Abstract
Oral cancer has emerged as one of the ubiquitous malignant tumors globally. Timely detection and treatment reduces the mortality rate of oral cancer. This study utilizes a vision transformer (ViT) framework to classify oral squamous cell carcinoma (OSCC) and healthy oral histopathology images. The proposed approach is implemented on a public database consisting of 4946 oral histopathology images. Although ViT architectures have been extensively used in the medical imaging field, they have not yet been explored in oral cancer detection. Though transformer architecture needs large dataset to attain better performance, our modified architecture accomplishes an accuracy, specificity and sensitivity of 97.78%, 96.72%, and 98.80%, respectively, on a relatively smaller medical dataset. The evaluation metrics of the proposed method have also been compared with eight pre-trained deep learning models, namely Xception, Resnet50, InceptionV3, InceptionResnetV2, Densenet121, Densenet169, Densenet201 and EfficientNetB7. It is observed that the modified ViT model performs better than the deep learning models, demonstrating the ability to extract various features from the histopathology images for the classification. The results of the proposed approach would aid the clinical community for detection of oral cancer in patients of diverse origin.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Oral cancer is indeed a fatal condition with a complex etiology and a high death rate. The world cancer research fund (WCRF) international claims that malignancies of the oral cavity and lip are one of the most prevalent type of cancers with more than 377,700 cases recorded globally in 2020. The malignancies of the oral cavity and lip are the 11th and 18th most frequently occurring in men and women, respectively. A well-formulated strategy is required for addressing oral cancer which includes early detection, risk factor management, and health literacy. Risk factors include contact with human papillomavirus (HPV), consuming alcohol, smoking, lack of dental hygiene, geographical location, lifestyle, and ethnicity [1].
Squamous cell carcinoma (SCC) may develop from precancerous lesions such as erythroleukoplakia, oral leukoplakia, and verrucous hyperplasia [2]. 90% of all oral cancers are SCCs [3]. The most accurate way to diagnose oral cancer is through biopsy, however, this method is painful, and in cases of extensive or many lesions, selecting the appropriate site and size for surgical treatment of the biopsy sample could be challenging [4]. Additionally, due to lesion variability, the prepared histology specimen may not accurately reflect the identification of the entire lesion. To achieve a successful cure, higher chances of survival, reduced death and morbidity rates, oral squamous cell carinoma (OSCC) must be detected early [5]. The average survival rate stands at 50% for OSCC [6, 7]. The accepted approach for diagnosing OSCC is tissue sample histopathological examination based on microscopy [8, 9]. However, the clinical value of this approach is constrained by the histopathologists interpretation, which is frequently laborious and prone to error [10]. Therefore, it is crucial to offer efficient diagnostic techniques to support pathologists in the evaluation and diagnosis of OSCC.
Recently, deep learning (DL) algorithms have become the state-of-the-art in field of computer vision and image processing owing to their strength in processing vast volumes of data [11,12,13]. As a result, numerous investigations have been conducted to aid pathologists through DL techniques specially convolutional neural networks (CNNs) in medical image classification, segmentation and localization [14,15,16]. Although CNNs excels at feature extraction, they are unable to encode the relative positions of distinct features. Convolution operations fails to recognize global information [17] and long-range relationships across an entire image [18]. Many researchers came up with different architectural changes for an effective solution in due course and eventually [19] proposed attention mechanism that learns the correlation between output and input patterns without relying on repetition. This enables efficient parallelization of Transformer implementations. In response to the popularity of Transformers in natural language processing (NLP) tasks, Transformer architecture was redesigned by [20], referred to as vision transformer (ViT). In the adapted version, the transformer accepts a series of fixed-size image patches as input to extricate complex features of the image. It pays global attention to the entire image overcoming the long-range dependency issue of CNNs. The potential of ViT has been explored by several researchers in diverse computer vision applications say point cloud classification, image enhancement, object detection and many more. In addition to success of ViT in NLP, it has made significant contribution in medical computer vision in a variety of medical imaging modalities.
In the realm of histopathological image classification, ViTs have demonstrated notable success in field of cancer diagnosis, i.e, renal cell carcinoma, breast cancer, cancerous esophagus tissues, glioblastoma, bladder urothelial carcinoma, lower grade glioma, and lung cancer [21, 22]. Despite the widespread utilization of ViT in various disease diagnoses, its potential in the domain of oral cancer has been underexplored. The application of ViTs to oral cancer classification introduces a novel dimension, emphasizing the distinct histopathological characteristics and clinical considerations unique to oral tissues. Oral cancer presents its own set of challenges, marked by specific cellular compositions, anatomical variations, and staining patterns that differentiate it from other cancers. The prevalence of oral cancer, often associated with risk factors like tobacco use, underscores the critical need for accurate diagnostic tools. While ViTs have been leveraged in other cancer types, the adaptation and application of ViTs to oral cancer represent a pioneering effort, addressing a notable gap in the existing literature. By recognizing the unique characteristics of oral cancer and harnessing the power of ViTs, this research contributes to advancing our understanding of oral cancer pathology and heralds a promising avenue for improved clinical outcomes. While Transformers outperform CNNs in interpreting contextual information, their computational demands and the necessity for extensive datasets present challenges in the medical imaging field. The scarcity of publicly accessible imaging datasets for oral cancer further intensifies these difficulties. Considering these constraints, the motivation emerges to employ a fine-tuned ViT for creating an automated diagnostic framework for the detection of oral cancer.
