Abstract
The Lateral Organ Boundaries Domain (LBD) proteins are a class of plant-specific transcription factor family, which participate in plant growth, development, and stress response. In present study, 61 PheLBD genes were identified in moso bamboo (Phyllostachys edulis) genome. These members clustered into two major classes (Class I and Class II) based on the previous study and phylogenetic analysis, and Class I was further divided into five subgroups (Class I–Class E). The gene architecture and conserved motifs suggested the members in one subgroup shared the structural similarities and highly conserved motif compositions. Scaffold position analysis showed PheLBDs were unevenly located on 19 moso bamboo scaffolds. Synteny analysis indicated segmental duplication and transposed duplication played significant roles in PheLBD gene expansion and some PheLBD genes have been and are undergoing markedly positive purifying selection during evolution. A large number of light-responsive elements, abiotic-stress and hormone-response elements were discovered in the promoter of PheLBDs. Public RNA-seq data helps to analyze the expression profile of PheLBD genes in 14 moso bamboo tissues. And we also found most genes in class II were significantly up-regulated under auxin naphthaleneacetic acid (NAA) treatment, but were sensitive after Gibberellins (GA)-treated. Moreover, quantitative real-time reverse transcription PCR (RT-qPCR) analysis showed that PheLBDs have different response to salt and drought stress as well as abscisic acid (ABA) and Methyl jasmonate (MeJA). Overall, these results paved a way for the further functional studies of PheLBDs.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Transcription factors (TFs) families play important roles in regulating the growth and development, signal transduction and environmental stress responses in higher plants. The lateral organ boundaries domain (LBD) is unique TFs to plants and may regulate plant particular growth and development processes (Shuai et al. 2002). They can be divided into two subclasses (Class I and Class II) based on the sequence characteristic of the LOB domain at the N-terminus. The LBD genes in Class I contain a C-motif, a leucine-zipper-like motif and a GAS block in between: C-motif is four conserved cysteine rich sequence (CX2CX6CX3C), which is presumably required for DNA-binding; A leucine-zipper-like coiled-coil motif (LX6LX3LX6L) allows the formation of coiled-coil protein interactions for LBD genes in class I who also possess a GAS block (Shuai et al. 2002; Majer and Hochholdinger 2011). However, class II LBD genes only have a conserved zinc finger-like domain (LX6LX3LX6L) (Semiarti et al. 2001). Until now, many LBD genes have been and are undergoing evolution, and some LBD genes have not fully met this standard (Majer and Hochholdinger 2011).
It is reported that the function of many LBD genes have been identified in a variety of plants. For example, ASYMMETRIC LEAVES2 (AS2) was initially characterized in Arabidopsis thaliana and expressed at the boundaries of lateral organs during plant development, indicating it might play a potential role in organ separation and lateral organ development (Iwakawa et al. 2002; Shuai et al. 2002). Subsequent studies reported AtLBD6/AS2 had defined lateral organ boundaries, and controlled leaf polarity and regulated flower development (Iwakawa et al. 2007). AtLBD16, AtLBD18 and AtLBD29 were the direct targets of AtARF7 and AtARF19, which triggered the regeneration of lateral roots and callus formation (Okushima et al. 2005; Yoko Okushima et al. 2007). And AtLBD37, AtLBD38 and AtLBD39 played negative roles in anthocyanin biosynthesis (Rubin et al. 2009). In poplar, PtaLBD1 had a positive role in secondary phloem growth, while PtaLBD15 and PtaLBD18 were peculiarly expressed in secondary xylem, implying that the LBD family was involved in secondary growth during xylem formation (Yordanov and Regan 2010). Besides these, recent studies demonstrated that the LBD gene family also participated in stress response. The expression of many StLBDs was responsive to mannitol and sodium chloride treatments, such as StLBD1-2, StLBD4-1, StLBD1-4 and StLBD9-1 (Liu et al. 2019b). And seven LBD genes in Physcomitrella patens (PpLBD1, PpLBD3, PpLBD12, PpLBD15, PpLBD22 PpLBD23 and PpLBD30) were up-regulated under ABA and mannitol treatments, indicating these LBD proteins might be play roles in stress response (Huang et al. 2020). A subfamily II gene SlLBD40 was proved to be a negative regulator of drought tolerance through overexpressing and knockout transgenic tomato plants (Liu et al. 2020). However, the overexpressed AtLBD15 plants exhibited ABA hypersensitivity and water tolerance, which showed opposite phenotypes to the loss-of-function mutant lbd15. Further analysis showed that LBD15 can directly bind to the CATTTAT motif sequence in the promoter region of the ABA signaling pathway gene ABI4, activated its expression and close the stomata, thereby reducing water loss and improving water-deficit stress tolerance (Guo et al. 2020). Moreover, the proteomic analysis in rice showed LBD proteins were down-regulated in rolled leaf mutant plant SRL1 and SRL2 whose drought tolerance was enhanced under drought stress compared with WT (Liao et al. 2019).
With the continuous development of sequencing technology, many plant species have done whole-genome sequencing, which has laid a solid foundation for the use of bioinformatics to study and identify gene functions. Many model plants, for example, 43 LBD genes in Arabidopsis (Matsumura et al. 2009), 35 in rice (Yang et al. 2006), 44 in maize (Zhang et al. 2014), 28 in Brachypodium (Gombos et al. 2017), 57 in poplar (Zhu et al. 2007) and 58 members in apple (Wang et al. 2013) have been identified and some of which participate in a variety of biological processes. Inconceivable, the reports concerning LBD TFs in moso bamboo are rare. Moso bamboo (Phyllostachys edulis), an important woody bamboo, is widely distributed in the subtropics of China and has high ecological, cultural and economic values (Peng et al. 2013; Zhao H et al. 2017). However, moso bamboo also faces different adverse environmental circumstances during growth and development stages, including low temperature, high salt and drought. The environmental stress will limit plant growth, development and yield (Gong et al. 2020). For example, a severe drought in southern China destroyed 13,733 ha of forest in 2013, causing the death of 6.18 million bamboo culms and the decrease in the rhizome bud and winter shoot yields of about 40% and 20%, respectively (Ge et al. 2018). Additionally, if sufficient water was provided to the moso bamboo in autumn, the yield per unit area may increase. Specifically, the number of bamboo shoots can be increased by 30.12%, the yield of bamboo shoots can be increased by 54.18%, and the weight of bamboo shoots can be increased by 18.75% in the coming year (Qi-Jiang et al. 2001; Zhao et al. 2019). Therefore, it is necessary to study and transform stress-related genes to improve the yield and quality of moso bamboo in harsh environment. Although moso bamboo has completed two whole-genome sequencing and the genome sequence has been published, LBD genes in moso bamboo have not been researched yet (Peng et al. 2013; Zhao et al. 2018). In the present study, we used various in silico approaches to identify and characterize systematically PheLBD gene family, including gene structure, conserved motif organization, synteny analysis, evolutionary pattern, cis-acting elements, tissue expression profile, induced expression level, and the subcellular localization analysis were carried out. This study provides a theoretical basis for further researching the function of moso bamboo LBD genes particularly in abiotic stress responses.
