functional oxygen and nitrogen atoms acted as hydrogen bond acceptors, establishing hydrogen bonds with the residues Gly-236, Gly-261, and Lys-238. The benzene ring and triazole core formed hydrogen carbon bonds with Arg-231. For the T01 ligand, the oxygen atom and sulfamide group nitrogen acted as a hydrogen bond acceptor by creating hydrogen interactions with Glu-236 and Lys-238, respectively. The triazole ring nitrogen established a hydrogen bond with Arg-231, while the benzene ring of ligand T02 established π-Alkyl interactions with residue Leu-234. These global interactions with Arg-231 and Gly-262 could be the reason for the high docking affinity and hence it promises to be potent and also selective for activity.
The accuracy of the docking technique was assessed by comparing the predicted bonding conformation generated by the molecular docking performed and the experimental bonding mode observed by X-ray crystallography. The reliability of the docking mode was confirmed by the estimate of the root mean square deviation (RMSD) between the predicted and observed X-ray crystallographic conformation of (PDB code: 4PK5), which was determined to be 0.529 Å. This value obtained from RMSD suggests that the parameter set for the Glide XP docking is reasonable to reproduce the X-ray crystal structure [56]. Moreover, as shown in the Fig. 12, we observed a similar orientation during the superimposition of the co-crystalized conformation original and re-docked.
Assessment of free binding energy
The free energy binding of protein with compound 25 and designed compounds was estimated using the MM-GBSA method. The MM-GBSA prime module implanted in Schrödinger, 2021, was used to calculate the average binding energy as shown in Table 10.
The average binding free energy of T01, T02, T03, T04, and T05 complexed with the protein were found to be − 49.44291, − 55.9892, − 51.2387, − 57.6433, and − 59.9874, respectively, which was more interesting than the free binding energy shown by the compound 25 (− 43.5170). In addition, the results obtained show that energetic components of de van der Waals (∆GvdW), the packing fraction (∆GPacking) and lipophilic (∆GLipo), and Coulomb (∆GCoulomb) energetically favored the formation of the protein–ligand complex, while Binding Solvation GB and covalent binding (∆GCovalent) showed more positive values opposing the formation of the complex. Free energy data further reveal that van der Waals interactions contribute significantly to protein–ligand complexes for all selected systems studied.
ADME results
ADME properties are of great interest to researchers in the development of new drugs due to their cost-effectiveness and their ability to be evaluated at high throughput [57].
Using QikProp, 16 relevant pharmacokinetic and physics-related parameters were determined for all designed ligands, as well as ligand 25 (Table 11). Some of these parameters include molecular weight, octanol/water partition coefficient (QPlog(Po/w), water/gas partition coefficient (QP log Kp), hydrogen acceptors and donors, percentage of oral absorption in humans, brain-blood partition coefficient (QP log BB), solubility, dipole, and SASA calculated values of drugs within a range that indicates similarity to existing drugs. These properties provide useful information on how similar the ligands are to existing drugs.
Conclusion
In this study, two practical methods, field-based 3D QSAR and atom-based 3D QSAR, were used to analyze the structural properties correlated with the experimental activity of 4,5-Disubstituted 1,2,3-triazole derivatives as IDO1 inhibitors. Gaussian-based 3D-QSAR models were generated to identify distinct pharmacophoric characteristics. The study revealed both favorable and unfavorable chemical structural characteristics for activity and provided useful information for the design of new triazole-based inhibitors. The results indicated that hydrophobic (38.4%), electrostatic (21.8%), steric (21.6%), and H-Bond acceptor (17.7%) characteristics play an interesting role in inhibiting IDO1. Additionally, a six-point pharmacophore model based on the AAADHR_1 hypothesis was generated to gain a better understanding of the favorable spatial fingerprints of triazole as IDO1 inhibitors. Using this structural information, four new inhibitors were designed and demonstrated improved activity. To evaluate the mode of binding of compound 25, and the designed compounds in the active site was used for molecular docking analysis. The results showed the stability of the designed molecule as an advantage over the compound 25. The calculation of free binding energy also revealed the interesting role of van der Waals, hydrophilic, and Columbic interactions in the inhibition of IDO1, which confirms the findings of the 3D QSAR models. Finally, this study provided valuable pharmacophore information for molecule design with favorable activities against the IDO1 enzyme.
Availability of data and materials
Not applicable.
