Abstract
Oral cancer is common cancer that appears in the mouth, posing a significant threat to public health due to its high mortality rate. Oral Squamous Cell Carcinoma (OSCC) is the most prevalent type of oral cancer, accounting for most cases, and it holds the seventh position among all types of cancers worldwide. Detecting OSCC early on is crucial to increase the chances of successful treatment and improve patients' survival rates. However, traditional diagnosis methods such as biopsy, where small tissue samples are extracted from the affected area and tested under a microscope, are time-consuming and require expert analysis. Moreover, due to the heterogeneity of OSCC, accurate diagnosis is challenging, and there is a need for alternative approaches to enhance the detection result of OSCC images. Therefore, this work develops two new approaches for segmenting and identifying OSCC with deep learning techniques named Mask Mean Shift CNN, named MMShift-CNN. The proposed MMShift-CNN approach attained the highest results in segmenting the OSCC region from the input image by retrieving color, texture, and shape features. The novel proposed method attained better performance with accuracy, F-measure, MSE, precision, sensitivity, and specificity of 0.9883, 0.9883, 0.0117, 0.999, 0.9867, and 0.99, respectively. These results reveal the efficiency of the proposed approach in accurately detecting oral cancer and potentially improving the efficiency of oral cancer diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Oral cancer is a prevalent form of cancer that affects the mouth, tongue, and throat, and can have reduced a patient's lifetime. OSCC is the predominant subtype of oral cancer, accounting for more than 90% of all cases (Du et al. 2020; Li et al. 2022; Warnakulasuriya and Greenspan 2020). Experts cannot accurately predict OSCC due to the absence of specific clinical vital signs, which is a main danger related with this type of disease. Early detection of OSCC is crucial for improving patient outcomes, as the prognosis is often better when the cancer is detected at an early stage (Perdomo et al. 2016). Nonetheless, there are various indicators that can be used to predict OSCC, including the lesion's location within the mouth, its size, colour, appearance, as well as the patient's history of tobacco and alcohol use. OSCC is typically diagnosed through a combination of clinical examination, imaging studies, and biopsy (Anwar et al. 2020). However, this approach is both time-consuming and labour-intensive, requiring a high level of expertise, and is prone to errors. Moreover, the accuracy of these diagnostic methods can be limited, and there is a need for more reliable and accurate methods of detecting OSCC (Chakraborty et al. 2019; Eckert et al. 2020; Sivakumar et al. 2023). Deep learning techniques have become a potential method for OSCC identification in current era (Deif and Hammam 2020; Kong et al. 2009; Santana and Ferreira 2017). Artificial neural networks are used in deep learning to evaluate enormous datasets and find patterns or features that are pertinent to a particular task. Deep learning may be employed to evaluate OSCC photos and find features that are specific to OSCC in the case of OSCC detection (Simla et al. 2023; Altaf et al. 2019; Deif et al. 2021). Deep learning algorithms have the benefit of being trained on big datasets, which can increase the model's accuracy (Leo et al. 2023; Duggento et al. 2021). Large datasets of OSCC photos are now accessible for OSCC identification, and these datasets can be utilised to train deep learning models. For instance, more than 10,000 photos of OSCC lesions are available in the Oral Cancer Imaging Database (OCID), which can be used to train and evaluate deep learning models (Wang et al. 2019; Goldenberg et al. 2019).
There are several deep learning approaches that have been used for OSCC detection, including convolutional neural networks (CNNs), recurrent neural networks (RNNs), and deep belief networks (DBNs). CNNs are a popular choice for OSCC detection, as they are well-suited for image analysis tasks. CNNs work by applying convolutional filters to the input image and identifying local features, which are then combined to identify more complex features. RNNs and DBNs are also well-suited for OSCC detection, as they can capture temporal or contextual information that may be relevant to the diagnosis. Among several deep learning models, CNN is widely recognized as one of the very effective deep learning approaches for histology diagnosis. These models are trained by analysing the features of all trial image to those stored in the training data, allowing them to learn the unique features of each disease. However, the accuracy of CNN models may be influenced by factors such as image noise, insufficient or imbalanced images in datasets, the quantity and type of layers used, and the choice of activation function. Therefore, the aim of this paper was to examine these challenges and propose solutions to improve the accuracy of CNN models in diagnosing OSCC, which is crucial for early detection.