Materials and Methods
Identification Analyses of PheLBD TFs
To obtain the putative PheLBD proteins in moso bamboo, 43 AtLBD proteins in Arabidopsis thaliana (Matsumura et al. 2009) and 28 BdLBD proteins in Brachypodium distachyon (Gombos et al. 2017) were used as query in local BLASTP program to seek out the LBD proteins in moso bamboo database download from the website (http://gigadb.org/dataset/view/tdsourcetag/s_pcqq_aiomsg/id/100498/File_sort/name/File_page/2) with a significant E-value (< lE-3) (Zhang et al. 2014; Yang et al. 2016; Liu et al. 2019b). Then, the Pfam database (http://pfam.xfam.org/search) and the NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi) were used to verify the candidate proteins contained the LOB domain (PF03195) (Marchler-Bauer et al. 2017; Finn et al. 2006, 2016). The information in regards to the coding sequence (CDS) length, amino acids number, molecular weight (MW), and the isoelectric point (pI) of PheLBD proteins were searched with the aid of ExPASy proteomics server (https://www.expasy.org/) (Wilkins et al. 1999). Plant-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) was employed in predicting subcellular location of PheLBD proteins (Chou and Shen 2010).
Multiple Sequence Alignment and Phylogenetic Analyses
To unravel the phylogenetic relationships of PheLBDs among different species (Phyllostachys edulis, Brachypodium distachyon, Oryza sativa and Zea mays), ClustalW software was used for multiple sequence alignments, and MEGA 6.0 was subsequently utilized to construct the phylogenetic tree based on the multiple alignment results using the neighbor-joining algorithm and Poisson model with a bootstrap analysis of 1000 replicates (Thompson et al. 1994; Tamura et al. 2013).
Exon–intron Structure Determination and Conserved Motif Analyses
The online Gene Structures Display Server (http://gsds.cbi.pku.edu.cn/) was utilized to identify and presented the exon–intron structures of the PheLBDs (Guo et al. 2007). The online tool MEME Version 5.3.3 (https://meme-suite.org/meme/tools/meme) was used to analyze the conserved motifs of PheLBD proteins (Default parameter setting: maximum number of motifs, 20; optimum motif length ranged between 6 and 200) and visualize with TBtools software (Bailey et al. 2006).
Gene Distribution and Synteny Analysis
The scaffold location information (GFF) of Phyllostachys edulis genes was downloaded from http://gigadb.org/dataset/view/tdsourcetag/s_pcqq_aiomsg/id/100498/File_sort/name/File_page/2, from which all PheLBD gene location was screened and visualized in Tbtools-Gaphics-Show Gene on Chomosome-Basic Circos. In addition, gene duplication analysis of PheLBDs was performed using the Multiple Collinearity Scan toolkit (MCScanX) using default parameters (Wang et al. 2012). The PheLBD homeology was presented together with the gene location mentioned above. However, the synteny relationship of LBD genes among Phyllostachys edulis and Arabidopsis thaliana, Oryza sativa, Brachypodium distachyon, Zea mays, and Sorghum bicolor was displayed via Dual Systeny Plotter software (https://github.com/CJ-Chen/TBtools) (Chen et al. 2020). Thereafter, the values of the non-synonymous (ka), synonymous (ks) and Ka/Ks of the duplicated gene pairs were calculated by Simple Ka/Ks Calculator (NG) in TBtools software (Gao et al. 2020). Generally, the ratio of Ka/Ks greater than 1 indicates the positive selection, equal to 1 means the neutral selection, and less than 1 represents the negative selection (Cannon et al. 2004). And the divergence time is counted with the following formula: T = Ks/2λ (λ = 6.5 × 10–9) (Peng et al. 2013).
Promoter Cis-acting Element Analysis
To survey the putative cis-acting elements in promoter region, the 2000 bp upstream sequences of the translation start site (TSS) of all PheLBDs were retrieved from the genomic sequences and then were uploaded to PlantCARE website (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (Lescot 2002).
Expression Pattern Analysis
The data of 14 moso bamboo tissues (0.1 cm root on shoot, 0.5 cm root on shoot, 2 cm root on shoot, 10 cm root on shoot, Blade leaf, Leaf sheath, Sheath sheet, New root with lateral roots, Root on rhizome, Bud on lower portion of 3 m shoot, Bud on middle portion of 3 m shoot, Bud on top portion of 3 m shoot, Bud on rhizome, Rhizome) was downloaded and processed in tbtools using high throughput RNA sequencing (RNA-seq) from NCBI database (Zhao et al. 2018). The TPM values were log2 with (1 +) conversion and presented as a heatmap using TBtool software (Chen et al. 2020). And the transcriptome data of hormone treatment, including NAA and GA in moso bamboo, was also obtained and performed in the same method, but the heat map was visualized with the TPM value (Wang et al. 2017; Zhang et al. 2018). In addition, the corresponding SRA number is list in Table S8.
Plant Materials and Stress Treatments
Moso bamboo seeds were collected from the Tianmu Mountain National Nature Reserve in Zhejiang Province, China, and were grown in plastic containers (top diameter: 20 cm; bottom diameter:13 cm; height: 16 cm) in a greenhouse with the 16-h light/8-h dark cycle at 22 °C. To investigate the expression patterns of PheLBDs under stress and stress-related phytohormone treatments, three-month-old seedlings were sprayed with 0.1 mM abscisic acid (ABA, 500 ml) and 0.1 mM methyl jasmonate (Me-JA, 500 ml) (Liu et al. 2018). 20% PEG-6000 (500 ml) and 200 mM NaCl solution (500 ml) were used to simulate drought and salt stress, respectively (Chen et al. 2017). All samples were collected at 0, 1, 3, 6, 12, and 24 h from seedlings after treatment. And untreated leaves (0 h) were used as a control and stored in liquid nitrogen immediately and then stored at − 80 °C for RNA extraction.
Quantitative Real-time PCR (qRT-PCR) Analysis
The seedling RNA under abiotic stress and phytohormone treatments for qRT-PCR experiment was extracted from the plant samples using TRIzol reagent (Invitrogen, Ca, USA) in accordance with the instructions and was then reverse transcribed into cDNA using a PrimeScriptTM RT Reagent Kit (TaKaRa, Dalian, China). Gene-specific primers and TIP41 (tonoplast intrinsic protein 41) as an internal control (Fan et al. 2013) were designed by Primer Version 5.0 for RT-qPCR. Each sample for RT-qPCR with TransStart® Tip Green qPCR Super Mix (TransGen Biotech, Beijing, China) was repeated at least three times on a CFX96 Real-Time System (Bio-Rad). The qRT-PCR parameters were as followed: 94 °C for 30 s; 39 cycles of 94 °C for 5 s and 60 °C for 30 s, followed by a melting curve.
