Abstract
Background and aims
Soil salinization with high pH condition is a major abiotic stress to plant growth and crop productivity. Helianthus tuberosus L. is an important stress tolerant plant and can survive in the saline-alkali soil and semiarid areas. The aim of this study is to identify the effect of alkali stress on H. tuberosus through global proteomics analysis and improve understanding of the alkalinity resistance of plants.
Methods
H. tuberosus seedlings were exposed to different level alkali stress for 7 days. Protein profiling was quantified by conducting MS-based comparative proteomics analysis. RT-PCR study was carried out to analyze the mRNA expression levels of candidate alkali stress response proteins.
Results
The response of H. tuberosus to alkali stress was detected at both physiological and molecular levels. 104 differentially expressed proteins from H. tuberosus leaves response to Na2CO3 treatment were successfully identified. Functional categorization of these identified proteins showed that the accumulation level of proteins involved in glycolysis, TCA cycle, PSI system, ROS scavenging and signal transduction increased under alkali stress.
Conclusions
Based on the observation of plant growth and the investigation of molecular regulation, H.tuberosus could resist certain alkali stress by modulating carbohydrate metabolism and redox homeostasis. These findings provide a new sight into the underlying molecular mechanisms of alkali resistance in plant.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Soil salinization is one of the most common abiotic stressors to plant growth and has become a highly important issue that impacts agricultural production and ecological environments worldwide (Zhu 2001; Tuteja 2007). Saline-alkali soil is becoming particularly widespread and may cover more than 50% of all arable lands by the year 2050 (Wang et al. 2003). In the northeast of China, alkalinized grasslands with high pH (>9.0) sodic alkaline soil have reached more than a 70% covering due to the presence of excessive sodium carbonate (Na2CO3) (Kawanabe and Zhu 1991; Zheng and Li 1999; Zhang et al. 2013b). Compared to neutral salt stress (NaCl), Na2CO3 stress combines the influence of high pH and excessive sodium ions. A wide range of research is dedicated to studying the adaptive mechanisms by which plants response to NaCl stress and three major regulatory approaches have been identified including maintenance of ion homeostasis, osmotic adjustment and homeostasis, and detoxification (Zhu 2002; Gupta and Huang 2014). Still, our knowledge of the modulation mechanisms by which plants response to alkali stress is insufficient.
Alkali stress with a high pH can directly reduce mineral element availability to plant cells by precipitating Ca2+, Mg2+, and HPO3 - (Yang et al. 2007). High pH can also immediately destroy root membrane structure and disrupt ionic homeostasis, which can then lead to negatively affected plant growth and development (Xue and Liu 2008; Guo et al. 2009). Countable studies of the basic physiological mechanisms of the alkali stress response were conducted in several plant species including crops and halophytes during recent years. The metal element (K, Ca, Mg, Fe, Cu, Zn) and free ions (NO3 -, Cl-, H2PO4 -, SO4 2-) content was decreased in alkali treated plants such as wheat, rice and seabuckthorn, suggesting that high pH stress inhibits the nutrient absorption and disturbs ion balance in plants (Chen et al. 2009; Wang et al. 2012; Guo et al. 2015). Studies have shown that alkali stress also significantly affects photosynthesis and photosynthetic electron transport. In rice, sunflower and thirty switchgrass, alkali stress caused a sharp reduction in photosynthetic parameters and chlorophyll fluorescence (Liu and Shi 2010; Bu et al. 2012; Hu et al. 2015). The same situation occurred in halophyte Chloris virgate except the net photosynthetic rate (PN) was increased slightly under moderate alkali stress (Yang et al. 2008a). Comparative metabolic analysis showed that the metabolites involved in glycolysis and the tricarboxylic acid cycle (TCA cycle) were clearly down-regulated, suggesting that alkali stress had a large negative effect on energy metabolism. Still, free amino acid content was higher compared to that in non-treated plant, which might indicate that high pH inhibits protein biosynthesis or stimulates protein degradation in plants (Zhang et al., 2012b; Guo et al. 2015). In the plant response to alkali stress, transcriptomics analysis revealed that genes with expression alterations were mainly enriched in pathways that included metabolic processes, anion transport, signaling transduction, transcription factors and reactive oxygen species metabolism (Zhang et al. 2011; Fan et al. 2013; Zhang et al. 2013a, b, c; Yu et al. 2014; Kobayashi et al. 2015). Genetic engineering studies showed that with ectopic overexpression of Glycine soja GsPPCK3 (PEPC kinase), GsJAZ2 (jasmonate zim-domain) and tomato SAMS1 (S-adenosyl-L-methionine synthetase), 14-3-3 protein can enhance alkaline stress tolerance in plants (Zhu et al. 2012; Xu et al. 2013; Gong et al. 2014a; Sun et al. 2014). Furthermore, according to proteomic analysis, plants can provide a higher ability to resist alkali stress by up-regulating proteins involved in carbohydrate and energy metabolism, signaling pathways and ROS scavenging pathways (Yu et al. 2013; Gong et al. 2014c).
Helianthus tuberosus L., an annual flowering plant that has been cultivated as a vegetable, is a fodder crop and has benn used as bioenergy material in many countries owing to its enrichment of polysaccharide, especially inulin (Wright et al. 1977, Barta and Pátkai 2007; Long et al., 2010a, b; Jung et al. 2014). The ability to survive in the saline-alkali soil and in semiarid areas is one of the most important characters of H. tuberosus (Zhao et al. 2006; Long et al., 2010a, b). Physiological research showed that H. tuberosus could enhance its resistance to salt stress by osmotic adjustment, selective ion absorption and antioxidant system reinforcement (Chen et al., 2011b; Huang et al. 2012; Huang et al. 2013). Still, a complete understanding of the molecular regulation network in the plants response to salt stress and particularly alkaline salt stress is lacking. In the present study, a comparative proteomic analysis was performed to identify the dynamic changes of proteins in H. tuberosus under Na2CO3 stress. This was coupled with annotation of protein functions and metabolic pathways to investigate the underlying molecular mechanism of alkali tolerance in H. tuberosus.
Materials and methods
Plant material, growth conditions, stress treatment
Seeds of H. tuberosus were kindly provided by Mr. Jiyu Jiang at Dalian Shenju Ecological Development Co., Ltd. The seeds were stripped off the seed coats and sown in plastic pots filled with aseptic vermiculite and fertile black soil (2:1). Seedlings were grown in a greenhouse at 25/20° (day/night) with a 8 h light/16 h dark photoperiod, photosynthetically active radiation 150 mol·m-2·s−1 and 50-70% relative humidity and were irrigated daily by half strength Hoagland’s solution (pH 6.21±0.10). Eight weeks-old seedlings were treated with half strength Hoagland’s solution containing 0, 20 mM (pH 11.15), 50 mM Na2CO3 (pH 11.28). More than three times biological replicates were independently carried out, one single plant from each treatments as one biological replicate. After exposure for 1 day, 3 days, 5 days, 7 days, the leaves of seedlings (n≧3) from control and exposed groups were randomly harvested and used for experiment, separately.
Measurement of biomass and water content
Plant leaves were harvested from five independent plants after stress exposure for 1 day, 3 days, 5 days and 7 days respectively. Fresh weights (FW) were determined and then remained the fresh samples. The remainders of the samples were dying for 10 minutes in 105°i and dried in an oven at 80°a to a constant weight, and then dry weights (DW) was determined. The water content (%) was calculated according to the formula (FW-DW)*100% FW-1. Five biological replicates of each treatment were independently performed. One-way ANOVA was used to test the differences between control and each treatment. The differences were considered significant when p<0.05.
Antioxidant enzyme activity assay
To determine superoxide dismutase (SOD, EC 1.15.1.1), catalase (CAT, EC 1.11.1.6), peroxidase (POD, EC 1.11.1.7) and ascorbate peroxidase (APX, EC:1.11.1.11) activity, the mature leaves were collected from three independent plants at each time point after alkali exposure. 0.3 g of fresh leaf tissue was ground to a fine powder in liquid nitrogen and homogenized in 2 mL 50 mM phosphate buffer (pH 7.8), containing 0.1 mM ethylenediaminetetraacetic acid (EDTA), 4% polyvinypolypyrrolidone (PVPP). After centrifugation at 15,000 g, 4 °, for 20 min, the supernatants were used for enzyme activity assays. SOD activity was determined by inhibiting the photochemical reduction of nitroblue tetrazolium (NBT) at 560 nm. The amount of enzyme needed to inhibit 50% of NBT photoreduction was defined as one unit of SOD activity. The activities of CAT and POD were monitored by measuring H2O2 consumption at 240 nm and guaiacol oxidation at 470 nm, respectively. CAT and POD activities were calculated as described previously (Yu et al., 2011). APX activity was measured spectrophotometrically at 290 nm based on the oxidation of ascorbic acid (AsA), applying an AsA extinction coefficient of 2.8 mM-1 cm-1.Total protein was determined using Bradford approach and made a standard curve with bovine serum albumin (BSA). Three biological replicates of each treatment were independently performed.
