Abstract
The regulation mechanism of bamboo height growth has always been one of the hotspots in developmental biology. In the preliminary work of this project, the function of LBD transcription factor regulating height growth was firstly studied. Here, a gene PheLBD12 regulating height growth was screened. PheLBD12-overexpressing transgenic rice had shorter internodes, less bioactive gibberellic acid (GA3), and were more sensitive to GA3 than wild-type (WT) plants, which implied that PheLBD12 involve in gibberellin (GA) pathway. The transcript levels of OsGA2ox3, that encoding GAs deactivated enzyme, was significantly enhanced in PheLBD12-overexpressing transgenic rice. The transcript levels of OsAP2-39, that directly regulating the expression of EUI1 to reduce GA levels, was also significantly enhanced in PheLBD12-overexpressing transgenic rice. Expectedly, yeast one-hybrid assays, Dual-luciferase reporter assay and EMSAs suggested that PheLBD12 directly interacted with the promoter of OsGA2ox3 and OsAP2-39. Together, our results reveal that PheLBD12 regulates plant height growth by modulating GA catabolism. Through the research of this topic, it enriches the research content of LBD transcription factors and it will theoretically enrich the research content of height growth regulation.
Key message
We investigated the functions in height growth of PheLBD12, encoding a LBD transcription factor in moso bamboo. PheLBD12 regulates plant height growth by modulating GA catabolism.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The regulation mechanism of bamboo height growth has always been one of the hotspots in developmental biology. At present, the research on bamboo height growth regulation mainly focuses on morphological and anatomical studies, physiological and biochemical changes and hormone regulation and molecular regulation (Li et al. 2023; Tao et al. 2018). Morphological and anatomical studies showed that during the high growth phase of moso bamboo, a single internode can be divided into cell division zones, cell elongation zones, and secondary cell wall thickening zones (Chen et al. 2022). The rapid growth of bamboo culms is due to a large number of cells dividing and elongating in the cell division and elongation zones between the nodes (Chen et al. 2022; Wei et al. 2019). Proteins and carbohydrates are essential components in the growth process of bamboo, including cell division, cell elongation, and cell wall thickening (Fang et al. 2019; Song et al. 2016). Plant hormones play a crucial role in regulating the growth and development of plants, including bamboo. The accumulation level of Gibberellin A3 (GA3) and 3-indoleacetic acid (IAA) were positively correlated with the growth rate of bamboo shoots (Cui et al. 2012), while zeatin was positively correlated with cell division speed and abscisic acid (ABA) was negatively correlated with it.
With the deepening of the study on the high growth of bamboo and the development of omics, molecular regulation of bamboo height growth has been widely studies, many functional genes related to the high growth of bamboo have been found. They participated in the synthesis and degradation of plant hormones by regulating the division and differentiation, elongation and maturity of cells, thus affecting the high growth process of bamboo. For example, Chen et al. (2010) have cloned 10 cDNA sequences encoding cellulose synthase protein (CesA), BoCesA1-10, from Bambusa oldhamii. Analysis of the tissue expression pattern of bamboo shoots at different heights and in different portions found that BoCesA5, BoCesA6 and BoCesA7 genes showed high expression in the middle portion of bamboo shoots, so it is speculated that these three genes may be involved in the high growth process of bamboo (Chen et al. 2010). BohLOL1, a C2C2 zinc finger protein came from Bambusa oldhamii, which was highly expressed in various bamboo shoot tissues, and also up-regulated in bamboo shoots during the high growth of bamboo, indicating that BohLOL1 may be related to the high growth of bamboo (Yeh et al. 2011). The Bamboo SINAT protein PpSINA, that an Arabidopsis SINAT5 homologous gene. PpSINA was expressed in all organs of bamboo shoots, and its expression in rhizome buds was significantly higher than that in shoot buds and bamboo shoots, which is the main site of auxin synthesis in bamboo shoots. Therefore, it is inferred that PpSINA may regulate the growth and development of bamboo shoots by participating in auxin signaling transduction (Wang et al. 2009). Huang et al. identified seven sucrose synthase protein (BeSUS1-BeSUS7) from bamboo genome, and found that BeSUS5 has a highly expression during the development of bamboo shoots, and was clearly induced by a cellulose synthesis inhibitor. Further experiments revealed that BeSUS5-overexpressing transgenic poplar showed high content of the cellulose in culms, while low content of total soluble sugar (TSS) and starch in leaves, these results indicated BeSUS5 participated in partitioning of carbon to cellulose (Huang et al. 2020). DsEXLA2, an expansion gene, that was cloned from Dendrocalamus sinicus, and DsEXLA2-overexpressing transgenic Arabidopsis exhibited higher plant height, thicker culm, larger leaf, higher cellulose content and less epidermal hair number and smaller stomatal aperture in the prophase and metaphase of growth. In addition, the cell wall was thickened significantly in the transgenic plant (Li et al. 2023).
Moso bamboo (Phyllostachys edulis), which has the largest planting area, the widest distribution range and the highest economic value in China’s bamboo forest, and has attracted much attention for its unique rapid growth mode. however, there are few studies on the mechanism of gene regulation in the process of high growth of bamboo.
Although LBD proteins play a variety of functions in the development of plant leaves and flowers, as well as the formation of lateral roots and branches (Di Mambro et al. 2017; Fan et al. 2012; Kim et al. 2015; Lee et al. 2013; Pandey et al. 2018; Ma et al. 2009; Okushima et al. 2007), little is known about the function of LBD regulating height growth in plant.