Subcellular Localization Analysis
The open reading frame without the stop codon of PheLBDs was cloned and sequenced by RT-PCR with gene-specific primers. Then, the correct PCR product was embedded in two restriction sites of the vector pCAMBIAI1305, which harbored a green fuorescent protein (GFP) sequence driven by 35S. The recombinant plasmid (PheLBD-GFP) and the empty control vector (GFP) were independently transferred into Agrobacterium tumefaciens cells GV3101 (Weidi, Shanghai, China). The corresponding Agrobacterium tumefaciens liquid was separately used to infect Nicotiana benthamiana cells with good growth condition. After 40 h, the leaves were observed with the LSM710 confocal laser scanning microscope (CarlZeiss).
Results
Identification and Phylogenetic Analysis of PheLBD Genes in Moso Bamboo
Sixty-one LBD genes were identified by BLASTP method in moso bamboo genome and were named from PheLBD1 to PheLBD61 based on the scaffold location. The detailed PheLBD characteristics, including the gene identifier, gene position, length of coding sequences (CDS) and the size of amino acids, pI (isoelectric point) and molecular weight (MW) were summarized in Table S1. The CDS length ranged from 288 bp (PheLBD15) to 1482 bp (PheLBD22) with an average length of 686 bp. Correspondingly, the amino acids varied from 95 (PheLBD15) to 493 aa (PheLBD22) with the protein molecular weight in range of 10.258 (PheLBD15) − 52.674 kDa (PheLBD22). The pI of LBD proteins was from 4.40 (PheLBD55) to 9.81 (PheLBD38). Consistently, 61 PheLBDs were all predicted to be located in the nucleus.
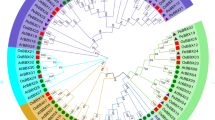
To elucidate the evolutionary relationships between 61 PheLBD proteins and other Poaceae species, we constructed a neighbor-joining (NJ) phylogenetic tree based on the alignment of 168 LBD protein sequences from moso bamboo (61), Brachypodium distachyo (28), Oryza sativa (35) and Zea mays (44). The phylogenetic tree showed that the predicted LBD proteins clustered into two groups, Class I and Class II, And Class I could be further divided into five (Class IA, IB, IC, ID and IE) and Class II has one subgroups in Fig. 1 and Fig. 2a, as described in the previous reports (Zhang et al. 2014). For moso bamboo, maize and Brachypodium distachyon, Class IB has the most LBD members, while class IE was the largest subfamily (10) in rice. Additionally, the LBD gene identifier from rice, maize and Brachypodium distachyon is listed in Table S2 and the results of multiple sequence alignment used for constructing evolutionary tree were shown in Attachment file 1 and file 2.
Phylogentic and distribution of LBD proteins from four plant species: Phyllostachys edulis, Brachypodium distachyon, Oryza sativa and Zea mays. a The phylogenetic tree was performed using the neighbor-joining method with a bootstrap analysis of 1000 replicates in MEGA6.0 from LBD protein sequences alignment by Clustal W software. b Statistics of LBD genes in each subfamily of Phyllostachys edulis, Brachypodium distachyon, Oryza sativa and Zea mays
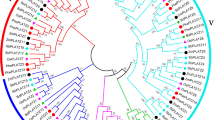
Phylogenetic relationships, gene structure and conserved motifs in 61 PheLBDs. a The phylogenetic tree was performed according to the full-length sequences of PheLBD protein using MEGA6.0 software. b Exon/intron structure of PheLBD genes. Yellow boxes represent exons, grey lines indicated introns and blue boxes represented untranslated 5′- and 3′-regions. c The motif composition of PheLBD proteins. Schematic representation of the 20 conserved motifs in PheLBD proteins. The motifs, numbered 1–20, were showed in different colored boxes. The detailed sequence information for each motif is provided in Table S3
Exon–intron Structure and Motif Compositions of PheLBD Genes
As shown Fig. 2b, the intro number varied from 0 to 5. Forty-six PheLBDs harbored one intro. Only two members (PheLBD41 and PheLBD43) harbored 4 intros and PheLBD2 contained 5 introns. Distinct exon–intron distribution patterns showed structural similarities and differences in the same branch. For instance, all PheLBDs in the Class II subfamily covered one intron and two exons. PheLBD43 in Class IE contained 4 introns with longest intron sequence, whereas three members (PheLBD20, PheLBD24 and PheLBD28) of this subfamily had no introns, indicating PheLBDs might have a high degree of divergence. Moreover, to further understand the compositions and diversification of motifs in the predicted PheLBD proteins, a total of 20 distinct motifs were identified and designated from motif 1 to motif 20 (Fig. 2c and Table S3). Motif 1 and 2 represented the LOB domains. Therefore, we also found almost all PheLBD members contained one of them or both except PheLBD23. Interestingly, the motif compositions in six subfamilies were not exactly the same. For example, in Class II subfamily, motif 1, 4, 2 and 6 existed in each member: PheLBD30 and PheLBD45 contained specifically motif 16, and motif 20 was unique to PheLBD31 and PheLBD52. These results showed the conservation and specificity of the PheLBD gene family in terms of gene structure and motif compositions.
Scaffold Location and Synteny Analysis of PheLBD Genes
A total of 61 PheLBDs were unevenly distributed on 19 moso bamboo genomic scaffolds except for scaffold 1, 2 8, 20 and 23 (Fig. 3a). Scaffold 14 and 19 both hold the most PheLBD members (9), followed by scaffold 15 (8) and scaffold 16 (7). For PheLBD gene evolution, 56 paralogous pairs (Phe-Phe) were detected, including 41 pairs of segmental duplication, 13 pairs of transposed duplication and 2 pairs of tandem duplication (Fig. 3a), suggesting segmental duplications and transposed duplications might be the main driving force of PheLBD gene family expansion. The Ks values of 26 pairs were concentrated on 0.1–0.4 and the corresponding divergence time was approximately 7.69–30.76 million years ago, which was very close to the timing of a putative whole-genome duplication event in moso bamboo (7–12 million years ago) (Peng et al. 2013). In addition, the Ka/Ks ratios of 20 pairs were greater than 0.5, of which 4 pairs were over 1 (Fig. 3b & Table S4). Regarding orthologous pairs, PheLBD genes had the most homologous gene pairs with the LBD genes of Zea mays (76), followed by Sorghum bicolor (63), Oryza sativa (58), Brachypodium distachyon (46), Arabidopsis thaliana (12) (Fig. 4). Meanwhile, we found some LBD genes on moso bamboo scaffold 5, 7, 9, 11, 13, 14, 15, 16, 18, 21 and 22 corresponded to two homologous genes on different chromosomes of Oryza sativa. This phenomenon was also observed in the collinear analysis between moso bamboo and Zea mays, Sorghum bicolor as well as Brachypodium distachyon. Moreover, the average values of Ks were 0.486 (Phe-Bd), 0.448 (Phe-Os), 0.480 (Phe-Sb) and 0.536 (Phe-Zm), which was basically in line with the divergence time between bamboo and the corresponding species (Peng et al. 2013). And the Ka/Ks ratios of all orthologous gene pairs among moso bamboo and other four Poaceae plants were all less than 1 (Table S5).