Photosynthetic parameter and chlorophyll fluorescence analysis
Net photosynthetic rate (PN) stomatal conductance (Gs), transpiration rate (E), and intercellular CO2 concentration (Ci) of leaves were determined at am 09:00-11:00 h, using a portable open flow gas exchange photosynthesis system LI-6400 XT (Li-Cor, Inc., Lincoln, NE, USA). The photo-synthetically active radiation (PAR) was 1000 μmol·m-2·s-1 (saturation irradiance).
Chlorophyll fluorescence were performed on intact, dark- and light-adapted leaves with a modulated chlorophyll fluorometer OS5p+ (Opti-Sciences, Inc., Hudson, NH, USA). Before taking biochemical assessment, plants were kept in the dark for at least 30 min. Based on the steady-state level of photosynthesis was reached and application of a saturating light pulse (5500 μmol·m-2·s-1), the maximal efficiency of PSII photochemistry (Fv/Fm), minimal fluorescence yield (F0), electron transport rate (ETR), photochemical quenching (qP) and non-photochemical quenching (qN and NPQ) were determined. In addition to the fluorescence parameters at steady-state photosynthesis, an estimation of ETR in light-adapted leaves was extracted from rapid light curve measurements. Rapid light curves were generated by sending out subsequent saturating light pulse at different time intervals (10, 20, 30, 40, 50, 60, 70, 80, 90, 100, 110 and 120 s after initiation). Measurements were repeated six times for each blade from same position of five plants in each treatment group and the averages were recorded. One-way ANOVA was used to test the differences between control and each treatment. The differences were considered significant when p < 0.05.
Proteomics analysis
Protein extraction
The proteins were extracted by the phenol method (Wang et al. 2010). Three replicates were used for each treatment. Approximately 3 g fresh sample were harvested after exposure for 7 days from each treatment and ground into fine powder in liquid nitrogen with a mortar and pestle, then homogenized in 15 mL of precooled homogenization buffer (phenol:extraction buffer 1:1). The extraction buffer contained 0.1 M Tris-HCl, 0.9 M sucrose, 10 mM EDTA, and 0.4% β-mercaptoethanol. After vortex mixing, the homogenate was centrifuged at 10,000 g for 20 min at 4°C, and the top phenol phase was collected and mixed with five volumes of precooled methanol with ammonium acetate and stored overnight at -20°C. Pellets were twice washed with cold methanol with ammonium acetate and acetone, respectively. Pellets were dried by vacuum centrifuge and dissolved in 7 M urea, 2 M thiourea, 4% (w/v) cholamidopropyl CHAPS, 40 mM dithiothreitol (DTT), 2% (v/v) pharmalyte 4-7 (GE Healthcare, Waukesha, WI, USA), and 1% (v/v) proteinase inhibitor (GE Healthcare, Waukesha, WI, USA) and shook vigorously for 2 h at room temperature before being centrifuged at 40,000g at 4° for 1 h. The subsequent supernatant was collected. The protein concentration was determined using the 2D Quant kit (GE Healthcare, Waukesha, WI, USA) with BSA as a standard. Sample were frozen in liquid nitrogen and kept at -80°0 for further use.
Two dimensional gel electrophoresis and image analysis
Two dimensional electrophoresis of protein extract was performed using a GE Healthcare 2-DE system according to the manufacturer’s manuals. Each 1300 μg protein sample was loaded by rehydration to immobiline Dry Strips (pH 4-7 linear, 24 cm) (GE Healthcare, Waukesha, WI, USA). The separation on an IPGphor II unit (GE Healthcare, Waukesha, WI, USA) was performed with the following parameters: 30 V for 8 h, 50 V for 4 h, 100 V for 1 h, 300 V for 1 h, 500 V for 1 h, 1,000 V for 1 h, and 8,000 V for 12 h using hydration buffer (8 M urea, 2% CHAPS, 20 mM DTT) containing 0.6% (v/v) IPG buffer. After isoelectric focusing, the strips were equilibrated with 10 ml equilibration buffer I containing 6 M urea, 2% SDS, 2.5 mM Tris-HCl (pH 8.8), 30% glycerol, and 1% DTT for 15 min, followed with 10 ml equilibration buffer II containing 6 M urea, 2% SDS, 2.5 mM Tris-HCl (pH 8.8), 30% glycerol, and 4% 2-iodoacetamide (IAA) for 15 min. The second dimension separation of proteins was performed on SDS-PAGE gel (12.5% polyacrylamide) using EttanTM Daltsix apparatus (GE Healthcare, Waukesha, WI, USA). The electrophoresis was carried out at 25°. and 3.5 w/gel for 30 min and then 17.5 w/gel for 4.5 h until the bromophenol blue dye front arrived at the bottom of the gels.
The protein samples were visualized by coomassie brilliant blue R250 staining, and gel images were acquired using an ImageScanner (GE Healthcare, Waukesha, WI, USA). Image analysis was performed with ImageMaster 2D Platinum Software Version 7.0 (GE Healthcare, Waukesha, WI, USA). After automated detection and matching, manual editing was carried out to correct the mismatched and unmatched spots. Spots were considered reproducible when they were well resolved in the three biological replicates. For each matched spot, a measurement was carried out for each biological replicate, and normalized volumes were computed using the total spot volume normalization procedure of the software. The normalized volume of each spot was assumed to represent its expression abundance. Spots were considered to be differentially expressed if they presented a percent volume (% vol) ratio ≥ 1.5 in the alkali treated samples versus the control samples and an ANOVA test value ≤ 0.05
MALDI-TOF/TOF analysis and database searching
Selected spots were excised from 2D gels, washed with sterile deionized water, and digested with trypsin as described previously (Chen et al. 2011a). For MALDI-TOF/TOF MS analysis, tryptic peptides were desalted with C18 Ziptips (Millipore) and spotted onto a MALDI plate by mixing 1:1 with the matrix solution (1% a-cyano-4-hydroxy-trans-cinnamic acid in 60% ACN containing 0.1% TFA). MS/MS spectra were acquired using a 4800 MALDI-TOF/TOF mass spectrometer (Applied Biosystems/MDS Sciex, USA). Mass spectrometric analysis was carried out in a data-dependent manner with full scans (350-4500 m/z) acquired using the MALDI/TOF/TOF mass analyzer at a mass resolution of 15,000 at m/z 400. From each MS scan, the 20 most intense precursor ions were selected for MS/MS fragmentation and detected at a mass resolution of 5,000 at m/z 400. Higher-energy Collision Dissociation (HCD) was used as fragmentation method, with 40% normalized collision energy. The peptide MS/MS spectra were searched against NCBI non-redundant fasta database (69,159,658 entries, downloaded on July 14, 2015) using MASCOT search engine (http://www.matrixscience.com). Mascot was set up to search green plants only, assume trypsin digestion and one allowed miscleavage. The mass tolerance for both parent ion and fragment ion mass was set to be 0.3 Da. Iodoacetamide derivatization of Cys, deamidation of Asn and Gln, and oxidation of Met were specified as variable modifications. Unambiguous identification was judged by the number of peptides, sequence coverage, MASCOT MOWSE score and the quality of MS/MS spectra. Individual ion scores of more than 47 indicate identity or extensive homology (p < 0.05). Identifications were validated manually with at least one identified peptide with a score above homology.
For the proteins identified by MASCOT, their accession numbers were directly searched against the NCBInr database (http://blast.ncbi.nlm.nih.gov/) to obtain the proteins corresponding blast information. Protein functional classification was performed by KEGG pathway analysis (http://www.genome.jp/kegg/pathway.html). Protein-protein association information was evaluated with the STRING database (version 9.05, http://string-db.org) against Arabidopsis thaliana database.