Overall, deep learning approaches which are discussed in this section, have shown great promise for OSCC detection using OSCC images. The availability of large datasets, advances in deep learning techniques, and the development of pre-processing methods have enabled the development of accurate and reliable OSCC detection models. However, there are still challenges that need to be addressed, such as the need for more diverse datasets and more research on the interpretability of the models. Addressing these challenges will require collaboration robust and reliable deep learning methods for OSCC identification and classification. In CNNs, neurons are scalar and additive, lacking any spatial relationships with neighbouring neurons within the kernel of the previous layer. However, the max pooling process can cause a loss of valuable information and fails to capture the relative spatial relationships between features. As a result, CNNs are not able to maintain invariance when presented with significant transformations in input data. So, this paper introduced two novel approaches for segmenting oral cancer and classifying oral cancer types based on deep learning model.
The following is a summary of this paper's significant contributions:
-
(i)
The pre-processing mechanism is used in this paper to improve efficiency of oral cancer detection and identification.
-
(ii)
A fast unsupervised improved CNN approach, named MMShift-CNN is developed for oral cancer segmentation.
-
(iii)
To classify the oral cancer, Support Vector Onion Network, named SV-OnionNet is used and the hyper-parameters of SV-OnionNet is trained by adaptive Coati optimization algorithm.
This paper is organised as follows: a summary of earlier studies is provided in Sect. 2. The methods used for histological image segmentation and diagnosis of OSCC are examined in Sect. 3 of this article. In Sect. 4, the experimental results of the newly developed approaches are addressed along with a comparison and explanation of the methodology adopted in this work. Finally, Sect. 5 provides the conclusion and future enhancement of this paper.
2 Literature review
The most pertinent research in the literature is critically reviewed in this section, with an emphasis on the trends and difficulties in OSCC diagnosis.
Leo and Kalapalatha Reddy (2021) identified keratin pearls in oral histopathology photos using CNN and RF. With 96.88% classification rate, the Random Forest method properly recognized keratin pearls, while the CNN model correctly segmented keratin areas with 98.05 percent accuracy. In order to categorize oral biopsy images using Broder's histological grading system, Das et al. employed DL. Another option was CNN, which had a classification value of 97.5% (Das et al. 2020). Active Learning (AL) was found to be 3.26% more accurate than Random Learning (RL) when oral cancer images were divided into 7 types by Folmsbee et al. using CNN (Folmsbee et al. 2018). Additionally, Martino et al. used a variety of deep learning models, such SegNet, and U-Net with encoder, to partition oral pictures into 3 groups. A deep learning model, like enhanced U-Net through ResNet50 as an encoder, has been exposed to be further perfect over the traditional U-Net (Martino et al. 2020).
Jubair et al. (2022) used a dataset of oral cancer images and developed a new deep convolutional neural network (DCNN) architecture called the Light-Weight Deep Convolutional Neural Network (LWDCNN). LWDCNN consists of 6 convolutional layers and 4 fully connected layers and was designed to be more efficient and faster than other DCNNs while maintaining high accuracy. The LWDCNN was trained using transfer learning, where pre-trained models were used as a starting point for training the new model.
Deif et al. (2022) applied a hybrid feature selection approach that combines a filter with wrapper techniques. The filter approach selected the most relevant features based on statistical analysis and correlation coefficients, while the wrapper method used a genetic algorithm to select the best subset of features. SVM classifier was trained using the chosen features categorizes the histology of colorectal cancer. Ghosh et al. (2022) developed a new deep-reinforced neural network (DRNN) model for oral cancer risk prediction. The DRNN model combines deep learning with reinforcement learning, where the model learns to select the most relevant features for prediction while receiving rewards for accurate predictions. The DRNN model was trained using a combination of cell images and cyto-spectroscopic data, which provided complementary information for more accurate prediction.
The concatenated model developed by Amin et al. (2021) combines the outputs of multiple pre-trained CNNs to enhance the performance of OSCC classification. It was trained by transfer learning, where the pre-trained CNNs were fine-tuned using the histopathological image dataset. Musulin et al. (2021) used a combination of CNNs and a conditional random field model, to accurately grade OSCC and segment epithelial and stromal tissue. This model was trained using a huge dataset of histopathological pictures with annotations for OSCC grading and tissue segmentation.
3 Methodology
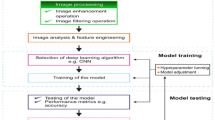
The objective of the this paper is to classify oral cancer images as either OSCC or normal. The block diagram of proposed approach is displayed in Fig. 1. The proposed oral cancer detection model comprises three main phases, namely image pre-processing, segmenting oral cancer region and oral cancer detection. Initially, the input image undergoes pre-processing to enhance contrast and reduce noise. After that, an innovative deep learning-based segmentation method is performed for fast and accurate oral cancer segmentation. The final stage involves deep feature extraction and classification of the trial images using adaptive COA to determine whether the image belongs to OSCC or normal. The Adaptive Coati Deep Convolutional Neural Network (ACDCNN) exhibits notable performance in accurately classifying oral cancer histopathological images when compared to existing diagnostic methods commonly employed in clinical practice. This study indicates that ACDCNN leverages its deep learning capabilities to discern intricate patterns and features within the images, leading to a heightened level of accuracy in classification tasks.