Scaffold location and gene duplication of PheLBD genes. a PheLBD genes were showed on 24 scaffolds and scaffold numbers were marked on the scaffold block. The paralogous pairs of tandem duplication were drawn by the orange frame, transposition duplication pairs were colored with green lines and segmental duplication pairs were connected with purple lines. b The Ka, Ks distribution of 56 paralogous pairs (Phe-Phe) was exhibited in a scatterplot
Synteny analysis of PheLBD genes between Phyllostachys edulis and other plant species (Arabidopsis thaliana, Brachypodium distachyon, Oryza sativa, Sorghum bicolor and Zea mays). Gray lines in the background represented the collinear blocks within the moso bamboo and other five model plant genomes, while the cyan lines highlighted the syntenic LBD gene pairs
Identification of Cis-acting Elements in Promoter Region of PheLBD Genes
The cis-regulatory elements in promoter region are closely related to gene expression and potential functions (Todeschini et al. 2014). To further investigate the regulatory function of PheLBDs, the cis-regulatory elements in promoter regions of plant growth and development and abiotic stress response were analyzed (Fig. 5 and Table S6). We found that light-responsive cis-acting elements (Sp1, AE-box, ATC-motif, Box 4, GA-motif, GATA-motif, G-box, GT1-motif, I-box and TCCC-motif) were the most common in plant growth and development, especially G-box, which appeared 15 times for PheLBD42 and 13 times for PheLBD22, followed by PheLBD30 and PheLBD37 (12). In addition, CAT-box related to meristem expression was identified 60 times in 35 PheLBDs. The cis-regulatory elements associated with seed-specific regulation (RY-element) and auxin responsiveness were also detected in 12 and 27 PheLBDs, respectively. In the second category, numerous hormone-related elements, including ABA response (ABRE), gibberellin response (TATC-box, P-box and GARE-motif), MeJA (TGACG-motif), salicylic acid (SA) response (TCA-element) were counted among 61 PheLBDs, of which ABRE and TGACG-motif involved in the MeJA-responsiveness ranked first and second in the total number of statistics, respectively. ABRE had the highest frequency (251 times) and PheLBD42 was the biggest holder (18). And TGACG-motif appeared 136 times in 53 PheLBD members. Moreover, the number of cis-acting regulatory elements related to abiotic stress, such as low-temperature responsiveness (LTR), defense and stress (TC-rich repeats), and drought stress (MYB, MYC and MBS) was very large, especially MYB and MYC with 475 and 227, respectively. To sum up, our findings suggested the number of cis-acting elements of light, hormones and stress response was quite high in PheLBDs (Table S6), implying they might play an important regulatory function in plant growth development and stress response.
Expression Profiles of PheLBDs in Various Tissues
To evaluate the expression patterns of PheLBD genes in various tissues, we used transcriptome data to found that a half of PheLBD genes were undetectable in expression, in accordance as the roles of transcription factor (Fig. 6). Notably, five genes (PheLBD22, PheLBD12, PheLBD42, PheLBD32 and PheLBD53) showed high expression levels in 14 moso bamboo organs. For bud on lower portion of 3 m shoot, eight PheLBDs displayed very high expression level (TPM > 100), such as PheLBD22, PheLBD12, PheLBD42, PheLBD32, PheLBD53, PheLBD36, PheLBD45 and PheLBD57. There were some genes that were highly expressed in leaf sheath, but cannot be detected in blade leaf and sheath sheet, including PheLBD45, PheLBD57, PheLBD35, PheLBD56, PheLBD1, PheLBD18, PheLBD15, PheLBD7, PheLBD10, PheLBD9 and PheLBD17. The phenomenon of tissue-specific expression was very common in other tissues, including in bud on lower/middle/top portion of 3 m shoot of PheLBD58, PheLBD30, PheLBD35, PheLBD56, PheLBD1, PheLBD18, PheLBD15, PheLBD7, PheLBD10, PheLBD9, PheLBD17 and PheLBD26. Overall, compared with other tissues, there were more highly expressed genes in leaf sheath, bud on lower, middle and top portion of 3 m shoot as well as bud on rhizome.
The expression analysis of PheLBD genes in 14 tissues: 0.1 cm root on shoot, 0.5 cm root on shoot, 2 cm root on shoot, 10 cm root on shoot, Blade leaf, Leaf sheath, Sheath sheet, New root with lateral roots, Root on rhizome, Bud on lower portion of 3 m shoot, Bud on middle portion of 3 m shoot, Bud on top portion of 3 m shoot, Bud on rhizome, Rhizome. The numerical value represented the TPM values. The legend was to show the relative high and low expression
Expression Profiles of Class II PheLBDs Under phytohormone Treatment
To show the position of regulatory elements related to stress and hormone more clearly, GSDS website was performed to exhibit the specific location in promoter region of 11 PheLBDs in Class II subgroup (Fig. 7a). Most cis-elements distributed during 1500 bp near the transcription start position, especially PheLBD22, PheLBD30, PheLBD31, PheLBD42 and PheLBD52. We found that ABRE was distributed in every gene, as were the elements of drought stress (MYB, MYC and MBS). TGACG-motif, related to MeJA stress, was detected in 10 PheLBDs except PheLBD13. Although the number of these elements of auxin responsiveness (TGA-element) and GA stress (P-box, GARE-motif) was very small among these 11 genes, we found these PheLBDs respond drastically to auxin NAA and GA treatments (Fig. 7b). All PheLBDs were upregulated after NAA treatment, especially PheLBD13, PheLBD31 and PheLBD53. On the contrary, most PheLBDs was downregulated under GA-treated, but the expression of PheLBD13 and PheLBD31 was rising. Moreover, we cloned the corresponding promoter sequences of the eight genes (PheLBD4, PheLBD12, PheLBD22, PheLBD30, PheLBD31, PheLBD42, PheLBD45 and PheLBD50) and sent them to the Biological company (Shenggong Bioengineering Co., Ltd) for sequencing to verify the correctness of these sequence (Fig. 7c).
Expression Levels of PheLBD Genes Under MeJA, ABA PEG6000 and NaCl Treatments
According to previous studies in other plants, members of Class I gene family are mainly involved in growth and development, while Class II family members were mainly participated in the process of stress response (Ariel et al. 2010; Liu et al. 2019b; Cao et al. 2016). Hence, the expression levels of 11 PheLBDs in Class II subfamily (PheLBD4, − 12, − 13, − 22, − 30, − 31, − 32, − 42, − 45, − 52 and − 53) were investigated using moso bamboo seedlings after MeJA, ABA, PEG6000 and salt treatments by qRT-PCR.