RT-PCR analysis
Based on the findings of proteomic analysis, we chose five alkali stress response proteins which might be the underlying regulator of alkali tolerance for RT-PCR analysis for the verification of proteomic data. To analyze the abundance of 14-3-3 (14-3-3 protein) (spot 358 in Table 1, spot 176 in Table 2), Cu/Zn-SOD (Copper/zinc superoxide dismutase) (spot 77), TRX (Thioredoxin) (spot 20), PRX (Peroxiredoxin) (spot 52), HPR (Hydroxylpyruvate reductase) (spot 220) transcripts in H. tuberosus, total RNA was extracted with Trizol reagent (Invitrogen) from eight-week-old seedlings under 0, 20 mM, 50 mM Na2CO3 treatment for 6 h, 12 h, 24 h and 7days, respectively. RNA samples treated with RNase-free DNase I (TaKaRa, Tokyo, Japan) to remove genomic DNA. 10 μg of RNA was used for reverse transcription with PrimeScript™ RT reagent Kit (Perfect Real Time) (TaKaRa, Tokyo, Japan) according to the manufacturer’s instructions and the cDNAs were used for RT-PCR analysis with specific primers. Quantitative RT-PCR was performed with the Applied Biosystems 7500 real-time PCR system using Power SYBR green chemistry (Takara, Tokyo, Japan). Actin was quantified as an internal control and 2-∆∆Ct method was use to analyze differential expression (Livak and Schmittgen 2001). Values represent the mean of three biological replicates and two technical replicates. Gene were considered to be differentially expressed if fold change ≥ 1.5 or ≤ 0.6 compared to control in each time of treatment and an ANOVA test value ≤ 0.05. All primer sequences used in this study are listed in Supplementary Table S3.
Results
Effect of alkali treatment on H. tuberosus growth
Following 1 day, 3 days, and 5 days of alkali treatment, there was no obvious phenotype change in the leaves of 20 mM Na2CO3 treated seedlings, but in 50 mM Na2CO3 treated plants, there was a little wilting with some necrotic patches on the leaves that were under stress for 5 days (Fig. 1a). After 5 and 7 days of alkali stresses, plants exhibited some yellow-green chlorotic areas (indicated by white arrows) in leaves with 20 mM stress, but most areas of the leaves remained fresh and green (Fig. 1a). We found that plants showed severe wilting and leaf curl outwardly in 50 mM Na2CO3 treated seedlings as a result of water depletion after 7 days of treatment (Fig. 1a). With the increase of treatment time, plant water content was reduced under both 20 mM and 50 mM Na2CO3 stress and it displayed a most significant reduction in 50 mM treated plants following 7 days (Fig. 1b). Given that in all observations, there was no visible effect on plant growth with short term alkali treatment, 7 days was selected for comparative 2-DE analysis under low and high Na2CO3 concentration.
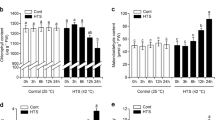
Effects of Na2CO3 treatment on plant growth. Eight weeks-old seedlings were treated with half strength Hoagland’s solution containing 0, 20 mM (pH 11.15), 50 mM Na2CO3 (pH 11.28). (a) Leaves phenotype of H. tuberosus under control and stress conditions. (b) After exposure for 1 day, 3 days, 5 days, 7 days, fresh and dry weight of leaves from each groups were measured (n≧3). Water content (%) was calculated according to the formula (FW-DW)*100 % FW−1. Error bars show the SE for biological replicates. Significant differences are indicated by different letters (p < 0.05 by one-way ANOVA analysis)
Effect of alkali treatment on antioxidant enzymes activities
With the increasing of treatment time, activity of SOD was obviously reduced after exposure to different Na2CO3 concentrations, the same situation in CAT except the plants under 20 mM Na2CO3 treatment for 3 days showed higher activity compared with control plants (Fig. 2a, b). However, higher POD activity were elicited by alkali condition after treatment for 1 day and 3 days, but the induction became slight under 5 and 7 days alkali treatment (Fig. 2c). Activity of APX was induced by alkali condition with an exception that the activity exhibited no significant difference under Na2CO3 stress for 7 days (Fig. 2d).
Effects of Na2CO3 treatment on antioxidant enzyme activity. Determination of SOD, CAT, POD and APX activity was carried out in H. tuberosus with or without 20 mM and 50 mM Na2CO3 treatment for 1 day, 3 days, 5 days and 7 days. Error bars show the SE for three biological replicates. Significant differences are indicated by different letters (p < 0.05 by one-way ANOVA analysis)
Comparative proteomic analysis in H. tuberosus leaves
To investgate the proteome response of H. tuberosus under alkali stress, eight-week-old seedlings were treated with 20 mM and 50 mM Na2CO3 for 7 days, 2-DE gels from three biological experiments were conducted to detect proteins that were extracted from leaves of control and treated seedlings (Fig. 3, Supplementary Figure S1 and S2). With the detection of ImageMaster 2D Platinum Software, we identified more than 1000 protein spots in the CBB staining 2D gels with good reproducibility (Fig. 3). 566 protein spots were matched in two experimental groups that contained three biological replicates, respectively. Based on more than a 1.5-fold change with a p-value <0.05 of variation in protein spot abundance (vol %), 101 and 40 protein spots were considered to be differentially expressed between control and 20 mM and 50 mM treated Na2CO3 plants, respectively. The identified proteins were indicated by black arrows and the ones failed to identify with blue arrows (Fig. 3b, c). A total of 104 proteins were successfully identified by MALDI-TOF/TOF analysis according to peptide matching results using MASCOT. Among these proteins, 79 proteins were detected in H. tuberosus leaves responding to 20 mM Na2CO3 (Table 1, Supplementary Table S4) and 25 proteins accumulated differentially in 50 mM Na2CO3 treated plants (Table 2, Supplementary Table S5). Compared with the control plants, 28 proteins were up-regulated and 51 proteins were down-regulated in 20 mM Na2CO3 stressed seedlings, 7 proteins were up-regulated and 18 proteins were down-regulated in 50 mM Na2CO3 stressed seedlings. The character of these proteins was determined based on BLASTp analyses of the NCBInr database.
Map of H. tuberosus leaf protein spots. Representative 2-DE gels of protein samples from H. tuberosus without Na2CO3 treatment (a) or 20 mM (b) and 50 mM (c) Na2CO3 treatment for 7 days. The statistically significant differential spots are labeled with arrows and match IDs, the successfully identified protein spots were indicated by black arrows and the ones failed to identify with blue arrows
KEGG pathway analysis classified function of proteins response to 20 mM Na2CO3 stress into nine groups involved in different metabolic pathways, which included carbohydrate and energy metabolism (18), photosynthesis (28), genetic information processing (13), reactive oxygen species metabolism (8), stress and defense (5), signal transduction (2), amino acid metabolism (3), methane metabolism (1) and other (1) (Fig. 4a), and classified the proteins response to 50 mM Na2CO3 stress into carbohydrate and energy metabolism (3), photosynthesis (7), genetic information processing (5), reactive oxygen species metabolism (1), stress and defense (2), signal transduction (1), nucleotide metabolism (1), amino acid metabolism (2), metabolism of cofactors and vitamins (1), transport and catabolism (1) and other (1) (Fig. 4b). The majority of these proteins were regulated by relatively moderate Na2CO3 treatment, and 37% of them displayed a strong accumulation in the leaves of H. tuberosus and were mainly involved in photosynthesis (31%) and carbohydrate metabolism (21%). On the basis of our observation, 14-3-3 protein (spot 176 and 417) was the only one protein overlapped in the different concentration alkali treated seedlings and showed a converse expression pattern. This indicates that H. tuberosus responds differently to moderate and aggravating alkali stress. Moreover, we observed that the differentially expressed proteins in response to 20 mM Na2CO3 stress interacted directly or indirectly and showed tight relationships in the functional network, but the proteins response to 50 mM Na2CO3 stress displayed an uncompact interaction (Fig. 5a, b).
Photosynthetic rate and chlorophyll fluorescence
Photosynthetic parameters were determined as a measure of steady-state photosynthesis in H. tuberosus under control and alkali condition (Table 3). In contrast with control plants, after 1 day and 3 days treatment, all the photosynthetic parameters were decreased in 50 mM Na2CO3 treated leaves but were not affected by 20 mM Na2CO3 treatment. Net photosynthetic rate (PN) and stomatal conductance (Gs) were significantly reduced by alkali stress with a long term of 5 or 7 days and showed dose dependence, the same situation in transpiration rate (E) and intercellular CO2 concentration (Ci) under alkali stress for 5 days with the exception of that the change of Ci did not show dose dependence. E was significantly impaired by 50 mM Na2CO3 stress with a period of 7 days exposure and dropped from 5.71 to 0.26. It is demonstrate that high level alkali stress may directly damage the structure of leaves. In addition, Ci showed a sudden rise in H. tuberosus under Na2CO3 stress for 7 days, perhaps because high pH condition reduced the capacity of CO2 fixation.