In our past studies, 61 PheLBD genes were identified in moso bamboo (Phyllostachys edulis) genome, the RNA-seq data revealed that the expression of PheLBD12 was changed by GA and NAA treatments. Further qRT-PCR results showed that PheLBD12 was induced significantly under Me-JA and ABA treatments (Gao et al. 2022). In this study, we investigated the function of PheLBD12 in plant growth and development, using overexpression transgenic rice lines, which reveals a molecular mechanism that PheLBD12 regulates plant height in transgenic rice.
Materials and methods
Plant materials and stress treatments
Moso bamboo seeds were collected from Guangxi Province, China, and were grown in a greenhouse (16-h light/8-h dark at 25 ◦C) for 3 months.
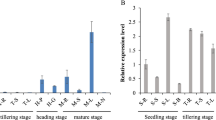
To examine the expression pattern of PheLBD12, the 3-month-old moso bamboo seedlings were sprayed with 50 μM GA or 10 μM PAC, respectively, then the treated leaves were collected after 1, 3, 6 and 12 h. The untreated leaves also need to be collected, and was regarded as the control (0 h). Each sample was collected with three biological replicates.
For GA3 and PAC treatments, after soaking and germinating the seeds of PheLBD12-overexpressing transgenic lines and WT, they were transferred to 1/2 MS solution for hydroponic cultivation. After growing for two weeks in the rice growth room (12-h light/12-h dark at 22 °C), they were treated with 50 μM GA3 or 50 μM PAC solution for 72 h, then plant height were investigated. Each box contained 40 seedlings. Every experiment was repeated three times.
For ABA treatments, the seeds of PheLBD12-overexpressing transgenic lines and WT were sown in 1/2 MS that containing 0, 30 and 50 μM ABA for seed germination assay in the rice growth room (12-h light/12-h dark at 28 °C), germination ratio was recorded after 6 d imbibition. Each plate contained 40 seeds. Every experiment was repeated three times.
RNA extraction and qRT-PCR
Total RNA from plants was extracted using TRI Reagent (Sigma, www.sigmaaldrich.com/). qRT-PCR analyses were performed as described by Wu et al. 2023. The primers used are listed in Table S1.
Yeast one-hybrid
For yeast one-hybrid assays, the CDS sequence of PheLBD12 was inserted into EcoRI–BamHI site of pGADT7 vector to generate a construct with activation domain and PheLBD12. The promoter OsGA2ox3 (OsGA2ox3pro) and OsAP2-39 (OsAP2-39pro) genes were inserted into pAbAi vector, respectively. The primers used are listed in Table S1. The transformation of yeast cells and screening of interactions was described by Wu et al. (Wu et al. 2023).
Quantification of endogenous GA and ABA
The quantification of endogenous GAs was measured by Gibberellin Content Kit (Mibio: ml077232). Endogenous ABA analysis according to the ABA detection kit (Mibio: ml077235).
Dual-luciferase assay
For the Dual-luciferase assays, the coding sequence of PheLBD12 was inserted into pGreenII 62-SK vector to generate the effector vector PheLBD12:62-SK. The promoter sequences and promoter fragments OsGA2ox3, (OsGA2ox3pro and P: −850 to −912 bp), the promoter sequences and promoter fragments of OsAP2-39 (OsAP2-39pro, P1: −183 to −293 bp and P2: −924 to −1035 bp), were inserted into pGreenII 0800-LUC vector to generate reporter vectors, and OsGA2ox3pro:0800-LUC, P:0800-LUC, OsAP2-39pro:0800-LUC, P1:0800-LUC and P2: 0800-LUC, respectively. The primers used are listed in Table S1. The N. benthamiana leaves were used for Dual-luciferase assays as described by Wu (Wu et al. 2023). The firefly luciferase (LUC) and renillia luciferase (REN) activities were measured by the Dual-Luciferase Reporter Assay System (Promega, www.promega.com).
EMSA assay
For EMSA assay, the coding sequence of PheLBD12 was inserted into EcoRI-XbaI site of pMAL-c2X vector to generate the recombinant vector, MBP::PheLBD12 were conducted as described (Wu et al. 2023). The primers used are listed in Table S1. The MBP-PheLBD29 fusion protein purified by the instructions of Anti-MBP Magnetic Beads (BeyoMag™). The promoter fragments of OsGA2ox3 and OsAP2-39 were synthesized and annealed as DNA probe for EMSA assay. EMSA assays were performed according to the manufacturer’s protocols of Lightshift Chemilumines-cent EMSA Kit (Thermo Scientific).
Result
PheLBD12 overexpression shortens plant height and cell length
In a previous study, we showed that PheLBD12 was changed under GA and NAA treatments by the RNA-seq data, and qRT-PCR results showed that PheLBD12 was induced significantly under Me-JA and ABA treatments (Gao et al. 2022). To further analyze the biological function of PheLBD12, we constructed PheLBD12-overexpressing transgenic rice lines. The positive independent T0 transgenic lines were examined by GUS stain and reverse-transcription PCR (RT-PCR), and PheLBD12 expression in T3 lines was also verified by Quantitative real time polymerase chain reaction (qRT-PCR) (Fig.S1). Four PheLBD12-overexpressing transgenic lines (OE-2, OE-5, OE-6 and OE-8) were used for subsequent experimental analysis.