Under MeJA treatment, apart from PheLBD22, PheLBD45 and PheLBD53, the relative expression level of the remaining 8 PheLBDs was upregulated at 1 h, whereas PheLBD32, PheLBD42 and PheLBD52 decreased rapidly, then increased slightly at 12 h, and finally decreased at 24 h (Fig. 8a). For ABA treatment, PheLBD4, PheLBD12, PheLBD13, PheLBD45 and PheLBD53 were strongly up-regulated > tenfold at the peak. However, PheLBD30 and PheLBD31 was also upregulated at 1 h (~ fourfold), the expression level tended to be stable as the control (0 h) at the later time point. Strangely, PheLBD42 performed stably before 12 h, but its expression level dropped sharply at 24 h (Fig. 8b). Notably, PheLBD4, PheLBD12 and PheLBD52 were gradually up-regulated, whereas PheLBD22, PheLBD30, PheLBD42 and PheLBD53 were downregulated, especially the latter two members significantly decrease after PEG6000 treatment (Fig. 9a). In the NaCl treatment, the expression of PheLBD4, -22, -45, -52 and -53 presented a sustained downward, but the relative expression level of PheLBD12, PheLBD31, PheLBD32 and PheLBD42 was up-regulated (Fig. 9b).
qRT-PCR expression analysis of 11 selected PheLBD genes following MeJA (a) and ABA (b) treatments. Relative expression levels of PheLBD genes were examined by qRT-PCR. PheTIP41 was used as an internal reference gene. Y-axes represent the scale of the relative expression levels. X-axes indicated time courses of MeJA stress treatments for each gene. Bars represented the standard deviations (SD) of three biological replicates
qRT-PCR expression analysis of 11 selected PheLBD genes following PEG6000 (a) and NaCl (b) treatments. Relative expression levels of PheLBD genes were examined by qRT-PCR. PheTIP41 was used as an internal reference gene. Y-axes represented the scale of the relative expression levels. X-axes indicated time courses of MeJA stress treatments for each gene. Bars represented the standard deviations (SD) of three biological replicates
Subcellular Localization Analysis
It is reported that some LBD proteins in other plants were located in nucleus, such as ASL11/LBD15 (Sun et al. 2013), AtLBD30 (Liu et al. 2019a), CsLOB_3, CsLBD36_2, CsLBD41_2 (Zhang et al. 2019), and PbrLBD20 (Song et al. 2020). To assess the location of LBD proteins in moso bamboo, PheLBD12, PheLBD31 and PheLBD45 were constructed on the vector pCAMBIAI1305 which contained a GFP gene sequence. As shown in Fig. 10, the green fluorescent signal of the empty protein (35S::GFP) as the control group filled throughout the whole cell, while the other three PheLBD proteins (35S:: PheLBD::GFP) were detected to be localized in the nucleus act as transcription factors, consistent with the prediction of subcellular localization in Plant-mPLoc website.
Discussion
Gene Subfamily Division and Evolutionary Relationships
In this study, a neighbor-joining (N-J) tree was generated from the full length LBD proteins sequences of 61 PheLBDs, 28 BdLBDs, 36 OsLBDs and 44 ZmLBDs. This method was also applicable to the identification of LBD genes in maize, potato, apple and Chinese white pear (Zhang et al. 2014; Liu et al. 2019b; Wang et al. 2013; Song et al. 2020). However, the conserved amino acid sequences (LOB domain motif) were used for multiple sequence alignment and phylogenetic analysis, which were the traditional subfamily classification of LBDs in Arabidopsis thaliana and Brachypodium distachyon (Gombos et al. 2017; Matsumura et al. 2009). Even, LOB domain nucleotide sequences of OsLBD genes were used to construct phylogenetic trees using both NJ and ML methods (Yang et al. 2006). However, we found the LOB domain of PheLBDs was not completely following the previous domain sequence form: many LBD members harbored the C-motif (CX2CX6CX3C), GAS block and leucine-zipper-like motif, but it was difficult for most genes to have the complete sequences for the three motifs (Fig. S1). In addition, the values of Ka/Ks of 20 paralogous pairs (Phe-Phe) were over 0.5, of which 4 pairs were greater than 1, suggesting these PheLBDs might be moving forward the direction of positive selection (Cheng et al. 2015). These results reflected the LBD genes in Phyllostachys edulis might evolve very quickly.
In terms of the number of LBD genes, moso bamboo (61) was slightly more than that of Brachypodium distachyon (28), rice (36), and maize (44), Arabidopsis thaliana (43), apple (58) and Chinese white pear (60) (Gombos et al. 2017; Zhang et al. 2014; Matsumura et al. 2009; Wang et al. 2013; Song et al. 2020). And the segmental duplication (42 pairs) and transposed duplication (13 pairs) events might contribute to the expansion of PheLBDs in the evolution process, which was also the same evolutionary model for the roles of LBD gene family in maize (Zhang et al. 2014). Furthermore, moso bamboo has undergone two whole-genome duplication (WGD) events (Qiao et al. 2019). Coincidentally, a recent gene duplication event has also occurred in maize and apple. In Chinese white pear, 76.67% (44) genes of the PbrLBD gene family were duplicated and retained from WGD/segmental duplication types, followed by dispersed duplications (6, 10%), tandem duplication (6, 10%) (Song et al. 2020). Therefore, there is little difference in the number of LBDs in these species. And we speculate that the number of LBD genes may be closely related to the evolutionary pattern of family genes.
Tissue-specific Expression
Up to now, researchers have studied and reported many biological functions of LBD genes in many plants, including lateral root growth, secondary woody growth, pollen development, auxin-induced callus formation, pathogen response and the regulation of abiotic stress (Okushima et al. 2007; Yordanov and Regan, 2010; Kim et al. 2015; Xu et al. 2018; Thatcher et al. 2012; Ariel et al. 2010). From the tissue expression profiles, some genes presented the high expression level in 14 moso bamboo tissues, including PheLBD22, PheLBD12, PheLBD42, PheLBD32 and PheLBD53, which all belong to Class II subgroup (Fig. 6). Some were tissue-specifically expressed. For instance, the TPM value of PheLBD30 in bud on lower portion of 3 m shoot reached 45.77, while it was nearly no expression values in moso bamboo other tissues. But the transcripts of PheLBD13 were accumulated in other ten organs apart from 10 cm root on shoot and new root with lateral roots. Surprisingly, nearly half of the LBD genes were not expressed in these moso bamboo tissues, similar as that of Brachypodium distachyon, Solanum tuberosum and Camellia sinensis, which might be one of the characteristics of LBD transcription factor (Gombos et al. 2017; Liu et al. 2019b; Zhang et al. 2019). From the other hand, the motif compositions in the one of six subgroups were not exactly the same, some subgroup genes contained unique motif. For example, motif 16 was only found PheLBD30 and PheLBD45. The motif 19 was peculiar to PheLBD36 and PheLBD57, and we found the tissues profiles of the two genes were very similar, whose transcription levels were quite high in leaf sheath and bud on lower portion of 3 m shoot (Fig. 6).
For moso bamboo, the fast growth rate is its biggest characteristic, which mainly due to the moso bamboo shoots will grow into moso bamboo rhizome (Lan et al. 2020). The LBD genes with the relative high expression level in three shoots (bud on low/ middle/top portion of 3 m shoot) were from PheLBD52 to PheLBD29 in Fig. 6. Combined with the cis-acting element analysis, CAT-box, linked to meristem expression, PheLBD22 and PheLBD45 who also contained one CAT-box showed the high expression in moso bamboo shoots (Fig. 6 & Table S6). These results indicated Class II PheLBDs might participate in the rapid growth process of moso bamboo.