References
Dunn GP, Old LJ, Schreiber RD (2004) The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2:137–148. https://doi.org/10.1016/j.immuni.2004.07.017
Zou W (2005) Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer 5:263–274. https://doi.org/10.1038/nrc1586
Hoos A (2016) Development of immuno-oncology drugs—from CTLA4 to PD1 to the next generations. Nat Rev Drug Discov 15:235–247. https://doi.org/10.1038/nrd.2015.35
He X, Xu C (2020) Immune checkpoint signaling and cancer immunotherapy. Cell Res 30:660–669. https://doi.org/10.1038/s41422-020-0343-4
Schoenfeld AJ, Hellmann MD (2020) Acquired resistance to immune checkpoint inhibitors. Cancer Cell 37:443–455. https://doi.org/10.1038/s41422-020-0343-4
van Baren N, Van den Eynde BJ (2015) Tumoral immune resistance mediated by enzymes that degrade tryptophan. Cancer Immunol Res 3:978–985. https://doi.org/10.1158/2326-6066.CIR-15-0095
Platten M, Wick W, Van den Eynde BJ (2012) Tryptophan catabolism in cancer: beyond IDO and tryptophan depletion tryptophan catabolism in cancer. Cancer Res 72:5435–5440. https://doi.org/10.1158/0008-5472.CAN-12-0569
Munn DH, Mellor AL (2013) Indoleamine 2, 3 Dioxygenase and metabolic control of immune responses. Trends Immunol 34:137–143. https://doi.org/10.1016/j.it.2012.10.001
Xiao Y, Freeman GJ (2015) The microsatellite instable subset of colorectal cancer is a particularly good candidate for checkpoint blockade immunotherapy. Cancer Discov 5:16–18. https://doi.org/10.1016/j.it.2012.10.001
Phan T, Nguyen VH, D’Alincourt MS, Manuel ER, Kaltcheva T, Tsai W, Blazar BR, Diamond DJ, Melstrom LG (2020) Salmonella-mediated therapy targeting Indoleamine 2, 3-Dioxygenase 1 (IDO) activates innate immunity and mitigates colorectal cancer growth. Cancer Gene Ther 27:235–245. https://doi.org/10.1038/s41417-019-0089-7
Alexandre JAC, Swan MK, Latchem MJ, Boyall D, Pollard JR, Hughes SW, Westcott J (2018) New 4‐amino‐1, 2, 3‐triazole inhibitors of Indoleamine 2, 3‐Dioxygenase form a long‐lived complex with the enzyme and display exquisite cellular potency. ChemBioChem 19:552–561. https://doi.org/10.1002/cbic.201700560
Siu L, Gelmon K, Chu Q, Pachynski R, Alese O, Basciano P, Walker J, Mitra P, Zhu L, Phillips P (2017) BMS-986205, an optimized Indoleamine 2, 3-Dioxygenase 1 (IDO1) inhibitor, is well tolerated with potent pharmacodynamic (PD) activity, alone and in combination with nivolumab (nivo) in advanced cancers in a phase 1/2a trial. In Cancer Res 77
Yue EW, Sparks R, Polam P, Modi D, Douty B, Wayland B, Glass B, Takvorian A, Glenn J, Zhu W (2017) INCB24360 (epacadostat), a highly potent and selective Indoleamine-2, 3-Dioxygenase 1 (IDO1) inhibitor for immuno-oncology. ACS Med Chem Lett 8:486–491. https://doi.org/10.1021/acsmedchemlett.6b00391
Kumar S, Waldo JP, Jaipuri FA, Marcinowicz A, Van Allen C, Adams J, Kesharwani T, Zhang X, Metz R, Oh AJ (2019) Discovery of clinical candidate (1 R, 4 r)-4-((R)-2-((S)-6-Fluoro-5 H-Imidazo [5, 1-a] Isoindol-5-Yl)-1-Hydroxyethyl) Cyclohexan-1-Ol (Navoximod), a potent and selective inhibitor of Indoleamine 2, 3-Dioxygenase 1. J Med Chem 62:6705–6733. https://doi.org/10.1021/acs.jmedchem.9b00662
Crosignani S, Bingham P, Bottemanne P, Cannelle H, Cauwenberghs S, Cordonnier M, Dalvie D, Deroose F, Feng JL, Gomes B (2017) Discovery of a novel and selective Indoleamine 2, 3-Dioxygenase (IDO-1) inhibitor 3-(5-Fluoro-1 H-Indol-3-Yl) Pyrrolidine-2, 5-Dione (EOS200271/PF-06840003) and its characterization as a potential clinical candidate. J Med Chem 60:9617–9629. https://doi.org/10.1021/acs.jmedchem.7b00974
Muller AJ, Manfredi MG, Zakharia Y, Prendergast GC (2019) Inhibiting IDO pathways to treat cancer: lessons from the ECHO-301 trial and beyond. Semin Immunopathol Springer 41:41–48. https://doi.org/10.1007/s00281-018-0702-0