Chlorophyll fluorescence is a well known indicator of the photosynthetic apparatus’ function. The maximal quantum efficiency (Fv/Fm) of photosystem II (P680), minimal fluorescence yield (FO), electron transport rate (ETR), photochemical quenching (qP) and non-photochemical quenching (qN and NPQ) was determined on dark-adapted leaves (Table 3). With the exception of qP increased in 3 days alkali treated plants, the parameters related PSII and photochemical quenching were unaffected by alkali stress with 1 to 3 days treatments. Fv/Fm and FO only changed in 7 days alkali treated leaves and exhibited significant reduction. This result suggests that the quantum yield of the primary photochemical reaction in the reaction center of PSII decreased. qP decreased and qN, NPQ increased significantly in H. tuberosus after 50 mM Na2CO3 stress for 7 days, this could be due to a retardation of oxygen-evolving complex and forming of inactive quenching fluorescence state of PSII. ETR was strongly inhibited by long term alkali stress (5 days and 7 days) and also showed dose dependence. To sum up, photosynthetic efficiency remained unaffected by alkali stress during 1 to 3 days treatments, but reduced by 20 mM and 50 mM Na2CO3 treatments at the time points of 5 days and 7 days, and the reduction showed more severe in 50 mM Na2CO3 treated plants.
Gene expression
Given that the changes on gene expression pattern were more rapid when plant response to environmental stress (Santos et al., 2011), 6 h, 12 h and 24 h treated seedlings were also chosen to determine mRNA abundance. Analysis of the expression levels of the six genes via qRT-PCR showed that Na2CO3 stress significantly changed the abundance of transcripts under different time treatments compared to control (Fig. 6). HPR (spot 220) and PRX (spot 52) were remarkably down-regulated by both 20 mM and 50 mM Na2CO3 stresses for 6 h and 12 h, but up-regulated their abundance under 20 mM Na2CO3 stress and recovered expression under 50 Na2CO3 stress for 7 days, which indicated that HPR and PRX genes negatively response to alkali stress with short term treatment (Fig. 6a, c). The expression of Cu/Zn-SOD (spot 77) was significantly induced by 6 h and 12 h 20 mM Na2CO3 stresses and suddenly kept parallel with control plants under 24 h and 7 days stresses (Fig. 6b). In addition, the transcripts of PRX, 14-3-3 2 (spot 176) and TRX (spot 20) were highly accumulated in H. tuberosus under 20 mM Na2CO3 stress for 7 days, the results of these three genes in transcript expression were consistent with the proteins expressed pattern from the results of proteomic analysis, but the other genes expression showed no change under 7 days treatment (Fig. 6).
qRT-PCR analysis of gene expression of alkali response proteins in H. tuberosus. RNA was extracted from eight-week-old seedlings under 0, 20 mM, 50 mM Na2CO3 treatment for 6 h, 12 h, 24 h and 7 days, respectively. Actin was quantified as an internal control and 2-∆∆Ct method was use to analyze differential expression. Values represent the mean of three biological replicates and two technical replicates. Each data point represents mean ± SE (n = 3). Asterisks indicated significant difference from control at p < 0.05, respectively
Discussion
A high pH environment surrounding plants can directly damage plant cell structure and cause the precipitation of nutrient elements, which further impacts plant growth and development (Shi and Zhao 1997; Yang et al. 2007). Physiological studies of halophytes subjected to alkali stress demonstrated that the growth rate and water content declined with increasing concentrations of Na2CO3 but did not show significant changes with a treatment time in limited to 1 day under a concentration of up to 150 mM Na2CO3 in Puccinellia tenuiflora (Zhang et al. 2012b). Similar to our observations, there was no visible phenotype change of H. tuberosus under alkali stress following 1 day and 3 days of treatment and no significant reduction in water content, implying H. tuberosus has a high-pH tolerance and can maintain essential growth under alkali stress conditions. Here, we provided a profile of H. tuberosus proteins that accumulate differentially in leaves between control and Na2CO3 treated plants. This provides an overview of the mode of regulations of the proteins response to alkali stress (Fig. 3, Table 1, 2). Based on the STRING analysis, the response of these differentially expressed proteins with 20 mM Na2CO3 stress revealed a well-connected network of directly or indirectly associated proteins (Fig. 5a).
Carbohydrate and energy metabolism
Previously, it had been reported that carbohydrate metabolism processes including glycolysis and the TCA cycle were significantly inhibited by alkali stress according to the reduction of relational metabolites (Guo et al. 2015). Based on our analysis, five enzymes involved in glycolysis including triosephosphate isomerase (TIM) (spot 298), fructose-bisphosphate aldolase (FBA) (spot 472, 935), glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (spot 525), phosphoglycerate kinase (PGK) (spot 614, 621) and enolase (spot 742, 771) were found to be differently expressed in the leaves of H. tuberosus following alkali stress (Table 1). TIM, FBA and GAPDH are principal enzymes that catalyzed the conversion of glucose to 1,3-bisphosphoglycerate, which is the first half of the glycolysis pathway (Plaxton 1996). These three proteins were down regulated under 20 mM Na2CO3 treatment. This might imply that the glycolysis process is inhibited by Na2CO3 treatment. To be supportive, we also found five proteins annotated as ATP synthase (spot 92, 165, 170, 539, 568) down regulated in 20 mM Na2CO3 treated plants, suggesting ATP synthesis were also reduced by alkali condition. However, the other two enzymes PGK and enolase, which were in charge of carrying out the reactions for the remaining part of glycolysis were significantly induced under Na2CO3 20 mM stress. PGK and enolase catalyze the formation of pyruvate from bisphosphoglycerate. The higher abundance of these two proteins in alkali treated plants might indicate that the accumulation of pyruvate was enhanced by high pH conditions, supplying more substrate to the metabolic pathway of the TCA cycle. Interestingly, we also found the key member of the TCA cycle, malate dehydrogenase (MDH) (spot 521), was significantly up regulated in the H. tuberosus response to 20 mM Na2CO3 treatment. MDH catalyze the interconversion of malate and oxaloacetate in a reversible reaction of the TCA cycle (Selinski et al. 2014). To summarize these results, these data suggest that proliferation of pyruvate may enhance the work of the TCA cycle in H. tuberosus. This might be a strategy for plant to enhance the ability of seedlings to survive moderate alkali stress, which makes it possible for the cell to adapt to its metabolic needs. Consistent with the findings in our study, Yu et al. (2013) also found that the abundance of MDH was induced by relatively low level alkali stress but reduced in 98 mM Na2CO3 treated halophyte Puccinellia tenuiflora.
In addition, our results showed another two proteins, transketolase (TK) (spot 834, 836), were significantly up regulated in H. tuberosus under 20 mM Na2CO3 condition, and TK is considered to participate in pentose phosphate pathway. The pentose phosphate pathway is important to maintain carbon homoeostasis, to provide precursors for nucleotide and amino acid biosynthesis, and to provide reducing molecules for defeating oxidative stress Stincone et al. (2014). TK is the key enzyme of the non-oxidative branch of the pentose phosphate pathway of carbohydrate transformation (Kochetov and Solovjeva 2014). Accumulation of TK would promote the enhancement of the non-oxidative branch, yielding ribose 5-phosphate for the synthesis of nucleic acids and amino acids accompanied by the production of NADPH, which is critical to maintain redox balance under stress situations.
Photosynthesis
Accumulating evidence suggests that photosynthesis is greatly inhibited under excessive sodium carbonate conditions via the decrease of chlorophyll biosynthesis and photosystem II efficiency (Yang et al. 2008a; Liu and Shi 2010; Bu et al. 2012). Our proteomic analysis showed 20 photosynthetic proteins were differentially expressed in H. tuberosus subjected to Na2CO3 stress. Under both 20 mM and 50 mM Na2CO3 treatment, photosystem II light harvesting chlorophyll a/b binding protein (spot 23, 272, 273, 275, 327, 428, 113, 175, 179) and photosystem II (PSII) oxygen-evolving enhancer protein (spot 190, 192, 222, 166) were distinctively affected by alkali stress. This may demonstrate that the photosynthetic capacity of PSII was sensitive to high pH conditions (Table 1, 2). Based on our determination of chlorophyll fluorescence, Fv/Fm and ETR significantly declined in response to alkali stress at 7 days (Table 3). In addition, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (spot 44, 47), which is an enzyme involved in the first major step of carbon fixation and RuBisCO activase (spot 639, 740) are both down regulated by 20 mM Na2CO3 stress. This was consistent with that the lower expression abundance of RuBisCO on the transcriptional level that was observed in the P. tenuiflora response to alkali stress (Zhang et al. 2012a). Environmental stress often induces a decrease of the photosynthetic rate, partially due to RuBisCO degradation (He et al. 2014). The determination of photosynthesis activity showed a dramatic reduction in several plants, as exemplified in barley, wheat, as well as the halophyte Chloris virgate (Yang et al. 2008a, b, 2009). Moreover, four carbonic anhydrases (CA) involved in Calvin cycle (spot 287 and 316 in Table 1; spot 130 and 148 in Table 2) were identified to be significantly down regulated by either 20 mM or 50 mM Na2CO3 stress. CAs play role in CO2 uptake and fixation, its can transmit CO2 to RuBisCO (Ludwig 2016). Overexpression of the mitochondrial CA in Arabidopsis resulted in an increase in plant biomass (Fabre et al. 2007). In our study, 7 days of alkali stress inhibited the growth of H. tuberosus (Fig. 1) and Ci suddenly increased in 7 days alkali treated leaves (Table 3), also the reduction of CAs and RuBisCO expression may suggest that high pH salt condition impact the carbon fixation and even the photosynthetic efficiency of plants.