This efficacy is particularly evident when benchmarked against conventional diagnostic methods, which often rely on more manual and subjective interpretations. The ACDCNN's ability to extract relevant information from images results in improved sensitivity and specificity, ultimately contributing to enhanced diagnostic outcomes for oral cancer.
3.1 Input image
The oral image dataset is considered for performing the classification process, which is expressed by,
where, \(T_{q}\) implies whole number of images, and \(T_{r}\) represents \(r^{th}\) image in a dataset, which is passed to pre-processing phase.
3.2 Pre-processing
The main purpose of pre-processing process is to enhance the quality of input images as well as reduces noise and artifacts from input image. Here, gaussian filter is applied to input image \(T_{r}\) for removing noises and improving image quality. Gaussian filter improves the ability for providing similar transition during frequency domain. Moreover, it produces smother transition eradication of redundant data from input image, which is denoted by,
where, \(\psi\) refers standard deviation of distribution, \(T_{r}\) signifies input image and output of pre-processing by means of gaussian filter is denoted as \(R_{r}\).
3.3 Data augmentation
The images of normal cell and OSCC instances are not comprehensive and hence insufficient for generalization deep learning models for classification. Once pre-processing is completed, data augmentation is carried out for enlarging the data size and it avoids over-fitting issues. As a result, data augmentation is used to increase the quantity of images, which will solve the overfitting problem by giving the deep learning models intense generalization during testing. For each type of dataset, which involves modifying images using a variety of approaches, the data augmentation makes it possible to produce a much greater quantity of training data. In this study, data augmentation includes rotating, flipping, resizing, cropping, adding noise, and altering contrast of the original image. The data augmentation outcome is denoted as \(H_{r}\) and it is passed to segmentation process.
3.4 Segmentation using MMShift-CNN
Segmentation process is more imperative for identifying the cancer regions from augmented image. It accurately identifying the tumour region based on their visual characteristics, such as colour, texture, shape, or intensity. Thus, a novel MMShift-CNN model is designed for performing segmentation. Since, it is unsupervised CNN, the devised deep learning approach quickly segments the oral cancer image. This network does not require pre-trained model and it comprises of four layers, such as input, masking, convolution, and segmentation layer.
3.4.1 Input layer
This layer considers the data augmented output \(H_{r}\) as input for training the network and the following layer is masking.
3.4.2 Masking layer
This layer effectively speeds up the segmentation process and it is employed to roughly segment the image, which extracts the ROI region. To perform ROI masking operation, thresholding method is enabled. Masked ROI from the oral image is carried out by thresholding pixel intensities and provide initial \({ROI}_{init}\) is described as,
After that, the qualitative variations between the original image and the \({ROI}_{init}\) is observed based on mean distance value to provide relative thresholding at \({th}_{ROI}<{m}_{d}\), which will separate the OSSC and normal epithelium of the oral cavity regions in an image.
3.4.3 Convolution layer
The above masking section is incorporated with convolution layer, which is composed of 256 filters along with 6 kernel size and it utilizes scalars for extracting deep feature map. It is given by,
3.4.4 Segmenting layer
This layer is final one and it is employed for generating binary segmentation. This layer is carried out by mean shift clustering approach (Deng et al. 2015). The basic idea behind mean shift clustering is to iteratively shift the location of each pixel towards the centre of its neighbouring pixels that have similar features, until it converges to a stable region or mode.
Let \(X\) be the set of pixels or regions in the image, and let \(xi\) be a specific pixel or region in \(X\). The mean shift algorithm can be defined as follows,
Initially, choose a kernel function K that defines the similarity between neighbouring pixels or regions. The kernel function should be positive and symmetric, and should decay as the distance between pixels or regions increases. A common choice is the gaussian kernel is given as,
where \(\Vert k-l\Vert\) is the Euclidean distance among k and l, and \(\partial\) is the bandwidth value that maintains the width of the W. Then select a set of initial seed points or regions R within K and for each seed point or region s in S, iteratively update its location using the mean shift vector, until convergence. It is described as,
where Q(k) is the set of neighbouring pixels or regions of k within d, and \(Q\left(k\right)\) includes k itself. The weight of each neighbour y is given by the kernel function \(W\left(k,l\right)W\left(k,l\right)\), and the mean shift vector \(P\left(k\right),\) represents the degree and magnitude of the shift that will maximize the kernel density function. Finally, stop iterating when the mean shift vector is smaller than a threshold value, indicating convergence to a stable mode or region. The layer information of proposed MMShift-CNN model is shown in below Table 1. The output of segmentation process using novel MMShift-CNN is denoted as \(V_{r}\). The overall framework of MMShift-CNN segmentation model is displayed in Fig. 2.