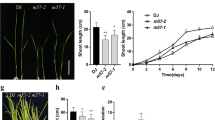
To determine the differences in phenotype between PheLBD12-overexpressing transgenic lines and WT, four-week old seedling OE-2, OE-5, OE-6, OE-8 and WT was investigated. As shown in Fig. 1, PheLBD12-overexpressing transgenic lines exhibited a phenotype of shortened seedling height during the seedling stage (Fig. 1). At maturity, as shown in Fig. 2A, the PheLBD12-overexpressing transgenic lines were still shorter than the WT. Significant differences were observed in the lengths of each internode of the main stem between the PheLBD12-overexpressing transgenic lines and WT (Fig. 2B, C).
Comparison of seedling height between PheLBD12-overexpressing rice and WT. A Image data of four-week old seedling. B Statistics analysis of seedling height of PheLBD12-overexpressing transgenic rice. Data shown represent the means (±SE) of three independent experiments (each with 30 seedlings for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01
Comparison of phenotype between PheLBD12-overexpressing transgenic rice and WT at maturity stage. A Image data of plant at maturity stage. B Statistics analysis of plant height at mature. Data shown represent the means (±SE) of three independent experiments (each with 20 seedlings for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01. C Statistics analysis of internode length. Data shown represent the means (±SE) of five independent experiments (each with 5 seedlings for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01. D Epidermal cells in the second internode of OE-2 and WT. (scale bar =200 μm). E Cell lengths in the second internode of OE-2 and WT. The average values were calculated from measurement of at least 30 cells. Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01
To determine the reason for shortened seedling height of PheLBD12-overexpressing transgenic rice, cell lengths were measured, and found that the cell length of PheLBD12-overexpressing transgenic rice, was significantly shorter than that of the WT (Fig. 2D, E). These results inform that the reduced internode length is the reason for shortened seedling height of PheLBD12-overexpressing transgenic lines.
Plant stem, an important agronomic trait, largely decides biomass, especially in moso bamboo. To verify the regulatory role of PheLBD12 genes in the biomass accumulation, the fresh and dry weight of PheLBD12-overexpressing transgenic and WT were measured. The fresh and dry weight of WT rice were higher than those of PheLBD12-overexpressing transgenic lines (Fig.S2A, B). The lignin content of stem in PheLBD12-overexpressing transgenic and WT rice was also measured. The lignin content in WT rice was about 417.27 ± 1.04mg͙*g−1, while the average lignin content in PheLBD12-overexpressing transgenic rice was about 223.31 ± 2.11mg͙*g−1 (Fig.S2C). Similarly, the cellulose content in WT rice was about 627.27 ± 10.12mg͙*g−1, while the average lignin content in PheLBD12-overexpressing transgenic rice was about 101.31 ± 12.09mg͙*g−1 (Fig. S2D).
Exogenous GA eliminates the dwarfing phenotype of PheLBD12-overexpressing transgenic rice
According to previous research, it has been shown that GA plays an important role in regulating the growth and development of plants, especially regulates cell elongation and determines plant height. To explore whether GA modulates the expression of PheLBD12, the 3-month-old moso bamboo seedlings were treated with GA3 and paclobutrazol (PAC). qRT-PCR showed that the expression of PheLBD12 was suppressed by GA3 (Fig. 3A), which was consistent with RNA-seq data (Gao et al. 2022). Whereas the expression of PheLBD12 was up-regulated by PAC (Fig. 3B). To further investigate whether dwarfing phenotype of PheLBD12-overexpressing transgenic rice is caused by GA deficiency or defects in GA signaling, we investigated the response of PheLBD12-overexpressing transgenic lines to exogenous GA3. The germinated seeds of PheLBD12-overexpressing transgenic rice and WT were incubated in 1/2 MS solution for two weeks, then were treated with 50 μM GA or 50 μM PAC, respectively (Fig. 3C). As showed in Fig. 3D, under 50 μM GA3 treatment, the seedling height of PheLBD12-overexpressing transgenic lines and WT was increased, while the seedling height of PheLBD12-overexpressing transgenic lines did not recover to the same height as the WT after 72 h. Under 50 μM PAC treatment, we found that the growth of PheLBD12-overexpressing transgenic rice and WT was all inhibited. Furthermore, the agar plate assay of α-amylase using half-seed method in PheLBD12-overexpressing transgenic lines and WT showed that there was no significant difference between PheLBD12-overexpressing transgenic lines seeds and WT seeds under GA treatment (Fig.S3).
Response of PheLBD12-overexpressing transgenic rice and WT to GA and PAC. A and B The expression analysis of PheLBD12 under different hormones treatment by qRT-PCR. Data shown represent the means (±SE) of three independent experiments. Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01. C Image data of seedling height of PheLBD12-overexpressing transgenic rice and WT under 0 μM, 50μM GA or 50 μM PAC treatments. D Statistics analysis of seedling height of PheLBD12-overexpressing transgenic rice and WT under 0 μM, 50μM GA or 50 μM PAC treatments. Data shown represent the means (±SE) of three independent experiments (each with 15 seedlings for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01
Together, these data indicate that PheLBD12-overexpressing transgenic rice enhanced GA3 sensitivity and reduced PAC sensitivity, which implies that GA signaling pathway in PheLBD12-overexpressing transgenic rice is not impaired.