Expression Patterns and Potential Ffunctions
According to the cis-acting element analysis, we have drawn the specific locations of abiotic stress and hormone regulatory factors in the corresponding promoter region for Class II PheLBD genes (Fig. 7a). And we analyzed the expression level of these genes under the treatment of auxin NAA and GA using public transcriptome data in heat map (Fig. 7b). The results showed the 11 PheLBDs were induced to respond positively under NAA treatment, suggesting these PheLBDs might play an important role in the early growth and development of moso bamboo roots (Wang et al. 2017). It is reported ARF7 and ARF19 regulate lateral root (LR) formation by activating the expression of LBD16/ASL18 and LBD29/ASL16 in Arabidopsis (Okushima et al. 2007). Based on many reports, LBD18 has been proved to be closely related to the formation of lateral roots. For instance, LBD18/ASL20 along with LBD16 regulated the formation of LR (Lee et al. 2009a, 2009b); LBD18 and LBD33/ASL24 regulated the initiation of LRs through transcriptional activation of E2Fa transcription factor, thereby regulating asymmetric cell division (Berckmans et al. 2011). And LBD18/ASL20 could not only directly combine with the EXPANSINA14 (EXPA14) promoter to enhance the appearance of lateral roots, but also could upregulate EXPA17 to promote LR formation during the auxin response in Arabidopsis (Lee and Kim 2013). GA, one of the most important kinds of growth-promoting phytohormones, plays crucial roles in growth promotion and flower induction (Zhang et al. 2018). However, the Class II genes were sensitive to GA-treated. Therefore, we speculated that the growth and development of moso bamboo need the coordination and balance of multiple hormones.
In Arabidopsis thaliana, LBD20 transcripts were enriched in roots, which were further induced by Fusarium oxysporum inoculation or methyl jasmonate treatment. And LBD20 had been confirmed that it was sensitive to Fusarium oxysporum via jasmonate (JA) signaling (Thatcher et al. 2012). And most PheLBDs were up-regulated by MeJA treatment, which might be related to the biotic stress. Under ABA, PEG600 and NaCl treatment, the expression level of Class II genes was slightly different. For example, the three nuclear localized genes (PheLBD12, PheLBD31 and PheLBD45) were actively induced expect PheLBD45 was down regulated under NaCl-treated. In recent study, a subfamily II gene, SlLBD40 (Solyc02g085910) was located in the nucleus and highly expressed in roots. And its expression was significantly induced by PEG, salt and MeJA treatments, similar to PheLBD12 and PheLBD31. Most importantly, SlLBD40 had been confirmed that it was a negative regulator of drought tolerance through its overexpressing and knockout transgenic tomato plants (Liu et al. 2020). In Arabidopsis, LBD15 could directly bind to the promoter of the ABI4 to activate its expression to optimally regulate ABA signaling-mediated plant growth and the tolerance of water-deficit (Guo et al. 2020). Therefore, we speculated that Class II PheLBDs in moso bamboo, especially PheLBD12 and PheLBD31, might play the important regulation function under biotic and abiotic stresses, which still required follow-up experiments to prove their specific functions.
Conclusion
In this study, 61 PheLBDs were identified in moso bamboo genome and classified into two classes (Class I and Class II), and Class I was further divided into five subclasses (Class IA, IB, IC, ID and IE) according to phylogenetic analyses and the previous reports. Subsequently, we performed bioinformatics analysis on the family genes, including exon–intron structure, conserved motifs, gene distribution on scaffolds, gene collinearity, cis-acting elements, tissue expression patterns, the expression level in different hormone and abiotic stress treatments as well as the subcellular localization analysis of three Class II genes. Our experimental findings suggest some PheLBDs might play roles in growth and development as well as environmental stresses response and adaptation in moso bamboo. The identification of LBD genes in moso bamboo provides a useful reference for further studies on the biological functions and related pathways and mechanisms of this gene family.
References
Ariel FD, Diet A, Crespi M, Chan RL (2010) The LOB-like transcription factor Mt LBD1 controls Medicago truncatula root architecture under salt stress. Plant Signal Behav 5(12):1666–1668
Bailey TL, Nadya W, Chris M, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373
Berckmans B, Vassileva V, Schmid SPC, Maes S, Parizot B, Naramoto S, Magyar Z, Kamei CLA, Koncz C, Bogre L, Persiau G, De Jaeger G, Friml J, Simon R, Beeckman T, De Veylder L (2011) Auxin-dependent cell cycle reactivation through transcriptional regulation of arabidopsis E2Fa by lateral organ boundary proteins. Plant Cell 23(10):3671–3683. https://doi.org/10.1105/tpc.111.088377
Cannon SB, Mitra A, Baumgarten A, Young ND, May G (2004) The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol 4:10
Cao H, Liu CY, Liu CX, Zhao YL, Xu RR (2016) Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J Genet 95(3):515–526. https://doi.org/10.1007/s12041-016-0660-z
Chen DM, Chen Z, Wu M, Wang Y, Wang YJ, Yan HW, Xiang Y (2017) Genome-wide identification and expression analysis of the HD-Zip gene family in moso bamboo (Phyllostachys edulis). J Plant Growth Regul 36(2):323–337. https://doi.org/10.1007/s00344-016-9642-x
Chen C, Chen H, Zhang Y, Thomas HR, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13(8):1194–1202. https://doi.org/10.1016/j.molp.2020.06.009
Cheng T, Fu B, Wu Y, Long R, Xia Q (2015) Transcriptome Sequencing and Positive Selected Genes Analysis of Bombyx mandarina. PLoS ONE 10(3):e0122837
Chou KC, Shen HB (2010) Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 5(6):e11335
Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, Eddy SR, Sonnhammer ELL, Bateman A (2006) Pfam: clans, web tools and services. Nucleic Acids Res 34:D247–D251. https://doi.org/10.1093/nar/gkj149
Finn RD, Penelope C, Eberhardt RY, Eddy SR, Jaina M, Mitchell AL, Potter SC, Marco P, Matloob Q, Amaia SV (2016) The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res D1:D279–D285