3.5 Classification using adaptive coati optimization algorithm
The segmentation output \(V_{r}\) is considered as input for classification process in which SV-OnionNet is used for classification of oral cancer. Furthermore, the hyperparameters employed in SV-OnionNet is trained by means of proposed adaptive COA, which enhances the classification performance.
3.5.1 SV-OnionNet structure
In CNNs, neurons are sum the input values from the preceding layer to produce a single output value, lacking any spatial relationships with neighboring neurons inside the kernel of the preceding layer. However, the max pooling process can cause a loss of valuable information and fails to capture the relative spatial relationships between features. As a result, CNNs are not able to maintain invariance when presented with significant transformations in input data and replace the fully connected layer by SVM to improve the efficiency of the OnionNet. To overcome this drawback, this work proposed deep learning network called SV-OnionNet. Within this innovative network, the neuron-level information includes spatial relationships with neighboring neurons within the kernel of the previous layer. Ther diagram of Onionnet is displayed in Fig. 3. It contains three layers, namely input primary onion, optimization and fully connected layer.
3.5.1.1 Input layer
This layer taken the segmented image output \(V_{r}\) as input to SV-OnionNet model for performing classification of oral cancer.
3.5.1.2 Primary onion layer
The sucessive layer is primary onion layer, which is composed of 256 filters with a kernel size of 6 that use scalars to extract the deep feature map. This is given by,
Zero padding method is used in this layer to preserve the size of the input features, while the RLU is utilized as the non-linear activation function. The RLU layer retains positive values and sets negative values to zero using Eq. (15).
3.5.1.3 Optimization layer
In SV-OnionNet max-pool layers are removed, SV-because of max-pool layer, CNN can not able to keep the unique feature values and spatial information from the given input. This is major disadvantage of traditional CNN approach. So SV-OnionNet uses a novel optimizer technique called adaptive COA instead of pooling. The novel optimization approach, named adaptive COA is already expalined detail in below Sect. 3.5.2.
3.5.1.4 Final onion layer
The traditional CNN model has a fully connected layer for classification as its final layer. But, for our proposed Onionnet, this layer is removed. Instead of this layer, Onionnet used an SVM (Cristianini and Shawe-Taylor 2000) classifier to predict the oral cancer label. The advantage of using an SVM instead of a fully connected layer is that it can better handle high-dimensional feature spaces and can lead to better generalization performance. To use an SVM for classification in place of a fully connected layer, the result of the final optimization layer is flattened and given to an SVM approach.
3.5.2 Adaptive COA for training process of MMShift-CNN
The SV-OnionNet is tuned by novel adaptive COA for improving the classification accuracy. Members of the Coati and Nasuella genera in the Procyonidae family are coatis, also known as coatimundis. Native to the southern United States, Mexico, Central America, and South America, they are nocturnal mammals. All Coati (coatis) share a thin head with a flexible, elongated, somewhat upward-turned nose, black paws, small ears, and a long non-prehensile tail used for signaling and balancing. From head to tail tip, an adult coati can measure up to 69 cm, which is as long as their body. At 30 cm tall at the shoulder and weighing between 2 and 8 kg, coatis are about the size of a large house cat. A green iguana is one of coati's preferred foods. Iguanas are huge reptiles that coati’s hunt in packs because they frequently live in trees. While others rapidly attack it, several of them climb trees to frighten the iguana into jumping to the ground. Nevertheless, coatis are vulnerable to predator attacks. Some of the coati's predators include jaguars, ocelots, tayras, dogs, foxes, boa constrictor snakes, maned wolves, anacondas, and jaguarundis. Large raptors including harpy eagles, black-and-chestnut eagles, and ornate hawk-eagles also pursue them (Dehghani et al. 2023). Based on the attacking and the escaping characteristics of the Coati the optimization algorithm is mathematically formulated in the below section. The position update of the Coati takes place based on the assaulting and savaging characteristics of the coati and the steps involved in the optimization is given as follows.
The proposed comprehensive evaluation demonstrates that the ACDCNN model exhibits strong generalization. It effectively learns and adapts to different staining techniques, accommodating variations in color and texture that can occur due to staining variations. The model's architecture, enriched by deep learning mechanisms, allows it to capture relevant features across different image resolutions, enhancing its robustness. Moreover, the ACDCNN model's ability to handle sample heterogeneity is noteworthy. It can adeptly identify key patterns despite variations in tissue structures and cellular appearances, contributing to its reliable performance across diverse samples. By employing techniques like data augmentation during training and leveraging the inherent feature extraction capabilities of deep learning, the ACDCNN model displays remarkable adaptability to variations commonly encountered in oral cancer histopathological images. This adaptability ensures its potential for broader clinical applicability and reinforces its efficacy in real-world scenarios.