To our surprise, we found the expression of several proteins, such as photosystem I (PSI) light harvesting chlorophyll a/b binding protein (spot 234, 251) and plastocyanin (spot 59), were up regulated in H. tuberosus under 20mM Na2CO3 stress, while no significant change was observed in 50mM Na2CO3 treated plants compared to control (Table 1, 2). PSI light harvesting chlorophyll binding proteins are the intrinsic transmembrane antenna proteins (Lhca’s) occurring in the reaction center of PSI. PSI is known to be the most efficient light converter in nature since pigments in the PSI are not being quenched and energy transfer to the electron donor is very rapid (Croce and Amerongen. 2013). Plastocyanin functions as an electron transfer agent between cytochrome f and P700+ from PSI (Farkas and Hansson 2011). It differed from the previous proteomic analysis of tomato and P. tenuiflora, which had not found any differentially expressed protein associated with PSI under alkali stress (Yu et al. 2013; Gong et al. 2014c). We speculated that as PSI light harvesting chlorophyll binding protein passed more excitation energy to the reaction center, the accumulation of plastocyanin can donate more electrons to PSI. This demonstrates that those two proteins with a higher abundance may help PSI to convert more light energy and minimize the energy loss caused by a reduction of PSII efficiency in the H. tuberosus response to moderate Na2CO3 stress.
Genetic information process
Quantitative proteomics studies have shown that the levels of DNA replication and transcription related proteins, as well as protein synthesis and fold involving proteins, are responsive to salt stress and play a pivotal role in salinity tolerance (Zhang et al., 2012a, b). Our results displayed that some genetic information processing proteins, such as DNA replication related proteins helicase (spot 642), translation related ribosomal protein L7/L12 (spot 131, 136, 139), elongation factor tu (EF-Tu) GTP-binding protein (spot 330, 640, 643), protein posttranslational processing proteins including chaperonin 10 Kd subunit (spot 110, 249, 269), groel-like type I chaperonin (spot 773,785, 791), and chaperonin-60 beta subunit (spot 407), were differently expressed in H. tuberosus under Na2CO3 stress.
Helicases are motor proteins that move directionally along a nucleic acid phosphodiester backbone, separating two annealed nucleic acid strands in plants (Knoll and Puchta 2011), which is induced by 20 mM Na2CO3 treatment in our study. Several works conducted previously have shown that helicases contribute to salinity-stress tolerance by improving photosynthesis and antioxidant machinery in plants due to the essential role of helicases in genome stability (Tuteja et al. 2013; Khan et al. 2014). The induction of helicases from our finding may provide a positive effect in H. tuberosus against high pH and salt conditions.
Protein biosynthesis is carried out by ribosomal machinery and requires the interaction of several translation factors with the ribosomal stalk complex. We found three ribosomal proteins, including L7/L12, were highly accumulated in the H. tuberosus response to Na2CO3 stress. However, elongation factor tu (EF-Tu) GTP-binding proteins from the EF-Tu subfamily were found to be down regulated by 20 mM and 50 mM Na2CO3 treatment in our study. There is evidence that the exchange of L7/L12 monomers and dimers can be affected by elongation factor binding, which has been proposed to modulate ribosomal activity during translation (Deroo et al. 2012). Overall, our results imply that down regulation of EF-Tu related proteins may intend to maintain the activity of ribosomal proteins and modulate translation stability in response to environmental stimuli. Chaperonins are universally conserved molecular machines that facilitate the proper folding of nascent and partially folded polypeptides into their respective three-dimensional structures (Horwich 2013). Previously, proteomic analysis demonstrated that chaperonins with different molecular weights were induced by salt stress to assist the folding of stress-denatured proteins in plants (Ndimba et al. 2005; Pang et al. 2010; Manaa et al. 2011). However, our results showed that molecular chaperons, including the chaperonin 10 Kd subunit, chaperonin-60 beta subunit and groel-like type I chaperonin, were significantly down regulated by alkali stress in H. tuberosus. This suggests that the high pH condition of Na2CO3 treatment may have a severe effect on the protein biosynthesis of chaperonin, which is different from the situation in plants under neutral salt stress based on the expressed abundance change of chaperonins. Certainly, this needs to be confirmed in future studies.
Reactive oxygen species metabolism
Environmental stresses can cause an increased production of ROS and bring oxidative damage to functional biomolecules (Mittler et al. 2004). For that, plants have evolved a highly efficient antioxidant defense system to scavenge excessive ROS (Miller et al., 2010). The balance of SOD and APX activities are crucial for suppressing toxic ROS levels. SOD alternately catalyzes the dismutation of the O2 .- radical into either ordinary molecular oxygen or H2O2 which is the first step of detoxification (Alscher et al. 2002). APX is one of the key members of the glutathione-ascorbate cycle (GSH-AsA cycle) and functions to reduce H2O2 to water using AsA as an electron donor (Miller et al., 2010). Peroxiredoxin (Prx) employ a thiol-based catalytic mechanism to reduce H2O2 and is regenerated using thioredoxin (Trx) as electron donor (Dietz 2011).
In Solanum lycopersicum, superoxide (O2 ._) and hydrogen peroxide (H2O2) were remarkably increased in roots and leaves exposed to NaHCO3 stress, and the ROS burst seriously injured the membrane system and chloroplast ultrastructure (Gong et al. 2014b). According to our findings, ROS scavenging related enzymes and proteins such as copper/zinc SOD (spot 77), APX (330, 404), typical 2-cys Prx (176), Trx (spot 20) were highly accumulated in H. tuberosus under 20 mM Na2CO3 stress. This was consistent with the change in transcript abundance, but we found a different situation in 50 mM Na2CO3 treated plants, where Prx (spot 52) was significantly down regulated. Distinctly from the response to NaCl as shown in a previous study, the activities of SOD, POD and CAT were decreased in leaves and caused oxidative damage to cell structures. This indicates that the antioxidant defense was impeded by salt stress in H. tuberosus (Xue et al. 2008; Long et al. 2010a). To sum up, the accumulation of ROS scavenging related proteins in our study suggests that the SOD pathway, APX dependent GSH-AsA cycle and Prx/Trx pathway of the antioxidant system play a major role in maintaining redox homeostasis to protect cellular components (Fig. 7).
Schematic view of differentially expressed proteins involved in key metabolic pathways in H. tuberosus under alkali stress. The related pathways and differentially expressed proteins were marked with bold, and the protein match IDs from 20 mM and 50 mM Na2CO3 treated plants were marked with black and blue, respectively. The changes in expression abundance of each protein were indicated by color code rectangle, up-regulated proteins were in red and down-regulated proteins in green. GAPHD, glyceraldehyde 3-phosphatedehydrogenase; PGK, phosphoglycerate kinase; FBA, fructose-bisphosphate aldolase; TIM, triosephosphate isomerase; OADAT, 2-oxoacid dehydrogenases acyltransferase; TK, Transketolase; MDH, malate dehydrogenase; RuBisCO, ribulose-1,5-bisphosphate carboxylase/oxygenase; RCA, RuBisCO activase; SOD, superoxide dismutase; APX, ascorbate peroxidase; Prx, peroxyredoxin; Trx, thioredoxin; OEC, oxygen evolving complex; CA, carbonic anhydrase; EF-Tu, elongation factor tu
Stress and defense
Stress response protein ribosome-associated inhibitor A (raiA) can inhibit translation at the aminoacyl-tRNA binding stage and is involved in general adaptation of the translation machinery to environmental stress (Agafonov et al. 2001). In the present work, we found two proteins of raiA (spot 174, 923) were significantly induced by both 20 mM and 50 mM Na2CO3 treatments. A relevant report has demonstrated that raiA owns anti-miscoding activity in vivo and is capable of strongly reducing mistranslation (Agafonov and Spirin 2004). On the basis of our finding, it is indicated that the raiA with a higher abundance in H. tuberosus may help the plant to acclimate to alkali stress by reducing mistranslation. Additionally, three protein spots of jacalin-like plant lectin domain (spot 51, 53, 65) were down regulated in H. tuberosus exposed to 20 mM Na2CO3.The jacalin-like lectin domain is a mannose-binding lectin domain and known to be important for many biological processes due to their ability to recognize cell surface carbohydrates with high specificity in plants. This is possibly important to protein-carbohydrate interactions in stress responses in plants (Zhang et al. 2000). In the present study, the abundance of jacalin-like lectin decreased, suggesting that the recognition of lectin may be inhibited by high pH and salt conditions in H. tuberosus.