PheLBD12 alters the transcription of genes involved in GA metabolism
To investigate whether the regulation of plant height growth by PheLBD12 is influenced by GA, the endogenous levels of GA in PheLBD12-overexpressing transgenic lines and WT was examined (Table 1). Compared with the WT, the endogenous active GA4 content of PheLBD12-overexpressing transgenic seedlings was significantly reduced, the content of other GA precursor substances: GA9, GA15 and GA24 were also reduced to varying degrees (Table 1). These results indicate that overexpression of the PheLBD12 gene can cause a decrease in endogenous active GA content, especially GA4 content, in rice.
Additionally, we used qRT-PCR analysis to investigate the expression of genes encoding GA biosynthesis, including OsKS1 (Helmut Aach et al. 1997), OsKO, OsKAO (Helliwell et al. 2001). Compared with WT, we found that the transcript levels of GA biosynthesis related genes were down-regulated in PheLBD12-overexpressing transgenic rice, except KS1 (Fig. 4A). Next we compared the expression of GA metabolism related genes in PheLBD12-overexpressing transgenic lines and WT, including OsGA3oxs, OsGA20oxs and OsGA2oxs. OsGA3ox1, OsGA20ox2 and OsGA20ox4 showed a low expression in PheLBD12-overexpressing transgenic lines (Fig. 4B). As expect, OsGA2ox2 and OsGA2ox3 showed a high expression in PheLBD12-overexpressing transgenic lines (Fig. 4C). EUI1, which was involved in GA inactivation, were up-regulated in PheLBD12-overexpressing transgenic lines compared with that in WT. In addition, some transcription factors that reported to be involved in GA metabolism, such as OsAP2-39 (Yaish et al. 2010), OsMADS57 (Chu et al. 2019), OsEATB (Qi et al. 2011) and HOX12 (Gao et al. 2016). The qRT-PCR results showed that these transcription factors were also up-regulated in PheLBD12-overexpressing transgenic lines compared with that in WT (Fig. 4D).
PheLBD12 regulate the expression of GA-related genes. A Expression of GA biosynthesis related genes. B and C Expression of GA metabolic related genes. D Expression of GA-related transcription factors. Data shown represent the means (±SE) of three independent experiments. Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01
Therefore, these results provided evidence that the up-regulated expression of genes involved in GA catabolism, which was mainly led to reduced accumulation in GA levels, and further resulted in the defect in the stem elongation in PheLBD12-overexpressing transgenic lines.
PheLBD12 regulates the transcription of OsGA2ox3 and OsAP2-39 by interacting with their promoters
LBD domain can then bind specifically to the ‘GCGGCG’ element, which in turn regulates target gene expression (Majer and Hochholdinger 2011; Shuai et al. 2002).
To investigate whether PheLBD12 regulates the transcription of down-genes by binding to GCGGCG element in their promoter regions. Firstly, we conducted yeast one-hybrid assay, and the results showed that yeast cells with GCGGCG element transformed with pGADT7-PheLBD12 grew well on SD/-Leu medium that containing up to AbA (700 ng/mL) (Fig.S4), whereas no growth was observed in yeast strains with GCGGCG element transformed with pGADT7 vector. In addition, PheLBD12 is located in the nucleus and exhibits yeast self-activation (Fig.S4).
To further investigate whether PheLBD12 directly affects the expression of GA metabolism genes, as shown in Fig.S5, the potential PheLBD12 binding sites, GCGGCG at −880 to −874 bp (P) from ATG position of OsGA2ox3, and at −228 to −233 bp (P1) and −986 to −991 bp (P2) from ATG position of OsAP2-39. The PheLBD12 full-length cDNA was fused in frame to the GAL4 activation domain in the pGADT7 vector. The promoter sequence regions of OsGA2ox3 and OsAP2-39 were ligated into the pAbAi vector. The yeast one-hybrid assays suggested that PheLBD12 directly interacts with the promoter sequences of OsGA2ox3 and OsAP2-39 (Fig. 5A, B).
OsGA2ox3 and OsAP2-39 are the direct target genes of PheLBD12. A Schematic diagram of recombinant vectors for yeast one-hybrid assays. B Yeast one-hybrid assays. C Schematic diagram of recombinant vectors for Dual-luciferase assays. D Dual-luciferase promoter activation assays in tobacco. E and F EMSA showed that PheLBD12 bound to GCGGCG element in the promoter regions of OsGA2ox3 (E) and OsAP2-39 (F)
The interaction of PheLBD12 with OsGA2ox3 and OsAP2-39 genes promoter were further tested by the dualluciferase reporter assays. We constructed different effector and reporter vectors, the reporter and effector were co-transformed into tobacco leaves (Fig. 5C). The LUC and REN activities were measured, after transiently expressed in tobacco leaves, and found that co-expression of 35S:PheLBD12 + OsGA2ox3Pro:LUC and 35S:PheLBD12 + P2:LUC significantly increased the LUC:REN ratio. As expected, the co-expression of 35S:PheLBD12 + OsAP2-39Pro:LUC and 35S:PheLBD12 + P2:LUC also significantly increased the LUC:REN ratio, and the LUC activity was about fourfold than that in the control (Fig. 5D). PheLBD12 was successfully cloned into procaryotic expression vector pMAL-c2X proved by DNA sequencing, and obtained MBP::PheLBD12 protein. Then, electrophoretic mobility shifts assays (EMSAs) were used to examine the interactions between PheLBD12 protein and the DNA motifs of the promoter region of OsGA2ox3 and OsAP2-39. The results were showed in Fig. 5E, F, as we expected, the probe was incubated with PheLBD12 protein, the shifted band was observed clearly. No shift band was detected when the sample only contains the probe. Thus, our results indicate that PheLBD12 is an upstream transcriptional regulator of OsGA2ox3 and OsAP2-39.