3.5.2.1 Initialization
At the beginning, the candidate solution for the optimization is generated and the solutions are represented by the number of Coati present in the search space. The values for the decision factors are based on where each coati is in the search space. The starting position of the coatis in the search space is determined at random and is notified by the below equations,
here, the position of the Coati is represented by \(P\) and the random Coati is represented by \(n\), which is in the range \(\left[ {1,\,2,\,3,...x} \right]\). \(R_{rand}\) is a random real number in the lower and the upper bound \(LB\) and \(UB\) in the range \(\left( {0,1} \right)\). The total population of the Coati is notated by the equation,
3.5.2.2 Fitness evaluation
The fitness evaluation is performed for determining the best optimal solution. Here accuracy is selected to evaluate the proposed model. The optimal solution is selected if the fitness value achieved is higher than the previous iteration. The fitness is expressed availing the equation,
here, \(F\) denotes the fitness function of the Coati that helps in obtaining the training at higher speed.
3.5.2.3 Assaulting stage
The initial stage of updating the number of Coati’s in the search area is modelled using a simulation of their attack method on iguanas. In this method, a pack of coati scales the tree to get close to an iguana and startle it. Other coati gathers around the iguana as it falls to the ground while they wait under a tree. The coati attack and hunt the iguana after it hits the ground. With the use of this method, coatis can migrate to various locations within the search area, showcasing the capacity for global search inside the problem-solving domain.
The algorithmic design assumes that the iguana occupies the position of the population's best member. Furthermore, it is believed that half of the coati ascend the tree while the other half waits for the iguana to fall to the ground. The below equation is therefore used to replicate the coati’s position when they emerge from the tree.
where, \(P_{n,d}^{new}\) denotes the new position of the coati and \(R_{rand}\) denotes the random real number in the interval \(\left( {0,1} \right)\). An integer selected in the range \(\left\{ {1,2} \right\}\) is designated by the variable \(Int\).
The iguana is dropped to the ground and then positioned at random somewhere inside the search area. The search space is mimicked using a random position, and coati on the ground move based on this position.
If the updated position for each coati increases the value of the objective function, it is acceptable for the update process; otherwise, the coati stays in its former position.
Here, \(O_{T}^{g}\) s its objective function value, \(P_{n}^{new}\) is the new position calculated for the \(n^{th}\) coati, and \(T\) represents the iguana's position in the search space, which actually refers to the position. Iguana \(g\) is the position of the iguana on the ground, which is randomly generated.
3.5.2.4 Escaping stage
Based on coati’s typical behaviour when confronting and evading predators, the second stage of the process of updating the position of coati in the search space is mathematically modelled. A coati flees from its place when a predator attacks it. Coati's actions in this strategy result in it being in a secure location close to where it is right now, which shows the algorithm capacity for local search exploitation. Based on this behaviour the position update takes place and is represented by the equation as follows,
Moreover, the newly computed location is adequate if it enhances the objective function rate, which is expressed by Eq. (10).
On the other hand, adaptive concept is included with COA for improving the computational cost with minimal time period. From Eq. (11), the term \(T\) is expressed as,
where, \(\alpha\) implies depth weight, which is made as adaptive, \(T_{\max }\) and \(T_{\min }\) symbolizes predefined maximal and minimal value of \(T\) and \(\alpha\) represents highest iteration.
3.5.2.5 Evaluating feasibility of solution
The best optimal solution is achieved through fitness function, which is already expressed in Eq. (6), and fitness function with least value is considered as optimum solution.
3.5.2.6 Termination
The above steps are executed repeatedly until the optimal solution is obtained. The pseudocode for the new adaptive optimization algorithm is interpreted in Table 2.
Thus, the adaptive COA effectively identifies the oral image as OSCC or normal with minimal time and cost.
4 Results and discussion
The results obtained by the proposed oral cancer detection approach and is displayed in this section.
4.1 Experimental setup
All the experiments are executed in a Personal Computer (PC) with i7-processor, 32 GB RAM, and 16 GB GPU and PYTHON software is used to implement oral cancer classification.