14-3-3 protein
Plant 14-3-3 proteins bind a range of transcription factors and other signaling proteins, and have roles regulating plant development and stress responses (Roberts 2003). Environmental stresses can impact 14-3-3s directly by altering the expression of specific isoforms, stimuli may activate signaling pathways that cause the phosphorylation of client proteins to which 14-3-3s then recognize and bind (Denison et al. 2011). In this study, 14-3-3 proteins (spot 358, 417) were clearly up regulated under 20 mM Na2CO3 treatment, but the expression of the same protein (spot 176) was inhibited by 50 mM Na2CO3 treatment. This was consistent with the results of qRT-PCR analysis. Under NaCl stress, 14-3-3 λ and κ negatively regulate salt tolerance by inhibiting the salt overly sensitive pathway in Arabidopsis (Zhou et al. 2014). In contrast, the study of tomato demonstrates that the 14-3-3 gene TOMATO 14-3-3 PROTEIN4 was overexpressed under alkaline stress and 14-3-3 protein acts as a regulator in the integration of H+ efflux, basipetal auxin transport, and the PROTEIN KINASE5 (PKS5)-chaperone DNAJHOMOLOG3 (J3) pathway and also coordinates root apex responses to alkali stress for the maintenance of primary root elongation (Xu et al. 2013). All of the evidence shows that it is different from the NaCl stress response and that members of the 14-3-3 protein family have a diverse influence on the regulation pathways relevant to plant responses to Na2CO3 stress.
Conclusions
Plants enact mechanisms to mitigate salt stress by modulating various biological processes, but the understanding of plant adaptation characteristic to alkali stress is lacking. In this study, a comparative proteomic analysis showed that a total of 104 proteins detected from H. tuberosus leaves were found to display significant changes under Na2CO3 stress. The significant induction of metabolic enzymes involved in the glycolysis pathway and the TCA cycle revealed that plants are likely to regulate carbohydrate and energy demand to overcome high pH condition. The fact that PSI proteins showed a higher abundance indicate that the energy losses that are caused by a reduction of photosystem II efficiency may be minimized in H. tuberosus under Na2CO3 stress. The accumulation of antioxidant system proteins suggested by the SOD pathway, APX dependent GSH-AsA cycle and Prx/Trx pathway facilitate the detoxification of ROS and maintain redox homeostasis in the plant. Altogether, our present results document that alkali stress entails numerous modulation in the metabolic processes of H. tuberosus, which provides a new sight for extending our understanding of the underlying molecular mechanisms of alkali resistance in plants (Fig. 7).
Abbreviations
- 2-DE :
-
Two dimensional electrophoresis
- MALDI-TOF/TOF :
-
Matrix-assisted laser desorption/ionization time-of-flight/time-of-flight
- IAA :
-
Iodoacetamide
- CHAPS :
-
Cholamidopropyl propanesulfonate
- DTT :
-
Dithiothreitol
- ACN :
-
Acetonitrile
- TFA :
-
Trifluoroacetic acid
References
Agafonov DE, Spirin AS (2004) The ribosome-associated inhibitor A reduces translation errors. Biochem Biophys Res Commun 320:354–358. doi:10.1016/j.bbrc.2004.05.171
Agafonov DE, Kolb VA, Spirin AS (2001) Ribosome-associated protein that inhibits translation at the aminoacyl-tRNA binding stage. EMBO Rep 2:399–402. doi:10.1093/embo-reports/kve091
Alscher RG, Erturk N, Heath LS (2002) Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. J Exp Bot 53:1331–1341. doi:10.1093/jexbot/53.372.1331
Barta J, Pátkai GY (2007) Chemical composition and storability of Jerusalem artichoke tubers. Acta Aliment Hung 36:257–267. doi:10.1556/AAlim.36.2007.2.13
Bu N, Li XM, Li YY, Ma CY, Ma LJ, Zhang C (2012) Effects of Na2CO3 stress on photosynthesis and antioxidative enzymes in endophyte infected and non-infected rice. Ecotox Environ Safe 78:35–40. doi:10.1016/j.ecoenv.2011.11.007
Chen WC, Cui PJ, Sun HY, Guo WQ, Yang CW, Jin H, Fang B, Shi DC (2009) Comparative effects of salt and alkali stresses on organic acid accumulation and ionic balance of seabuckthorn (Hippophae rhamnoides L.). Ind Crop Prod 30:351–358. doi:10.1016/j.indcrop.2009.06.007
Chen YZ, Pang QY, Dai SJ, Wang Y, Chen SX, Yan XF (2011a) Proteomic identification of differentially expressed proteins in Arabidopsis in response to methyl jasmonate. J Plant Physiol 168:995–1008. doi:10.1016/j.jplph.2011.01.018
Chen Q, Zhang MD, Shen SH (2011b) Effect of salt on malondialdehyde and antioxidant enzymes in seedling roots of Jerusalem artichoke (Helianthus tuberosus L.). Acta Physiol Plant 33:273–278. doi:10.1007/s11738-010-0543-5
Croce R, Amerongen HV (2013) Light-harvesting in photosystem II. Photosynth Res 116:153–166. doi:10.1007/s11120-013-9838-x
Denison FC, Paul AL, Zupanska AK, Ferl RJ (2011) 14-3-3 proteins in plant physiology. Semin Cell Dev Biol 22:720–727. doi:10.1016/j.semcdb.2011.08.006
Deroo S, Hyung SJ, Marcoux J, Gordiyenko Y, Koripella RK, Sanyal S, Robinson CV (2012) Mechanism and rates of exchange of L7/L12 between ribosomes and the effects of binding EF–G. ACS Chem Biol 7:1120–1127. doi:10.1021/cb300081s
Dietz KJ (2011) Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15:1129–1159. doi:10.1089/ars.2010.3657
Fabre N, Reiter IM, Becuwe-Linka N, Genty B, Rumeau D (2007) Characterization and expression analysis of genes encoding a and b carbonic anhydrases in Arabidopsis. Plant Cell Environ 30:617–629
Fan XD, Wang JQ, Yang N, Dong YY, Liu L, Wang FW, Wang N, Chen H, Liu WC, Sun YP, Li HY (2013) Gene expression profiling of soybean leaves and roots under salt, saline–alkali and drought stress by high-throughput Illumina sequencing. Gene 512:392–402. doi:10.1016/j.gene.2012.09.100
Farkas D, Hansson Ö (2011) Thioredoxin-mediated reduction of the photosystem I subunit PsaF and activation through oxidation by the interaction partner plastocyanin. FEBS Lett 585:1753–1758. doi:10.1016/j.febslet.2011.04.073
Gong B, Li X, Vanden Langenberg KM, Wen D, Sun SS, Wei M, Li Y, Yang FJ, Shi QH, Wang XF (2014a) Overexpression of S-adenosyl-L-methionine synthetase increased tomato tolerance to alkali stress through polyamine metabolism. Plant Biotechnol J 12:694–708. doi:10.1111/pbi.12173
Gong B, Li X, Bloszies S, Wen D, Sun S, Wei M, Yang FJ, Shi QH, Wang XF (2014b) Sodic alkaline stress mitigation by interaction of nitric oxide and polyamines involves antioxidants and physiological strategies in Solanum lycopersicum. Free Radic Biol Med 71:36–48. doi:10.1016/j.freeradbiomed.2014.02.018
Gong B, Zhang CJ, Li X, Wen D, Wang SS, Shi QH, Wang XF (2014c) Identification of NaCl and NaHCO3 stress responsive proteins in tomato roots using iTRAQ-based analysis. Biochem Biophys Res Commun 446:417–422. doi:10.1016/j.bbrc.2014.03.005
Guo R, Shi LX, Yang YF (2009) Germination, growth, osmotic adjustment and ionic balance of wheat in response to saline and alkaline stresses. Soil Sci Plant Nutr 55:667–679. doi:10.1111/j.1747-0765.2009.00406.x