El Mchichi L, Tabti K, Kasmi R, El-Mernissi R, El Aissouq A, En-nahli F, Belhassan A, Lakhlifi T, Bouachrine M (2022) 3D-QSAR study, docking molecular and simulation dynamic on series of benzimidazole derivatives as anti-cancer agents. J Indian Chem Soc 99:100582. https://doi.org/10.1016/j.jics.2022.100582
Tabti K (2020) QSAR studies of new compounds based on thiazole derivatives as pin1 inhibitors via statistical methods. RHAZES: Green and Applied Chemistry 9:70–91. https://doi.org/10.48419/IMIST.PRSM/rhazes-v9.21394
El Masaoudy Y, Tabti K, Koubi Y, Maghat H, Lakhlifi T, Bouachrine M (2023) In Silico Design of new Pyrimidine-2, 4-Dione derivatives as promising inhibitors for HIV reverse transcriptase-associated RNase H using 2D-QSAR modeling and (ADME/Tox) properties. Moroc J Chem 11:300–317. https://doi.org/10.48317/IMIST.PRSM/morjchem-v11i2.35455
Tabti K, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2020) 2D and 3D-QSAR/CoMSIA Comparative study on a series of thiazole derivatives as SDHI Inhibitors. Maghreb J Pure & Appl Sci 6:73–90. https://doi.org/10.48383/IMIST.PRSM/mjpas-v6i2.23108
Kranthi RK, Manohar S, Talluri VR, Rawat DS (2015) Insights into activity enhancement of 4-aminoquinoline-based hybrids using atom-based and field-based QSAR studies. Med Chem Res 24:1136–1154. https://doi.org/10.1007/s00044-014-1195-6
Al-Sha’er MA, Taha M, Alelaimat MA (2023) Development of Phosphoinositide 3-Kinase Delta (PI3Kδ) inhibitors as potential anticancer agents through the generation of ligand-based pharmacophores and biological screening. Med Chem Res 17:30. https://doi.org/10.1007/s00044-023-03057-3
Zhang L, Lai F, Chen X, Xiao Z (2020) Identification of potential Indoleamine 2, 3-Dioxygenase 1 (IDO1) inhibitors by an FBG-based 3D QSAR pharmacophore model. J Mol Graph Model 99:107628. https://doi.org/10.1016/j.jmgm.2020.107628
Jain S, Bhardwaj B, Amin SA, Adhikari N, Jha T, Gayen S (2020) Exploration of good and bad structural fingerprints for inhibition of Indoleamine-2, 3-Dioxygenase enzyme in cancer Immunotherapy Using Monte Carlo Optimization and Bayesian Classification QSAR Modeling. J Biomol Struct Dyn 38:1683–1696. https://doi.org/10.1080/07391102.2019.1615000
Tabti K, Elmchichi L, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2022) Molecular modelling of antiproliferative inhibitors based on SMILES descriptors using Monte-Carlo method, docking, MD simulations and ADME/Tox studies. Mol Simul 48:1575–1591. https://doi.org/10.1080/08927022.2022.2110246
Hajji H, Tabti K, En-nahli F, Bouamrane S, Lakhlifi T, Ajana MA, Bouachrine M (2021) In silico investigation on the beneficial effects of medicinal plants on diabetes and obesity: molecular docking, molecular dynamic simulations, and ADMET studies. Biointerface Res Appl Chem 11:6933–6949. https://doi.org/10.33263/BRIAC115.69336949
Tabti K, Abdessadak O, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2023) Design and development of novel spiro-oxindoles as potent antiproliferative agents using quantitative structure activity based Monte Carlo method, docking molecular, molecular dynamics, free energy calculations, and pharmacokinetics/toxicity studies. J Mol Struct 1284:135404. https://doi.org/10.1016/j.molstruc.2023.135404
Tabti K, Ahmad I, Zafar I, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2023) Profiling the structural determinants of pyrrolidine derivative as gelatinases (MMP-2 and MMP-9) inhibitors using in silico approaches. Comput Biol Chem 107855. https://doi.org/10.1016/j.compbiolchem.2023.107855
Panda S, Pradhan N, Chatterjee S, Morla S, Saha A, Roy A, Kumar S, Bhattacharyya A, Manna D (2019) 4, 5-Disubstituted 1, 2, 3-Triazoles: effective inhibition of Indoleamine 2, 3-Dioxygenase 1 enzyme regulates T cell activity and mitigates tumor growth. Sci Rep 9:18455. https://doi.org/10.1038/s41598-019-54963-9