4.2 Dataset description
This dataset (Available online 2023) comprises of 1224 images, which is separated into two groups with two dissimilar resolutions. First set includes 89 histopathological images along with regular epithelium of oral cavity as well as 439 images of OSCC in 100× magnification. Another set encompasses of 201 images with normal epithelium of oral and 495 histopathological images of OSCC in 400× magnification. The subsection of 269 images from second set is utilized for identifying OSCC based on textural features. In this database, images were taken by Leica ICC50HD microscope from Haematoxylin and Eosin (H&E) stained tissue slides gathered, arranged, and categorized by medical experts from 230 patients.
4.3 Data augmentation
In this work, to generalization of MMShift-CNN, data augmentation is employed and leading to 3000 images. Data augmented details in Table 3.
4.4 Dataset split-up
The total volume of image patches from Table 2 is divided into two groups: training and testing in the proportion of 80% and 20% respectively, as per the train-test split technique widely adopted. The training dataset is again divided into train and validation set in the proportion of 90% and 10% respectively. The number of training, validation and testing images are 2160, 240 and 600, respectively.
4.5 Experimental results
The experimental outcomes for the devised oral cancer detection and classification model are displayed as below Fig. 4. Here, input, pre-processing, segmentation, and classification images are specified.
4.6 Performance metrics
To evaluate the effectiveness of proposed approach, various performance measures, such as accuracy, Mean Square Error (MSE), precision, specificity, F-measure, and sensitivity are considered in this research. Accuracy is evaluated for detecting true positive and true negative of all images. Precision is referred as the number of positive images exactly classified to whole amount of positive predicted images. MSE is computed by cumulative squared error among detected and original image. Sensitivity is a percentage of the precise rate of true positives in oral cancer detection. Specificity is calculated for predicting the accurate detection rate of true positive rate. The F-measure metric is defined as the harmonic mean of precision and recall.
4.7 Accuracy and loss curves
The loss and accuracy curves of deep learning methods are deliberated in which every curve contains validation and training curves in Fig. 5. The validation curve is attained from a validation set, which reveals how well the method generalizes itself, whereas training curve refers how well the method is capable to learn. Moreover, error on training database is specified as training loss, although error after running validation database by trained network is termed as validation loss. This experiment has been performed for 0 epoch and it is increased to 50 epochs. Here, the accuracy curve of proposed approach varies from 0.990306 to 0.99646, whereas the loss curve rate changes from 0.009693 to 0.003533.
4.8 Confusion matrix
Confusion matrix represents the comprehensive illustration if prediction outcomes after classification process. The confusion matrix for all networks and proposed approach is displayed in Fig. 6. Here, true positive, false positive, true negative and false negative values are deliberated. Generally, in biomedical field, maximal true negative and true positive rates are essential, even though false negative and positive rates are also needed. The values in matrices reflect predictable proportion for respective classes in row and column for classifier. The evaluated true positives for the class are specified by diagonal values and other rates symbolizes error rates.
4.9 ROC curve
Figure 7, represents the ROC curve for the proposed method with other existing techniques and here true positive and false positive rates are varied from 0 to 1 to analyse the Area Under Curve (AUC),. The proposed technique attained a higher AUC of 0.9889, while CNN has less AUC of 0.93025, which reflects the proposed method to provide better model performance at distinguishing between the normal and OSCC classes.
4.10 Comparative discussion
Figure 8 presents the comparative analysis of proposed method with other existing techniques. Here, various performance measures, like F-measure, accuracy, specificity, sensitivity, MSE, and precision is considered for analysis. Moreover, the analysis is carried out by means of varying training data percentage from 60 to 90.
The comparative discussion for various methods with proposed approach based on different performance measures is presented in Table 4.
The proposed technique attained higher accuracy of 0.9883, while CNN has less accuracy of 0.93 for 90% training data. Moreover, least MSE rate 0.0117 achieved by proposed method and F-measure is high as 0.9883, while training data percentage is 90. In addition, precision specificity and sensitivity are high in proposed approach by 0.999, 0.99, and 0.9867, when 90% of training data. The ACDCNN model revolutionizes oral cancer diagnosis by enhancing efficiency through rapid analysis, improving accuracy by leveraging deep learning capabilities, maintaining consistency in interpretation, offering valuable quantitative insights, and optimizing resource utilization. Its integration stands to reshape the landscape of histopathological analysis, leading to more precise and timely diagnoses.