Guo R, Yang ZZ, Li F, Yan CR, Zhong XL, Liu Q, Xia X, Li H (2015) Comparative metabolic responses and adaptive strategies of wheat (Triticum aestivum) to salt and alkali stress. BMC Plant Biol 15:170. doi:10.1186/s12870-015-0546-x
Gupta B, Huang B (2014) Mechanism of salinity tolerance in plants: physiological, biochemical, and molecular characterization. Int J Genomics 2014:701596. doi:10.1155/2014/701596
He Y, Yu CL, Zhou L, Chen Y, Liu A, Jin JH, Hong J, Jiang D (2014) Rubisco decrease is involved in chloroplast protrusion and Rubisco-containing body formation in soybean (Glycine max.) under salt stress. Plant Physiol Biochem 74:118–124. doi:10.1016/j.plaphy.2013.11.008
Horwich AL (2013) Chaperonin-mediated protein folding. J Biol Chem 288:23622–23632. doi:10.1074/jbc.X113.497321
Hu GF, Liu YM, Zhang XZ, Yao FJ, Huang Y, Ervin EH, Zhao BY (2015) Physiological evaluation of alkali-salt tolerance of thirty Switchgrass (Panicum virgatum) lines. PLoS ONE 10:e0125305. doi:10.1371/journal.pone.0125305
Huang ZR, Long XH, Wang L, Kang J, Zhang ZH, Zed R, Liu ZP (2012) Growth, photosynthesis and H+-ATPase activity in two Jerusalem artichoke varieties under NaCl-induced stress. Process Biochem 47:591–596. doi:10.1016/j.procbio.2011.12.016
Huang ZR, Zhao L, Chen DD, Liang MX, Liu ZP, Shao HB, Long XH (2013) Salt stress encourages proline accumulation by regulating proline biosynthesis and degradation in Jerusalem artichoke plantlets. PLoS ONE 8:e62085. doi:10.1371/journal.pone.0062085
Jung WY, Lee SS, Kim CW, Kim HS, Min SR, Moon JS, Kwon SY, Jeon JH, Cho HS (2014) RNA-Seq analysis and de novo transcriptome sssembly of Jerusalem artichoke (Helianthus tuberosus Linne). PLoS ONE 9:e111982. doi:10.1371/journal.pone.0111982
Kawanabe S, Zhu TC (1991) Degeneration and conservation of Aneurolepidium chinense grassland in Northern China. J Jap Grassland Sci 37:91–99
Khan A, Garbelli A, Grossi S, Florentin A, Batelli G, Acuna T, Zolla G, Kaye Y, Paul LK, Zhu JK, Maga G, Grafi G, Barak S (2014) The Arabidopsis STRESS RESPONSE SUPPRESSOR DEAD-box RNA helicases are nucleolar- and chromocenter-localized proteins that undergo stress-mediated relocalization and are involved in epigenetic gene silencing. Plant J 79:28–43. doi:10.1111/tpj.12533
Knoll A, Puchta H (2011) The role of DNA helicases and their interaction partners in genome stability and meiotic recombination in plants. J Exp Bot 62:1565–1579. doi:10.1093/jxb/erq357
Kobayashi S, Satone H, Tan E, Kurokochi H, Asakawa S, Liu SK, Takano T (2015) Transcriptional responses of a bicarbonate-tolerant monocot, Puccinellia tenuiflora, and a related bicarbonate-sensitive species, Poa annua, to NaHCO3 stress. Int J Mol Sci 16:496–509. doi:10.3390/ijms16010496
Kochetov GA, Solovjeva ON (2014) Structure and functioning mechanism of transketolase. BBA-Proteins Proteom 1844:1608–1618. doi:10.1016/j.bbapap.2014.06.003
Liu J, Shi DC (2010) Photosynthesis, chlorophyll fluorescence, inorganic ion and organic acid accumulations of sunflower in responses to salt and salt-alkaline mixed stress. Photosynthetica 48:127–134. doi:10.1007/s11099-010-0017-4
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25:402–408
Long XH, Huang ZR, Huang YL, Kang J, Zhang ZH, Liu ZP (2010a) Response of two Jerusalem artichoke (Helianthus tuberosus) cultivars differing in tolerance to salt treatment. Pedosphere 20:515–524. doi:10.1016/S1002-0160(10)60041-0
Long XH, Huang ZR, Zhang ZH, Li Q, Zed R, Liu ZP (2010b) Seawater stress differentially affects germination, growth, photosynthesis, and ion concentration in genotypes of Jerusalem artichoke (Helianthus tuberosus L.). J Plant Growth Regul 29:223–231. doi:10.1007/s00344-009-9125-4
Ludwig M (2016) Evolution of carbonic anhydrase in C4 plants. Curr Opin Plant Biol 31:16–22. doi:10.1016/j.pbi.2016.03.003
Manaa A, Ben Ahmed H, Valot B, Bouchet JP, Aschi-Smiti S, Causse M, Faurobert M (2011) Salt and genotype impact on plant physiology and root proteome variations in tomato. J Exp Bot 62:2797–2813. doi:10.1093/jxb/erq460
Miller G, Suzuki N, Ciftic-Yilmaz S, Mittler R (2010) Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ 33:453–467. doi:10.1111/j.1365-3040.2009.02041.x
Mittler R, Vanderauwera S, Gollery M, Van Breusegem F (2004) Reactive oxygen gene network of plants. Trends Plant Sci 9:490–498. doi:10.1016/j.tplants.2004.08.009
Ndimba BK, Chivasa S, Simon WJ, Slabas AR (2005) Identification of Arabidopsis salt and osmotic stress responsive proteins using two-dimensional difference gel electrophoresis and mass spectrometry. Proteomics. 5:4185–4196. doi:10.1002/pmic.200401282
Pang QY, Chen SX, Dai SJ, Chen YZ, Wang Y, Yan XF (2010) Comparative proteomics of salt tolerance in Arabidopsis thaliana and Thellungiella halophila. J Proteome Res 9:2584–2599. doi:10.1021/pr100034f
Plaxton WC (1996) The organization and regulation of plant glycolysis. Annu Rev Plant Physiol Plant Mol Biol 47:185–214. doi:10.1146/annurev.arplant.47.1.185
Roberts MR (2003) 14-3-3 Proteins find new partners in plant cell signalling. Trends Plant Sci 8:218–223. doi:10.1016/S1360-1385(03)00056-6
Santos AP, Serra T, Figueiredo DD, Barros P, Lourenço T, Chander S, Oliveira MM, Saibo NJ (2011) Transcription regulation of abiotic stress responses in rice: a combined action of transcription factors and epigenetic mechanisms. OMICS 15:839–857. doi:10.1089/omi.2011.0095
Selinski J, König N, Wellmeyer B, Hanke GT, Linke V, Neuhaus HE, Scheibe R (2014) The plastid-localized NAD-dependent malate dehydrogenase is crucial for energy homeostasis in developingArabidopsis thaliana seeds. Mol Plant 7:170–186. doi:10.1093/mp/sst151
Shi DC, Zhao KF (1997) Effects of NaCl and Na2CO3 on growth of Puccinellia tenuiflora and on present state of mineral elements in nutrient solution. Acta Pratacu Sin 6:51–61
Stincone A, Prigione A, Cramer T, Wamelink MMC, Campbell K, Cheung E, Olin-Sandoval V, Grüning NM, Krüger A, Alam MT, Keller MA, Michael Breitenbach B, Brindle KB, Rabinowitz JD, Ralser M (2014) The return of metabolism: biochemistry and physiology of the pentose phosphate pathway. Biol Rev Camb Philos Soc 90:927–963. doi:10.1111/brv.12140
Sun MZ, Sun XL, Zhao Y, Zhao CY, DuanMu HZ, Yu Y, Ji W, Zhu YM (2014) Ectopic expression of GsPPCK3 and SCMRP in Medicago sativa enhances plant alkaline stress tolerance and methionine content. PLoS ONE 9:e89578. doi:10.1371/journal.pone.0089578
Tuteja N (2007) Mechanisms of high salinity tolerance in plants. Method Enzymol 428:419–438. doi:10.1016/S0076-6879(07)28024-3
Tuteja N, Sahoo RK, Garg B, Tuteja R (2013) OsSUV3 dual helicase functions in salinity stress tolerance by maintaining photosynthesis and antioxidant machinery inrice (Oryza sativa L. cv. IR64). Plant J 76:115–127. doi:10.1111/tpj.12277