Gao Y, Liu H, Zhang K, Li F, Xiang Y (2020) A moso bamboo transcription factor, Phehdz1, positively regulates the drought stress response of transgenic rice. Plant Cell Reports 40(1):187–204. https://doi.org/10.1007/s00299-020-02625-w
Ge X, Zhou B, Wang X, Li Q, Cao Y, Gu L (2018) Imposed drought effects on carbon storage of moso bamboo ecosystem in southeast China: results from a field experiment. Ecol Res 33:393–402
Gombos M, Zombori Z, Szecsenyi M, Sandor G, Kovacs H, Gyorgyey J (2017) Characterization of the LBD gene family in Brachypodium: a phylogenetic and transcriptional study. Plant Cell Rep 36(1):61–79. https://doi.org/10.1007/s00299-016-2057-0
Gong Z, Xiong L, Shi H, Yang S, Zhu JK (2020) Plant abiotic stress response and nutrient use efficiency. Sci China Life Sci 63(5):635–674. https://doi.org/10.1007/s11427-020-1683-x
Guo AY, Zhu QH, Chen X, Luo JC (2007) GSDS: a gene structure display server. Hereditas 29(8):1023–1026
Guo Z, Xu H, Lei Q, Du J, Li C, Wang C, Yang Y, Yang Y, Sun X (2020) The Arabidopsis transcription factor LBD15 mediates ABA signaling and tolerance of water-deficit stress by regulating ABI4 expression. Plant J 104(2):510–521. https://doi.org/10.1111/tpj.14942
Huang X, Yan H, Liu Y, Yi Y (2020) Genome-wide analysis of lateral organ boundaries domain-in Physcomitrella patens and stress responses. Genes & Genomics 42(6):651–662. https://doi.org/10.1007/s13258-020-00931-x
Iwakawa H, Ueno Y, Semiarti E, Onouchi H, Kojima S, Tsukaya H, Hasebe M, Soma T, Ikezaki M, Machida C, Machida Y (2002) The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol 43(5):467–478. https://doi.org/10.1093/pcp/pcf077
Iwakawa H, Iwasaki M, Kojima S, Ueno Y, Soma T, Tanaka H, Semiarti E, Machida Y, Machida C (2007) Expression of the ASYMMETRIC LEAVES2 gene in the adaxial domain of Arabidopsis leaves represses cell proliferation in this domain and is critical for the development of properly expanded leaves. Plant J 51(2):173–184. https://doi.org/10.1111/j.1365-313X.2007.03132.x
Kim MJ, Kim M, Lee MR, Park SK, Kim J (2015) LATERAL ORGAN BOUNDARIES DOMAIN (LBD) 10 interacts with SIDECAR POLLEN/LBD27 to control pollen development in Arabidopsis. Plant J 81(5):794–809. https://doi.org/10.1111/tpj.12767
Lan YG, Wu L, Wu M, Liu HL, Gao YM, Zhang KM (2020) Xiang Y Transcriptome analysis reveals key genes regulating signaling and metabolic pathways during the growth of moso bamboo (Phyllostachys edulis) shoots. Physiol Plant. https://doi.org/10.1111/ppl.13296
Lee HW, Kim J (2013) EXPANSINA17 Up-Regulated by LBD18/ASL20 promotes lateral root formation during the auxin response. Plant Cell Physiol 54(10):1600–1611. https://doi.org/10.1093/pcp/pct105
Lee DJ, Park JW, Lee HW, Kim J (2009a) Genome-wide analysis of the auxin-responsive transcriptome downstream of iaa1 and its expression analysis reveal the diversity and complexity of auxin-regulated gene expression. J Exp Bot 60(13):3935–3957. https://doi.org/10.1093/jxb/erp230
Lee HW, Kim NY, Lee DJ, Kim J (2009b) LBD18/ASL20 regulates lateral root formation in combination with LBD16/ASL18 downstream of ARF7 and ARF19 in arabidopsis. Plant Physiol 151(3):1377–1389. https://doi.org/10.1104/pp.109.143685
Lescot M (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30(1):325–327
Liao S, Qin X, Luo L, Han Y, Wang X, Usman B, Nawaz G, Zhao N, Liu Y, Li R (2019) CRISPR/Cas9-induced mutagenesis of semi-rolled leaf 1,2 confers curled leaf phenotype and drought tolerance by influencing protein expression patterns and ROS scavenging in rice (Oryza sativa L.). Agronomy-Basel. https://doi.org/10.3390/agronomy9110728
Liu HL, Ww M, Li F, Gao YM, Chen F, Xiang Y (2018) TCP transcription factors in moso bamboo (Phyllostachys edulis): genome-wide identification and expression analysis. Front Plant Sci. https://doi.org/10.3389/fpls.2018.01263
Liu H, Cao M, Chen X, Ye M, Zhao P, Nan Y, Li W, Zhang C, Kong L, Kong N, Yang C, Chen Y, Wang D, Chen Q (2019b) Genome-wide analysis of the lateral organ boundaries domain (LBD) gene family in solanum tuberosum. Int J Mol Sci. https://doi.org/10.3390/ijms20215360
Liu C, Yu H, Li LG (2019a) SUMO modification of LBD30 by SIZ1 regulates secondary cell wall formation in Arabidopsis thaliana. PLoS Genet. https://doi.org/10.1371/journal.pgen.1007928
Liu L, Zhang J, Xu J, Li Y, Guo L, Wang Z, Zhang X, Zhao B, Guo Y-D, Zhang N (2020) CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. https://doi.org/10.1016/j.plantsci.2020.110683
Majer C, Hochholdinger F (2011) Defining the boundaries: structure and function of LOB domain proteins. Trends Plant Sci 16(1):47–52. https://doi.org/10.1016/j.tplants.2010.09.009
Marchler-Bauer A, Bo Y, Han LY, He JE, Lanczycki CJ, Lu SN, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lu F, Marchler GH, Song JS, Thanki N, Wang ZX, Yamashita RA, Zhang DC, Zheng CJ, Geer LY, Bryant SH (2017) CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1):D200–D203. https://doi.org/10.1093/nar/gkw1129
Matsumura Y, Iwakawa H, Machida Y, Machida C (2009) Characterization of genes in the ASYMMETRIC LEAVES2/LATERAL ORGAN BOUNDARIES (AS2/LOB) family in Arabidopsis thaliana, and functional and molecular comparisons between AS2 and other family members. Plant J 58(3):525–537. https://doi.org/10.1111/j.1365-313X.2009.03797.x
Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D (2005) Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17(2):444–463
Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19(1):118–130. https://doi.org/10.1105/tpc.106.047761
Peng Z, Lu Y, Li L, Zhao Q, Feng Q, Gao Z, Lu H, Hu T, Yao N, Liu K, Li Y, Fan D, Guo Y, Li W, Lu Y, Weng Q, Zhou C, Zhang L, Huang T, Zhao Y, Zhu C, Liu X, Yang X, Wang T, Miao K, Zhuang C, Cao X, Tang W, Liu G, Liu Y, Chen J, Liu Z, Yuan L, Liu Z, Huang X, Lu T, Fei B, Ning Z, Han B, Jiang Z (2013) The draft genome of the fast-growing non-timber forest species moso bamboo (Phyllostachys heterocycla). Nat Genet 45(4):456–461. https://doi.org/10.1038/ng.2569
Qiao X, Li QH, Yin H, Qi KJ, Li LT, Wang RZ, Zhang SL, Paterson AH (2019) Gene duplication and evolution in recurring polyploidization-diploidization cycles in plants. Genome Biol. https://doi.org/10.1186/s13059-019-1650-2
Qi-Jiang HE, Wang KH, Weng PJ, Rong WU (2001) Preliminary report on experiment of watering bamboo stand for shoot. J Zhejiang Forest Sci Technol 2001(05):18–20
Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR (2009) Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in arabidopsis. Plant Cell 21(11):3567–3584
Semiarti E, Ueno Y, Tsukaya H, Iwakawa H, Machida C, Machida Y (2001) The asymmetric leaves2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 128(10):1771–1783
Shuai B, Reynaga-Pena CG, Springer PS (2002) The LATERAL ORGAN BOUNDARIES gene defines a novel, plant-specific gene family. Plant Physiol 129(2):747–761. https://doi.org/10.1104/pp.010926
Song BB, Tang ZK, Li XL, Li JM, Zhang MY, Zhao KJ, Liu HN, Zhang SL, Wu J (2020) Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC Genomics. https://doi.org/10.1186/s12864-020-06999-9
Sun XD, Feng ZH, Meng LS, Zhu J, Geitmann A (2013) Arabidopsis ASL11/LBD15 is involved in shoot apical meristem development and regulates WUS expression. Planta 237(5):1367–1378. https://doi.org/10.1007/s00425-013-1844-x
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Thatcher LF, Powell JJ, Aitken EAB, Kazan K, Manners JM (2012) The lateral organ boundaries domain transcription factor LBD20 functions in fusarium wilt susceptibility and jasmonate signaling in arabidopsis. Plant Physiol 160(1):407–418. https://doi.org/10.1104/pp.112.199067
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):1673–1680
Wang Y, Tang H, Debarry JD, Tan X, Li J, Wang X, Tae-Ho L, Jin H, Barry M, Guo H (2012) MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res 40(7):e49–e49
Wang X, Zhang S, Su L, Liu X, Hao Y (2013) A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES Domain) gene family in malus domestica with a functional characterization of MdLBD11. Plos One 8(2):e57044. https://doi.org/10.1371/journal.pone.0057044
Wang W, Gu L, Ye S, Zhang H, Cai C, Xiang M, Gao Y, Wang Q, Lin C, Zhu Q (2017) Genome-wide analysis and transcriptomic profiling of the auxin biosynthesis, transport and signaling family genes in moso bamboo (Phyllostachys heterocycla). BMC Genomics 18(1):870
Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Hochstrasser DF (1999) Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol Biol 112(112):531–552
Xu C, Cao HF, Zhang QQ, Wang HZ, Xin W, Xu EJ, Zhang SQ, Yu RX, Yu DX, Hu YX (2018) Control of auxin-induced callus formation by bZIP59-LBD complex in Arabidopsis regeneration. Nature Plants 4(2):108–115. https://doi.org/10.1038/s41477-017-0095-4
Yang Y, Yu XB, Wu P (2006) Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol Phylogenet Evol 39(1):248–262. https://doi.org/10.1016/j.ympev.2005.09.016
Yang TQ, Fang GY, He H, Chen JH (2016) Genome-wide identification, evolutionary analysis and expression profiles of LATERAL ORGAN BOUNDARIES DOMAIN gene family in Lotus japonicus and Medicago truncatula. Plos One. 11(8):e0161901. https://doi.org/10.1371/journal.pone.0161901
Yoko Okushima HF, Onoda M, Theologis A, Tasaka M (2007) ARF7 and ARF19 Regulate Lateral Root Formation via Direct Activation of LBD/ASL Genes in Arabidopsis. Plant Cell 19(1):118–130
Yordanov YS (2010) Regan, Busov (2010) Members of the LATERAL ORGAN BOUNDARIES DOMAIN transcription factor family are involved in the regulation of secondary growth in populus. Plant Cell 22(11):3662–3677
Zhang YM, Zhang SZ, Zheng CC (2014) Genomewide analysis of LATERAL ORGAN BOUNDARIES Domain gene family in Zea mays. J Genet 93(1):79–91. https://doi.org/10.1007/s12041-014-0342-7
Zhang H, Wang H, Qiang Z, Gao Y, Wang H, Zhao L, Wang Y, Xi F, Wang W, Yang Y (2018) Transcriptome characterization of moso bamboo (Phyllostachys edulis) seedlings in response to exogenous gibberellin applications. BMC Plant Biol 18(1):125
Zhang X, He Y, He W, Su H, Xu P (2019) Structural and functional insights into the LBD family involved in abiotic stress and flavonoid synthases in Camellia sinensis. Sci Rep 9(1):15651. https://doi.org/10.1038/s41598-019-52027-6
Zhao H, Gao Z, Wang L, Wang J, Wang S, Fei B, Chen C, Shi C, Liu X, Zhang H, Lou Y, Chen L, Sun H, Zhou X, Wang S, Zhang C, Xu H, Li L, Yang Y, Wei Y, Yang W, Gao Q, Yang H, Zhao S, Jiang Z (2018) Chromosome-level reference genome and alternative splicing atlas of moso bamboo (Phyllostachys edulis). Gigascience. https://doi.org/10.1093/gigascience/giy115
Zhao J, Gao P, Li C, Lin X, Guo X, Liu S (2019) PhePEBP family genes regulated by plant hormones and drought are associated with the activation of lateral buds and seedling growth in Phyllostachys edulis. Tree Physiol 39:1387–1404
Zhao H, Zhao S, Fei B, Liu H, Yang H, Dai H, Wang D, Jin W, Tang F, Gao Q, Xun H, Wang Y, Qi L, Yue X, Lin S, Gu L, Li L, Zhu T, Wei Q, Su Z, Wan TBWA, Ofori DA, Muthike GM, Mengesha YM, de Castro E Silva RM, Beraldo AL, Gao Z, Liu X, Jiang Z, International Network for B, Rattan (2017) Announcing the Genome Atlas of Bamboo and Rattan (GABR) project: promoting research in evolution and in economically and ecologically beneficial plants. GigaScience 6(7):1–7. https://doi.org/10.1093/gigascience/gix046
Zhu QH, Guo AY, Gao G, Zhong YF, Xu M, Huang MR, Luo JC (2007) DPTF: a database of poplar transcription factors. Bioinformatics 23(10):1307–1308. https://doi.org/10.1093/bioinformatics/btm113
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31670672), Postdoctoral Science Foundation of China (2019M652166), the Graduate Innovation Fund from Anhui Agricultural University (2020ysj-55) and Project of introducing and stabilizing talents in Anhui Agricultural University (yj2019-10).
Author information
Authors and Affiliations
Contributions
YMG participated in the revision of the manuscript, KW wrote the draft manuscript and conceived main frame of this study, RJW processed the experimental data, LNW and HXL designed and performed experiments, MW assisted to complete the writing of this paper, YX, the correspondence author, provided financial support for the article and designed the way and frame of this research. All authors read and approved the revised manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors have no conflicts of interest to declare.
Additional information
Handling Editor: Václav Motyka.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Gao, Y., Wang, K., Wang, R. et al. Identification and Expression Analysis of LBD Genes in Moso Bamboo (Phyllostachys edulis). J Plant Growth Regul 41, 2798–2817 (2022). https://doi.org/10.1007/s00344-021-10475-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10475-3