5 Conclusion
The model proposed in this research has the potential to bring about a revolutionary change in the medical field. By accurately identifying cancerous patients, it can help prevent unnecessary treatments and tests, potentially saving lives. Additionally, the model can assist paramedic staff in efficiently treating such patients, leading to improved healthcare outcomes. For effective diagnosis of oral histopathological images is essential for accurate diagnosis and treatment planning, and the model can provide doctors with a dependable second opinion on the presence of oral lesions. To achieve this goal, a novel MMShift-CNN is used to segment the oral cancer region from input images. Additionally, classification of OSCC and normal oral tissues is performed through SV-OnionNet from and it is trained by novel adaptive COA. The proposed approach was effectively evaluated using the performance metrics, like accuracy, F-measure, MSE, precision, sensitivity, and specificity for evaluating the effectiveness of proposed approach. The results of this research are promising, with an accuracy rate of 0.9883, MSE of 0.0117, F-measure of 0.9883, sensitivity of 0.9867, specificity of 0.99 and precision of 0.999. In future, real time oral cancer images will be used for analysing the performance of the proposed method.
Data availability
Data will be made available on request.
Change history
09 September 2024
This article has been retracted. Please see the Retraction Notice for more detail: https://doi.org/10.1007/s11082-024-07437-w
Abbreviations
- OSCC:
-
Oral squamous cell carcinoma
- OCID:
-
Oral cancer imaging database
- CNNs:
-
Convolutional neural networks
- RNNs:
-
Recurrent neural networks
- AL:
-
Active learning
- RL:
-
Random learning
- DCNN:
-
Deep convolutional neural network
- LWDCNN:
-
Light-weight deep convolutional neural network
- DRNN:
-
Deep reinforced neural network
- CAM:
-
Class activation map
- SCC:
-
Squamous cell carcinoma
- OC:
-
Oral cavity
- PCA:
-
Principal component analysis
- COA:
-
Coati optimization algorithm
- SVM:
-
Support vector machine
- AUC:
-
Area under curve
References
Altaf, F., Islam, S.M.S., Akhtar, N., Janjua, N.K.: Going deep in medical image analysis: concepts, methods, challenges, and future directions. IEEE Access 7, 99540–99572 (2019)
Amin, I., Zamir, H., Khan, F. F.: Histopathological image analysis for oral squamous cell carcinoma classification using concatenated deep learning models. medRxiv. (2021) http://medrxiv.org/content/early/2021/05/14/2021.05.06.21256741. Abstract
Anwar, N., Pervez, S., Chundriger, Q., Awan, S., Moatter, T., Ali, T.S.: Oral cancer: clinicopathological features and associated risk factors in a high-risk population presenting to a major tertiary care center in Pakistan. PLoS ONE 15(8), e0236359 (2020)
Available online: https://www.kaggle.com/ashenafifasilkebede/dataset?select=val Accessed on 5 Jan 2023.
Chakraborty, D., Natarajan, C., Mukherjee, A.: Advances in oral cancer detection. Adv. Clin. Chem. 91, 181–200 (2019)
Cristianini, N., Shawe-Taylor, J.: An Introduction to Support Vector Machines and other Kernelbased Learning Methods. Cambridge University Press, Cambridge (2000)
Das, N., Hussain, E., Mahanta, L.B.: Automated classification of cells into multiple classes in epithelial tissue of oral squamous cell carcinoma using transfer learning and convolutional neural network. Neural Netw. 128, 47–60 (2020)
Dehghani, M., Montazeri, Z., Trojovská, E., Trojovský, P.: Coati optimization algorithm: a new bio-inspired metaheuristic algorithm for solving optimization problems. Knowl.-Based Syst. 259, 110011 (2023)
Deif, M.A., Solyman, A.A.A., Alsharif, M.H., Uthansakul, P.: Automated triage system for intensive care admissions during the COVID-19 pandemic using hybrid XGBoost-AHP approach. Sensors 21(19), 6379 (2021)
Deif, M.A., Attar, H., Amer, A., Issa, H., Khosravi, M.R., Solyman, A.A.A.: A new feature selection method based on hybrid approach for colorectal cancer histology classification. Wirel. Commun. Mob. Comput. 2022, 1–14 (2022)
Deif, M.A., Hammam, R.E.: Skin lesions classification based on deep learning approach. J. Clin. Eng. 45(3), 155–161 (2020)
Deng, C., Li, S., Bian, F., Yang, Y.: Remote sensing image segmentation based on mean shift algorithm with adaptive bandwidth. In: Bian, F., Xie, Y. (eds.) Geo-Informatics in Resource Management and Sustainable Ecosystem. GRMSE 2014. Communications in Computer and Information Science, vol. 482. Springer, Berlin, Heidelberg (2015)