Wang WX, Vinocur B, Altman A (2003) Plant responses to drought, salinity and extreme temperatures: towards genetic engineering for stress tolerance. Planta 218:1–14. doi:10.1007/s00425-003-1105-5
Wang XN, Chen SX, Zhang H, Shi L, Cao FL, Guo LH, Xie YM, Wang T, Yan XF, Dai SJ (2010) Desiccation tolerance mechanism in resurrection fern-ally Selaginella tamariscina revealed by physiological and proteomic analysis. J Proteome Res 9:6561–6577. doi:10.1021/pr100767k
Wang H, Wu ZH, Han JY, Zheng W, Yang CW (2012) Comparison of ion balance and nitrogen metabolism in old and young leaves of alkali-stressed rice plants. PLoS ONE 7:e37817. doi:10.1371/journal.pone.0037817
Wright L, Wrench P, Hinde RW, Brady CJ (1977) Proline accumulation in tubers of Jerusalem artichoke. Aust J Plant Physiol 4:51–60. doi:10.1071/PP9770051
Xu WF, Jia LG, Shi WM, Baluška F, Kronzucker HJ, Liang JS, Zhang JH (2013) The tomato 14-3-3 protein TFT4 modulates H+ efflux, basipetal auxin transport, and the PKS5-J3 pathway in the root growth response to alkaline stress. Plant Physiol 163:1817–1828. doi:10.1104/pp.113.224758
Xue YF, Liu ZP (2008) Effects of NaCl and Na2CO3 stresses on photosynthesis and parameters of chlorophyll fluorescence in Helianthus tuberosus seedlings. J Plant Ecol 32:161–167. doi:10.3773/j.issn.1005-264x.2008.01.018
Xue YF, Liu L, Liu ZP, Mehta SK, Zhao GM (2008) Protective role of Ca against NaCl toxicity in Jerusalem artichoke by up-regulation of antioxidant enzymes. Pedosphere 18:766–774. doi:10.1016/S1002-0160(08)60072-7
Yang CW, Chong JN, Li CY, Kim C, Shi DC, Wang DL (2007) Osmotic adjustment and ion balance traits of an alkali resistant halophyte Kochia sieversiana during adaptation to salt and alkali conditions. Plant Soil 294:263–276. doi:10.1007/s11104-007-9251-3
Yang CW, Jianaer A, Li CY, Shi DC, Wang DL (2008a) Comparison of the effects of salt-stress and alkali-stress on photosynthesis and energy storage of an alkali-resistant halophyte Chloris virgata. Photosynthetica 46:273–278. doi:10.1007/s11099-008-0047-3
Yang CW, Wang P, Li CY, Shi DC, Wang DL (2008b) Comparison of effects of salt and alkali stresses on the growth and photosynthesis of wheat. Photosynthetica 46:107–114. doi:10.1007/s11099-008-0018-8
Yang CW, Xu HH, Wang LL, Liu J, Shi DC, Wang DL (2009) Comparative effects of salt-stress and alkali-stress on the growth, photosynthesis, solute accumulation, and ion balance of barley plants. Photosynthetica 47:79–86. doi:10.1007/s11099-009-0013-8
Yu JJ, Chen SX, Zhao Q, Wang T, Yang CP, Diaz C, Sun GR (2011) Physiological and proteomic analysis of salinity tolerance in Puccinellia tenuiflora. J Proteome Res 10:3852–3870. doi:10.1021/pr101102p
Yu JJ, Chen SX, Wang T, Sun GR, Dai SJ (2013) Comparative proteomic analysis of Puccinellia tenuiflora leaves under Na2CO3 Stress. Int J Mol Sci 14:1740–1762. doi:10.3390/ijms14011740
Yu Y, Huang WG, Chen HY, Wu GW, Yuan HM, Song XX, Kang QH, Zhao DS, Jiang WD, Liu Y, Wu JZ, Cheng LL, Yao YB, Guan FZ (2014) Identification of differentially expressed genes in flax (Linum usitatissimum L.) under saline–alkaline stress by digital gene expression. Gene 549:113–122. doi:10.1016/j.gene.2014.07.053
Zhang WL, Peumans WJ, Barre A, Astoul CH, Rovira P, Rougé P, Proost P, Truffa-Bachi P, Jalali AAH, Van Damme EJMV (2000) Isolation and characterization of a jacalin-related mannose-binding lectin from salt-stressed rice (Oryza sativa) plants. Planta 210:970–978. doi:10.1007/s004250050705
Zhang CX, Bian MD, Yu H, Liu Q, Yang ZM (2011) Identification of alkaline stress-responsive genes of CBL family in sweet sorghum (Sorghum bicolor L.). Plant Physiol Bioch 49:1306–1312. doi:10.1016/j.plaphy.2011.08.010
Zhang H, Han B, Wang T, Chen SX, Li HY, Zhang YH, Dai SJ (2012a) Mechanisms of plant salt response: insights from proteomics. J Proteome Res 11:49–67. doi:10.1021/pr200861w
Zhang PP, Fu JM, Hu LX (2012b) Effects of alkali stress on growth, free amino acids and carbohydrates metabolism in Kentucky bluegrass (Poa pratensis). Ecotoxicology 21:1911–1918. doi:10.1007/s10646-012-0924-1
Zhang LM, Liu XG, Qu XN, Yu Y, Han SP, Dou Y, Xu YY, Jing HC, Hao DY (2013a) Early transcriptomic adaptation to Na2CO3 stress altered the expression of a quarter of the total genes in the maize genome and exhibited shared and distinctive profiles with NaCl and high pH stresses. J Integr Plant Biol 55:1147–1165. doi:10.1111/jipb.12100
Zhang X, Wei LQ, Wang ZZ, Wang T (2013b) Physiological and molecular features of Puccinellia tenuiflora tolerating salt and alkaline-salt stress. J Integr Plant Biol 55:262–276. doi:10.1111/jipb.12013
Zhang YH, Lin XY, Ou XF, Hu LJ, Wang JM, Yang CW, Wang SC, Liu B (2013c) Transcriptome alteration in a rice introgression line with enhanced alkali tolerance. Plant Physiol Bioch 68:111–117. doi:10.1016/j.plaphy.2013.04.012
Zhao GM, Liu ZP, Chen MD, Kou WF (2006) Effect of saline aquaculture effluent on salt-tolerant Jerusalem artichoke (Helianthus tuberosus L.) in a semi-arid coastal area of China. Pedosphere 16:762–769. doi:10.1016/S1002-0160(06)60112-4
Zheng HY, Li JD (1999) Form and dynamic trait of halophyte community. In: Saline plants in Songnen plain and restoration of alkaline-saline grass. Science Press, Beijing, pp. 137–138
Zhou HP, Lin HX, Chen S, Becker K, Yang YQ, Zhao JF, Kudla J, Schumaker KS, Guo Y (2014) Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26:1166–1182. doi:10.1105/tpc.113.117069
Zhu JK (2001) Plant salt tolerance. Trends Plant Sci 6:66–71. doi:10.1016/S1360-1385(00)01838-0
Zhu JK (2002) Salt and drought stress signal transduction in plants. Annu Rev Plant Biol 53:247–273. doi:10.1146/annurev.arplant.53.091401.143329
Zhu D, Cai H, Luo X, Bai X, Deyholos MK, Chen Q, Chen C, Ji W, Zhu YM (2012) Over-expression of a novel JAZ family gene from Glycine soja, increases salt and alkali stress tolerance. Biochem Biophys Res Commun 426:273–279. doi:10.1016/j.bbrc.2012.08.086
Acknowledgments
This work was supported by the National Natural Science Foundation of China (No. 31470467) and Ph.D. Student Innovation Fund of Northeast Forestry University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Frans J.M Maathuis.
A. Zhang and W. Zang contributed equally to this work.
Electronic supplementary material
Supplementary Figure S1
2-DE gel images of proteomes extracted from H. tuberosus under control and 20 mM Na2CO3 stress. (PPTX 1.58 mb)
Supplementary Figure S2
2-DE gel images of proteomes extracted from H. tuberosus under control and 50 mM Na2CO3 stress. (PPTX 1.66 mb)
Supplementary Table S3
(XLSX 10 kb)
Supplementary Table S4
(XLSX 25.9 kb)
Supplementary Table S5
(XLSX 14 kb)
Rights and permissions
About this article
Cite this article
Zhang, A., Zang, W., Zhang, X. et al. Global proteomic mapping of alkali stress regulated molecular networks in Helianthus tuberosus L.. Plant Soil 409, 175–202 (2016). https://doi.org/10.1007/s11104-016-2945-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-016-2945-7