PheLBD12-overexpressing transgenic rice show more sensitivity to ABA
Our data confirmed PheLBD12 can directly interact with the promoter sequences of OsAP2-39, Yaish et al. have been proved that OsAP2-39 can control not only the GA deactivation protein (EUI), but also the ABA biosynthesis gene (OsNCED-1) (Yaish et al. 2010). Thus, we checked seed germination and post-germinated growth of PheLBD12-overexpressing transgenic rice under different concentration ABA. In the treatment with 0 µM ABA, the relative germination rate of the WT was about 97%, but the relative germination rate of PheLBD12-overexpressing transgenic rice was about 80% (Fig. 6A). With the addition of ABA, the germination rate of PheLBD12-overexpressing transgenic rice was significantly reduced. For example, with 30 µM ABA, the relative germination rate of the WT was about 77.58%, but the relative germination rate of PheLBD12-overexpressing transgenic rice was about 43.25% (Fig. 6A). When ABA concentration was increased to 50 µM, the relative germination rate of the WT was about 37.54%, but the relative germination rate of PheLBD12-overexpressing transgenic lines was about 15.76% (Fig. 6A). The WT and PheLBD12-overexpressing transgenic rice were also assessed for their responses to ABA during the post-germination growth stage. The seedlings were germinated on 1/2 MS medium for 3 d, and then transferred to medium supplemented with different concentrations of ABA. The PheLBD12-overexpressing transgenic rice exhibited a slow growth compared with the WT plants, when the nutrient solution contains 0 mM ABA (Fig. 6B). Similarly, with the addition of ABA, the plant height of PheLBD12-overexpressing lines was significantly inhibited (Fig. 6B, C). Together, these data suggested that the overexpression of PheLBD12 increased the sensitivity of the transgenic rice to ABA.
PheLBD12-overexpressing transgenic rice responds to ABA treatment. A Statistics analysis of germination rate. Data shown represent the means (±SE) of five independent experiments (each with 50 seeds for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01. B Image data of PheLBD12-overexpressing transgenic rice and WT grown on 0, 30 or 50 μM ABA. C Statistics analysis of plant height. Data shown represent the means (±SE) of five independent experiments (each with 15 seeds for each line). Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01
Increased expression of marker genes involved in the ABA biosynthesis and ABA accumulation in PheLBD12-overexpressing transgenic rice
As we all know that ABA plays a negative function in seed germination and seedling growth, combined the above experimental results, suggesting that PheLBD12 may play a key role in ABA accumulation in transgenic plants, so the endogenous ABA contents were measured. The ABA level in the OE-2 was about twofold than that in the WT plants (Fig. 7A). In addition, Dihydrophaseic acid (DPA) (Fig. 7B) and Abscisic acid glucose ester (ABAGE) (Fig. 7C), those of ABA derivative compounds, their levels are also increased in the PheLBD12-overexpressing transgenic lines. The reason for increased ABA levels in PheLBD12-overexpressing transgenic lines was investigated, the genes that involved in ABA biosynthesis and catabolism were studied, such as NCED1, NCED4, NCED5, OsABA8ox1, OsABA8ox2 and OsABA8ox3 (Iuchi et al. 2001; Qin and Zeevaart 1999; Saika et al. 2007). Interestingly, the results showed that the expression level of genes involved in ABA biosynthesis and catabolism were changed due to the PheLBD12 overexpression. As showed in Fig. 7D, the expression levels of NCED1, NCED4 and NCED5, ABA biosynthesis enzymes, was up-regulated in the PheLBD12-overexpressing transgenic rice; while the expression levels of OsABA8ox1, OsABA8ox2 and OsABA8ox3, main ABA catabolism genes, were down-regulated in OE. Taken together, the above data show that PheLBD12 involved in the ABA biosynthesis.
ABA accumulation and expression analysis of ABA biosynthesis and catabolism genes in PheLBD12-overexpressing transgenic rice and WT. A–C Determination of endogenous ABA content (ng/gDW) in PheLBD12-overexpressing transgenic rice and WT. D Expression analysis of ABA biosynthesis and catabolism genes in PheLBD12-overexpressing transgenic rice and WT. Data shown represent the means (±SE) of three independent experiments. Significant differences were determined using Student’s t-test. *P < 0.05, **P < 0.01.