Tabti K, Elmchichi L, Sbai A, Maghat H, Bouachrine M, Lakhlifi T, Ghosh A (2022) In silico design of novel PIN1 inhibitors by combined of 3D-QSAR, molecular docking, molecular dynamic simulation and ADMET studies. J Mol Struct 1253:132291. https://doi.org/10.1016/j.molstruc.2021.132291
Shivakumar D, Williams J, Wu Y, Damm W, Shelley J, Sherman W (2010) Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput 6:1509–1519. https://doi.org/10.1021/ct900587b
Kalva S, Vinod D, Saleena LM (2013) Field-and Gaussian-based 3D-QSAR studies on barbiturate analogs as MMP-9 inhibitors. Med Chem Res 22:5303–5313. https://doi.org/10.1007/s00044-013-0479-6
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) Epik: a software program for PK a prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21:681–691. https://doi.org/10.1007/s10822-007-9133-z
Dixon SL, Smondyrev AM, Knoll EH, Rao SN, Shaw DE, Friesner RA (2006) PHASE: a new engine for pharmacophore perception, 3D QSAR model development, and 3D database screening: 1. Methodology and preliminary results. J Comput Aided Mol Des 20:647–671. https://doi.org/10.1007/s10822-006-9087-6
Mohamadi F, Richards NG, Guida WC, Liskamp R, Lipton M, Caufield C, Chang G, Hendrickson T, Still WC (1990) Macromodel—an integrated software system for modeling organic and bioorganic molecules using molecular mechanics. J Comput Chem 11:440–467. https://doi.org/10.1002/jcc.540110405
Dietrich M, Heinze J, Krieger C, Neugebauer FA (1996) Electrochemical oxidation and structural changes of 5, 6-Dihydrobenzo [c] Cinnolines. J Am Chem Soc 118:5020–5030. https://doi.org/10.1021/ja952722+
Sindhu T, Srinivasan P (2014) Pharmacophore modeling, 3D-QSAR and molecular docking studies of benzimidazole derivatives as potential FXR agonists. J Recept Signal Transduct Res 34:241–253. https://doi.org/10.3109/10799893.2014.885048
Tabti K, Elmchichi L, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2022) HQSAR, CoMFA, CoMSIA docking studies and simulation MD on quinazolines/quinolines derivatives for DENV virus inhibitory activity. Chem Afr 5:1937–1958. https://doi.org/10.1007/s42250-022-00484-4
Teli MK (2012) Pharmacophore generation and atom-based 3D-QSAR of N-Iso-Propyl pyrrole-based derivatives as HMG-CoA reductase inhibitors. Bioorganic Med Chem Lett 2:1–10. https://doi.org/10.1186/2191-2858-2-25
Yao K, Liu P, Liu H, Wei Q, Yang J, Cao P, Lai Y (2019) 3D-QSAR, molecular docking and molecular dynamics simulations study of 3-Pyrimidin-4-Yl-Oxazolidin-2-One derivatives to explore the structure requirements of mutant IDH1 inhibitors. J Mol Struct 1189:187–202. https://doi.org/10.1016/j.molstruc.2019.03.092
Palafox MA, Kattan D, de Pedraza Velasco ML, Isasi J, Posada-Moreno P, Rani K, Singh SP, Rastogi VK (2022) Base pairs with 4-Amino-3-Nitrobenzonitrile: comparison with the natural WC pairs. Dimer and tetramer forms, infrared and Raman spectra, and several proposed antiviral modified nucleosides. J Biomol Struct Dyn 1–23. https://doi.org/10.1080/07391102.2022.2069864
Tojo S, Kohno T, Tanaka T, Kamioka S, Ota Y, Ishii T, Kamimoto K, Asano S, Isobe Y (2014) Crystal structures and structure–activity relationships of imidazothiazole derivatives as IDO1 inhibitors. ACS Med Chem Lett 5:1119–1123. https://doi.org/10.1021/ml500247w
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank (Www. Rcsb. Org). Nucleic Acids Res 28:235–242
Madhavi SG, Adzhigirey M, Day T, Annabhimoju R, Sherman W (2013) Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des 27:221–234. https://doi.org/10.1007/s10822-013-9644-8
Tabti K, Baammi S, ElMchichi L, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2022) Computational investigation of pyrrolidin derivatives as novel GPX4/MDM2–P53 inhibitors using 2D/3D-QSAR, ADME/Toxicity, molecular docking, molecular dynamics simulations, and MM-GBSA free energy. Struct Chem 33:1019–1039. https://doi.org/10.1007/s11224-022-01903-5
Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA (2004) A hierarchical approach to all‐atom protein loop prediction. Proteins: Struct Funct Bioinfo 55:351–367. https://doi.org/10.1002/prot.10613
Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL (2016) OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput 12:281–296. https://doi.org/10.1021/acs.jctc.5b00864
Li J, Abel R, Zhu K, Cao Y, Zhao S, Friesner RA (2011) The VSGB 2.0 Model: a next generation energy model for high resolution protein structure modeling. Proteins: Struct. Funct. Bioinfo. 79:2794–2812. https://doi.org/10.1002/prot.23106
Rajagopal K, Varakumar P, Baliwada A, Byran G (2020) Activity of phytochemical constituents of Curcuma Longa (turmeric) and Andrographis Paniculata against coronavirus (COVID-19): an in silico approach. Future J Pharm Sci 6:1–10. https://doi.org/10.1186/s43094-020-00126-x
Ioakimidis L, Thoukydidis L, Mirza A, Naeem S, Reynisson J (2008) Benchmarking the reliability of QikProp. correlation between experimental and predicted values. QSAR Comb Sci 27:445–456. https://doi.org/10.1002/qsar.200730051
Onguéné PA, Ntie-Kang F, Mbah JA, Lifongo LL, Ndom JC, Sippl W, Mbaze LM (2014) The potential of anti-malarial compounds derived from African medicinal plants, part III: an in silico evaluation of drug metabolism and pharmacokinetics profiling. organic Med Chem Lett 4:1–9. https://doi.org/10.1186/s13588-014-0006-x
Kirchmair J, Markt P, Distinto S, Wolber G, Langer T (2008) Evaluation of the performance of 3D virtual screening protocols: RMSD comparisons, enrichment assessments, and decoy selection—What can we learn from earlier mistakes? J Comput Aided Mol Des 22:213–228. https://doi.org/10.1007/s10822-007-9163-6
Dessalew G, Beyene A, Nebiyu A, Astatkie T (2018) Effect of brewery spent diatomite sludge on trace metal availability in soil and uptake by wheat crop, and trace metal risk on human health through the consumption of wheat grain. Heliyon 4:e00783. https://doi.org/10.1016/j.heliyon.2018.e00783
Truchon J-F, Bayly CI (2007) Evaluating virtual screening methods: good and bad metrics for the “Early Recognition” problem. J Chem Inf Model 47:488–508. https://doi.org/10.1021/ci600426e
Halgren TA (2009) Identifying applications, and recent advances of protein-ligand docking in structure-based drug design. J Chem Inf Model 49:377–389
Tabti K, Baammi S, Sbai A, Maghat H, Lakhlifi T, Bouachrine M (2023) Molecular modeling study of pyrrolidine derivatives as novel myeloid cell leukemia-1 inhibitors through combined 3D-QSAR, molecular docking, ADME/Tox and MD simulation techniques. J Biomol Struct 1–17. https://doi.org/10.1080/07391102.2023.2183032
Tabti K, Hajji H, Sbai A, Maghat H, Bouachrine M, Lakhlifi T (2023) Identification of a potential thiazole inhibitor against biofilms by 3D QSAR, molecular docking, DFT analysis, MM-PBSA binding energy calculations, and molecular dynamics simulation. Phys Chem Res 11:369–389. https://doi.org/10.22036/PCR.2022.335657.2068
Acknowledgements
We are grateful to the “Association Marocaine des Chimistes Théoriciens” (AMCT) for its pertinent help concerning the programs.
Author information
Authors and Affiliations
Contributions
Kamal Tabti: Data curation, Writing—original draft. Abdelouahid Sbai: Conceptualization, Methodology, Software. Hamid Maghat: Visualization, Investigation, Supervision. Tahar Lakhlifi: Writing—review and editing.Mohammed Bouachrin: Software, Validation. All authors commented on previous versions of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
This chapter does not contain any studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tabti, K., Sbai, A., Maghat, H. et al. Discovery of novel indoleamine 2,3-dioxygenase-1 (IDO-1) inhibitors: pharmacophore-based 3D-QSAR, Gaussian field-based 3D-QSAR, docking, and binding free energy studies. Struct Chem 35, 135–160 (2024). https://doi.org/10.1007/s11224-023-02213-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-023-02213-0