Du, M., Nair, R., Jamieson, L., Liu, Z., Bi, P.: Incidence trends of lip, oral cavity, and pharyngeal cancers: global burden of disease 1990–2017. J. Dent. Res. 99(2), 143–151 (2020)
Duggento, A., Conti, A., Mauriello, A., Guerrisi, M., Toschi, N.: Deep computational pathology in breast cancer. Semin. Cancer Biol. 72, 226–237 (2021)
Eckert, A.W., Kappler, M., GroBe, I., Wickenhauser, C., Seliger, B.: Current understanding of the HIF-1-dependent metabolism in oral squamous cell carcinoma. Int. J. Mol. Sci. 21(17), 6083 (2020)
Folmsbee, J., Liu, X., Brandwein-Weber, M., Scott, D.: Active deep learning: improved training efficiency of convolutional neural networks for tissue classification in oral cavity cancer. In: Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), pp. 770–773. Washington (2018)
Ghosh, A., Chaudhuri, D., Adhikary, S., Das, A.K.: Deep reinforced neural network model for cyto-spectroscopic analysis of epigenetic markers for automated oral cancer risk prediction. Chemom. Intell. Lab. Syst. 224, 104548 (2022)
Goldenberg, S.L., Nir, G., Salcudean, S.E.: A new era: artificial intelligence and machine learning in prostate cancer. Nat. Rev. Urol. 16(7), 391–403 (2019)
Jubair, F., Al-karadsheh, O., Malamos, D., Al Mahdi, S., Saad, Y., Hassona, Y.: A novel lightweight deep convolutional neural network for early detection of oral cancer. Oral Dis. 28(4), 1123–1130 (2022)
Kong, J., Sertel, O., Shimada, H., Boyer, K., Saltz, J., Gurcan, M.: Computer-aided evaluation of neuroblastoma on whole-slide histology images: classifying grade of neuroblastic differentiation. Pattern Recogn. 42(6), 1080–1092 (2009)
Leo, L.M., Reddy, T.K.: Learning compact and discriminative hybrid neural network for dental caries classification. Microprocess. Microsyst. 82, 103836 (2021)
Leo, L. M., Reddy, T. K., Simla, A. J.: Impact of Selective median filter on dental caries classification system using deep learning models. J. Auton. Intell. 6(2) (2023)
Li, L., Yin, Y., Nan, F., Ma, Z.: Circ_LPAR3 promotes the progression of oral squamous cell carcinoma (OSCC). Biochem. Biophys. Res. Commun. 589, 215–222 (2022)
Martino, F., Bloisi, D.D., Pennisi, A., et al.: Deep learningbased pixel-wise lesion segmentation on oral squamous cell carcinoma images. Appl. Sci. 10(22), 8285 (2020)
Musulin, J., Štifanić, D., Zulijani, A., Ćabov, T., Dekanić, A., Car, Z.: An enhanced histopathology analysis: an ai-based system for multiclass grading of oral squamous cell carcinoma and segmenting of epithelial and stromal tissue. Cancers 13(8), 1784 (2021)
Perdomo, S., Roa, G.M., Brennan, P., Forman, D., Sierra, M.S.: Head and neck cancer burden and preventive measures in central and South America. Cancer Epidemiol. 44, 43–52 (2016)
Santana, M.F., Ferreira, L.C.L.: Diagnostic errors in surgical pathology. J. Bras. Patol. Med. Lab. 53, 124–129 (2017)
Simla, A.J., Chakravarthi, R., Leo, L.M.: Agricultural intrusion detection (AID) based on the internet of things and deep learning with the enhanced lightweight M2M protocol. Soft Comput. 1–12 (2023)
Sivakumar, M.S., Leo, L.M., Gurumekala, T., Sindhu, V., Priyadharshini, A.S.: Deep learning in skin lesion analysis for malignant melanoma cancer identification. Multimed. Tools Appl. 1–21 (2023)
Wang, S., Yang, D.M., Rong, R., et al.: Artificial intelligence in lung cancer pathology image analysis. Cancers 11(11), 1673 (2019)
Warnakulasuriya S., Greenspan, J.S.: Epidemiology of oral and oropharyngeal cancers. In: Textbook of Oral Cancer, Springer, Berlin (2020)
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
DR: responsible for the design, implementation, and evaluation of the proposed approaches. Developed the Mask Mean Shift CNN (MMShift-CNN) approach for segmenting and identifying OSCC using deep learning techniques. Conducted experiments and analyzed the results to demonstrate the efficiency of the proposed approach in accurately detecting oral cancer. RS: providing guidance and expertise in the field of oral cancer diagnosis and deep learning. Contributed to the interpretation and analysis of the experimental results.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article has been retracted. Please see the retraction notice for more detail: https://doi.org/10.1007/s11082-024-07437-w"
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dharani, R., Revathy, S. RETRACTED ARTICLE: Industry 4.0 transformation: adaptive coati deep convolutional neural network-based oral cancer diagnosis in histopathological images for clinical applications. Opt Quant Electron 56, 152 (2024). https://doi.org/10.1007/s11082-023-05716-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11082-023-05716-6