Discussion
Up to now, the research on the regulatory mechanism of plant height has always been a hot topic for breeders. Moso bamboo, which is famous for its height growth. However, in moso bamboo, the molecular regulatory mechanism of height growth has been still poorly studied. In our previous research, 61 PheLBD genes were identified in moso bamboo genome. Public RNA-seq data showed that PheLBD12 was induced significantly under GA and NAA treatments (Gao et al. 2022). Further qRT-PCR results showed that PheLBD12 was induced significantly under Me-JA and ABA treatments (Gao et al. 2022). Therefore, in this study, PheLBD12 was further studied as a candidate gene. The expression of PheLBD12 was induced by PAC but repressed by GA3 (Fig. 3A, B), which was consistent with previous research (Gao et al. 2022). The PheLBD12-overexpressing transgenic rice displayed semi-dwarf phenotype in seedlings and maturity stages, and the semi-dwarf phenotype can be rescued by applying GA3 (Fig. 3C). Examination of the GA content also confirimed that bioactive GA were reduced in PheLBD12-overexpressing transgenic rice compared with that of the WT. The main forms of bioactive GA in plants are GA1, GA3, GA4 and GA7, and GA4 via GA15, GA24, and GA9 to convert (Reinecke et al. 2013; Tudzynski et al. 2003). Compared with the WT, the endogenous active GA4 content of PheLBD12-overexpressing transgenic seedlings was significantly reduced, so the content of other GA precursor substances: GA15, GA24, and GA9 were also reduced to varying degrees (Table 1). And the agar plate assay of α-amylase using half-seed method (Fig.S3) further proved that semid-warf phenotype in PheLBD12-overexpressing transgenic rice result from GA deficiency but not malfunction in GA signaling.
Endogenous GA levels are modulated by the expression of genes involved in GA biosynthesis and deactivation, and also fine-tuned by feedback control of GA metabolism (Boden et al. 2014; Fukazawa et al. 2011, 2017; Hedden and Thomas 2012; Thomas et al. 2005; Yamaguchi 2008). GA20oxs (GA 20-oxidase), GA3oxs (GA 3-oxidase) and GA2oxs (GA 2-oxidase), which were the main GA metabolism genes. GA20oxs and GA3oxs can catalyze the formation of bioactive GAs, while GA2oxs encoded GAs deactivated enzyme (Yamaguchi 2008). In rice, overexpressing genes encoding GA2-oxidase increases tiller numbers but inhibits stem elongation, which is coupled with GA deficiency (Lo et al. 2008). In PheLBD12-overexpressing transgenic lines, the transcript levels of GA biosynthesis genes were reduced and GA catabolism genes were increased, resulting in less bioactive GA accumulation than WT (Fig. 4). Interestingly, the transcription of OsGA3ox2 and OsGA20 × 1 was up-regulated in PheLBD12-overexpressing transgenic rice, which may account for a feedback mechanism due to the reduced levels of bioactive GA in PheLBD12-overexpressing rice. Thus, it was likely that PheLBD12-overexpressing transgenic rice showed reduced transcription of OsGA2ox9. Furthermore, some transcription factors that reported to be involved in GA metabolism were also up-regulated in PheLBD12-overexpressing transgenic lines compared with that in WT (Fig. 4D), especially OsAP2-39. OsAP2-39 directly controls the gene that codes for a GA deactivation protein (EUI), EUI1 is also involved in GA deactivation reaction by catalyzing 16α, 17-epoxidation reaction of GA4, GA9 and GA12 (Ma et al. 2006; Zhu et al. 2006), which regulates internode elongation by modulating GA responses in rice (Luo et al. 2006). Meanwhile, EUI1 was up-regulated in PheLBD12-overexpressing transgenic rice in our studies. Given that, it perfectly explains the measurement of endogenous active GA content in PheLBD12-overexpressing transgenic rice. By scanning the promoter region of OsGA2ox3 and OsAP2-39, we found that the GCGGCG element in the promoter regions of OsGA2ox3 and OsAP2-39, bound by PheLBD12 protein (Fig.S4). Data from yeast one-hybrid, Dual-luciferase reporter assays and EMSA proved that OsGA2ox3 and OsAP2-39 are the direct target of PheLBD12 (Fig. 5). PheLBD12 directly increased the expression of OsGA2ox3 and OsAP2-39, resulting in low levels of bioactive GA, followed by the reduced plant height of the PheLBD12-overexpressing transgenic rice.
Yaish et al. (2010) also found that overexpression of OsAP2-39 increased the ABA content in rice plants, making seed germination and growth of transgenic lines were inhibited in the transgenic lines (Yaish et al. 2010). Molecular evidence suggests, in the process of plant growth and development, ABA and GA usually act through a complicated network of antagonistic interactions, a high endogenous level of ABA causes a reduction in the endogenous level of GA (Oh et al. 2007; Seo et al. 2006). Previous studies have shown that the growth rate of bamboo could positively correlate with GA and negatively with ABA concentration during internode elongation (Cui et al. 2012). Consistent with the observation of OsAP2-39, using of ABA delays seed germination in the PheLBD12-overexpressing transgenic rice and shows more drastic effect on the growth of PheLBD12-overexpressing transgenic rice than in the WT (Fig. 6). Additionally, the active endogenous ABA level of the PheLBD12-overexpressing transgenic lines was found to be 1.5-fold higher than the WT level (Fig. 7A). Plant endogenous ABA level is controlled by the equilibrium between ABA biosynthesis and catabolism (Zeevaart et al. 1988). In many cases, expressions of ABA biosynthesis genes and ABA catabolism genes were co-regulated in plant development. To obtain supporting evidence, gene expression analysis of NCEDs and ABA8ox was investigated. OsNCED1, OsNCED4 and OsNCED5, which encoding 9-cis-epoxycarotenoid dioxygenase, that is considered as the main ABA biosynthesis enzyme (Iuchi et al. 2001; Qin and Zeevaart 1999). OsABA8ox1, OsABA8ox2 and OsABA8ox3, which coding ABA 8’-hydroxylase, that is considered as the main ABA catabolism enzyme (Saika et al. 2007). qRT-PCR results revealed the up-regulation of NCEDs in the PheLBD12-overexpressing transgenic rice. Conversely, OsABA8ox1, OsABA8ox2 and OsABA8ox3, were down-regulated in PheLBD12-overexpressing transgenic rice. Collectively, these results provided evidence that PheLBD12 inhibited the growth of PheLBD12-overexpressing transgenic rice by increased contents of endogenous ABA.
In a conclusion, overexpression PheLBD12 in rice, resulting in reduced plant height of the PheLBD12-overexpressing transgenic rice. On the one hand, it is due to the low levels of bioactive GA. On the other hand, the high contents of endogenous ABA also play a key role in height growth. However, the regulation network of GA and ABA in PheLBD12-overexpressing transgenic rice is needed to further dissect.
Data availability
Enquiries about data availability should be directed to the authors.
References
Boden SA, Weiss D, Ross JJ, Davies NW, Trevaskis B, Chandler PM, Swain SM (2014) Early flowering 3 regulates flowering in spring barley by mediating gibberellin production and flowering Locus T expression. Plant Cell 26:1557–1569
Chen CY, Hsieh MH, Yang CC, Lin CS, Wang AY (2010) Analysis of the cellulose synthase genes associated with primary cell wall synthesis in Bambusa oldhamii. Phytochemistry 71:1270–1279
Chen M, Guo L, Ramakrishan M, Fei ZJ, Vinod KK, Ding YL, Jiao C, Gao ZP, Zha RF, Wang CY, Gao ZM, Yu F, Ren GD, Wei Q (2022) Rapid growth of Moso bamboo (Phyllostachys edulis): cellular roadmaps, transcriptome dynamics, and environmental factors. Plant C 34:3577–3610
Chu YL, Xu N, Wu Q, Yu B, Li XX, Chen RR, Huang JL (2019) Rice transcription factor OsMADS57 regulates plant height by modulating gibberellin catabolism. Rice. https://doi.org/10.1186/s12284-019-0298-6
Cui K, He CY, Zhang JG, Duan AG, Zeng YF (2012) Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J Proteome Res 11:2492–2507
Fan M, Xu C, Xu K, Hu Y (2012) Lateral Organ boundaries domain transcription factors direct callus formation in Arabidopsis regeneration. Cell Res 22:1169–1180. https://doi.org/10.1038/cr.2012.63
Fang D, Mei TT, Roll A, H¨olscher D (2019) Water transfer between bamboo culms in the period of sprouting. Front Plant Sci 10:786
Fukazawa J, Nakata M, Ito T, Matsushita A, Yamaguchi S, Takahashi Y (2011) bZIP transcription factor RSG controls the feedback regulation of NtGA20ox1 via intracellular localization and epigenetic mechanism. Plant Signal Behav 6:26–28
Fukazawa J, Mori M, Watanabe S, Miyamoto C, Ito T, Takahashi Y (2017) DELLAGAF1 complex is a Main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiol 175:1395–1406
Gao S, Fang J, Xu F, Wang W, Chu C (2016) Rice HOX12 regulates panicle exsertion by directly modulating the expression of elongated uppermost internode1. Plant Cell 28:680–695
Gao YM, Wang K, Wang RJ, Wang LN, Liu HX, Wu M, Xiang Y (2022) Identification and expression analysis of LBD Genes in Moso Bamboo (Phyllostachys edulis). J Plant Growth Regul 41:2798–2817
Hedden P, Thomas SG (2012) Gibberellin biosynthesis and its regulation. Biochemical Journal 444:11–25
Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci 98:2065–2070
Huang Y, Wang LL, Hu SL, Luo XG, Cao Y (2020) Overexpression of the bamboo sucrose synthase gene (BeSUS5) improves cellulose production, cell wall thickness and fiber quality in transgenic poplar. Tree Genet Genomes 16:75
Iuchi S, Kobayashi M, Taji T, Naramoto M, Seki M, Kato T, Tabata S, Kakubari Y, Yamaguchi-Shinozaki K, Shinozaki K (2001) Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J 27:325–333
Kim MJ, Kim M, Kim J (2015) Lateral organ boundaries domain (LBD) 10 interacts with sidecar pollen/LBD27 to control pollen development in Arabidopsis. The Plant J 81:794–809
Lee HW, Kim MJ, Kim NY, Lee SH, Kim J (2013) LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J 73:212–224
Li L, Yu MH, Yao WJ, Ding YL, Lin SY (2023) Research advance in growth and development of bamboo organs. Ind Crops Prod 205:117428
Lo SF, Yang YY, Chen KT, Hsing YI, Zeevaart J, Chen LJ, Yu SM (2008) A novel class of gibberellin 2-oxidases control semidwarfism, tillering, and root development in rice. Plant Cell 20:2603–2618.
Luo AD, Qian Q, Yin HF, Liu XQ, Yin CX, Lan Y, Tang JY, Tang ZS, Cao SY, Wang XJ, Xia K, Fu XD, Luo D, Chu CC (2006) EUI1, encoding a putative cytochrome P450 monooxygenase, regulates internode elongation by modulating gibberellin responses in rice. Plant Cell Physiol 47:181–191
Ma H, Zhang S, Ji L, Zhu H, Yang S, Fang X, Yang R (2006) Fine mapping and in silico isolation of the EUI1 gene controlling upper internode elongation in rice. Plant Mol Biol 60:87–94
Ma Y, Wang F, Guo J, Zhang XS (2009) Rice OsAS2 gene, a member of LOB domain family, functions in the regulation of shoot differentiation and leaf development. J Plant Biol 52:374–381
Majer C, Hochholdinger F (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16:47–52
Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Marée AFM, Costantino P, Grieneisen VA, Sabatini S (2017) Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci 114:E7641–E7649
Oh E, Yamaguchi S, Hu JH, Yusuke J, Jung B, Paik I, Lee HS, Sun TP, Kamiya Y, Choi G (2007) PIL5, a phytochromeinteracting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19:1192–1208
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130
Pandey SK, Lee HW, Kim MJ, Cho C, Oh E, Kim J (2018) LBD18 uses a dual mode of a positive feedback loop to regulate ARF expression and transcriptional activity in Arabidopsis. Plant J 95:233–251
Qi WW, Sun F, Wang QJ, Chen ML, Huang YQ, Feng YQ, Luo XJ, Yang JS (2011) Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol 157:216–228
Qin X, Zeevaart JA (1999) The 9-cis-epoxycarotenoid cleavage reaction is the key regulatory step of abscisic acid biosynthesis in water-stressed bean. Proc Natl Acad Sci 96:15354–15361
Reinecke DM, Wickramarathna AD, Ozga JA, Kurepin LV, Jin AL, Good AG, Pharis RP (2013) Gibberellin 3-oxidase gene expression patterns influence gibberellin biosynthesis, growth, and development in pea. Plant Physiol 163:929–945
Saika H, Okamoto M, Miyoshi K, Kushiro T, Shinoda S (2007) Ethylene promotes submergence-induced expression of OsABA8ox1, a gene that encodes ABA 8′-hydroxylase in rice. Plant Cell Physiol 48:287–298
Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366
Shuai B, Reynaga-Peña CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129:747–761
Song XZ, Peng CH, Zhou GM, Gu HH, Li Q, Zhang C (2016) Dynamic allocation and transfer of non-structural carbohydrates, a possible mechanism for the explosive growth of Moso bamboo (Phyllostachys heterocycla). Sci Rep 6:25908
Tao G, Fu Y, Zhou M (2018) Advances in studies on molecular mechanisms of rapid growth of bamboo species. J Agric Biotechnol 26:871–887
Thomas SG, Rieu I, Steber CM (2005) Gibberellin metabolism and signaling. Vitam Horm 72:289–338
Tudzynski B, Mihlan M, Rojas MC, Linnemannstons P, Gaskin P, Hedden P (2003) Characterization of the final two genes of the gibberellin biosynthesis gene cluster of Gibberella fujikuroi: des and P450–3 encode GA4 desaturase and the 13-hydroxylase, respectively. J Biol Chem 278(31):28635–28643
Wang KH, Peng HZ, Lin EP, Jin QY, Hua XQ, Yao S, Bian HW, Han N, Pan JW, Wang JH, Deng MJ, Zhu MY (2009) Identification of genes related to the development of bamboo rhizome bud. J Exp Bot 61:551–561
Wei Q, Guo L, Jiao C, Fei ZJ, Chen M, Cao JJ, Ding YL, Yuan QS (2019) Characterization of the developmental dynamics of the elongation of a bamboo internode during the fast growth stage. Tree Physiol 39:1201–1214
Wu M, He W, Wang LN, Zhang XY, XiangY, WK (2023) PheLBD29, an LBD transcription factor from Moso bamboo, causes leaf curvature and enhances tolerance to drought stress in transgenic Arabidopsis. J Plant Physiol 280:153865
Yaish MW, El-kereamy A, Zhu T, Beatty PH, Good AG, Bi YMR, SJ (2010) The APETALA-2-Like Transcription Factor OsAP2–39 Controls Key Interactions between Abscisic Acid and Gibberellin in Rice. Plos Genetics 6:e1001098
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251
Yeh SH, Lin CS, Wu FH, Wang AY (2011) Analysis of the expression of BohLOL1, which encodes an LSD1-like zinc finger protein in Bambusa oldhamii. Planta 234:1179–1189
Zeevaart J, Creelman R (1988) Metabolism and physiology of abscisic acid. Annu Rev Plant Biol 39:439–473
Zhu YY, Nomura T, Xu YH, Zhang YY, Peng Y, Mao BZ, Hanada A, Zhou HC, Wang RX, Li PJ, Zhu XD, Mander LN, Kamiya Y, Yamaguchi S, He ZH (2006) ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell 18:442–456
Funding
National Natural Science Foundation of China,32201640,Min Wu,Scientific Research Foundation of Education Department of Anhui Province of China,KJ2021A0164,Min Wu,Postdoctoral Science Foundation of Anhui Province,2021B549,Min Wu
Author information
Authors and Affiliations
Contributions
All authors conceived and designed the experiments. Min Wu designed the experiment, analyzed the data, conducted all the experiments and wrote the manuscript. Yufang Wang provided assistance in collecting plant materials. Shunran Zhang helped to handle figures and tables. Yan Xiang supervised the whole project, and provided financial support for the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have not disclosed any competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, M., Wang, Y., Zhang, S. et al. A LBD transcription factor from moso bamboo, PheLBD12, regulates plant height in transgenic rice. Plant Mol Biol 114, 95 (2024). https://doi.org/10.1007/s11103-024-01487-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-024-01487-0