The contributions of the paper are listed as:
-
1.
The performance of the proposed fine-tuned ViT model is either superior or comparable to that of state-of-the-art models in binary-class oral cancer classification across various publicly available oral cancer histopathology datasets.
-
2.
We have performed a comparative analysis of the deep learning (DL) models with the fine-tuned ViT, and it is inferred that ViT model performs better in comparison to DL models for classification of oral cancer.
-
3.
The fine-tuned ViT performs well with a smaller dataset, challenging the assumption that transformer models require large datasets for optimal performance.
The rest part of this manuscript is organized as follows: Sect. 2 discusses prior art of oral cancer classification and ViT in medical domain. Section 3 discusses the methodology utilized in the work. Section 4 presents the results of the proposed methodology and eight pre-trained deep learning models. Section 5 summarizes the work and outlines the future scope of our proposed approach.
2 Related works
Various approaches based on both machine learning and deep learning have been introduced in the literature for the diagnosis of oral cancer through the analysis of medical images. OSCC image databases involve hyperspectral imaging, autofluorescence imaging, computed tomography (CT), magnetic resonance imaging (MRI), and histopathological imaging. Tables 1 and 2 details some of the earlier recommended approaches to oral cancer classification implemented using machine learning and DL neural networks. For machine learning applications on OSCC images, [27] used SVM classifier to attain 91.64% accuracy. For CNN applications on OSCC images, [28] created a DL method that takes patient hyperspectral images into account for advanced computer-aided oral cancer diagnosis. The performance of the proposed regression-based partitioned DL strategy was assessed against other methods in terms of classifier accuracy, sensitivity, and specificity. [23] used CNN models to attain 91.13%. [29] developed an automated ensemble DL method that combines the benefits of Resnet-50 and VGG-16 to examine oral lesions achieving accuracy of 96.2%. [15] developed a lightweight EfficientNet-B0 DL model for classification of oral lesions images, separating benign from malignant or potentially malignant lesions. [24] explored a tailored AlexNet model designed for the detection of OSCC in histopathological images. [25] introduced a ten-layer CNN model, demonstrating superior performance in diagnosing OSCC from histopathological images compared to pre-trained CNN models. A hybrid optimization algorithm [26] was created combining particle swarm optimization (PSO) with Al-Biruni Earth Radius Optimization. This hybrid approach was employed to optimize the design parameters of Deep Belief Networks and CNNs in the context of identifying malignant oral lesions.
Based on the preceding discussion, it is evident that CNNs have proven to be highly effective in classifying oral cancer, showcasing remarkable accuracy, and establishing their significance in this domain. While CNNs with deep architectures excel at extracting features for numerous small objects within an image, identifying the truly critical regions may pose a challenge. To address this challenge, the utilization of the vision transformer (ViT) model has become prevalent in medical image classification which includes CT scans, X-rays, OCT/Fundus images, MRI Scans, PET, Histopathology images, Endoscopy, and Microscopy. [31] performed a multi-class colorectal cancer tissue classification using ViT and Compact Convolutional Transformer achieving accuracy of 93.3% and 95%, respectively. [32] developed an IL-MCAM framework. It employs interactive learning with attention techniques. [33] carried out a comprehensive analysis and review of the ViT framework for emphysema classification. [34] utilized ViT for Covid-19 detection using CT scans. They employed different ViTB-16, ViTB-32, ViTL-16, ViTL-32, and ViTH-14 for image classification. [35] compared the performance of pneumonia classification using ViT, CNN and VGG16 model. It was demonstrated that ViT achieved highest classification accuracy of 96.45%. [18] put forward an integrated Transformer model for multimodal image classification. The hybrid model comprised of a CNN to learn low-level features, followed by Transformers for global information. [36] classified normal and abnormal fundus images using Tranformer model achieving accuracy of 85.7%. [37] put forth a model that can interpret visual neural activities induced by natural images in form of descriptive text. In [30], a deep-learning methodology utilizing the Swin-Transformer attained a classification accuracy of 0.986 and an AUC of 0.99 in the task of classifying OSCC on clinical photographs. [22] provides an extensive overview of cutting-edge ViTs investigated in histopathological image analysis, covering applications such as segmentation, classification, and survival risk regression.
In our comprehensive review, it is evident that researchers strive to achieve promising diagnostic accuracy through diverse methods. Consequently, we have tailored the ViT framework for enhanced oral cancer detection.
3 Proposed methodology
Figure 3 shows the workflow of the classification methodology. We used the Vision Transformer architecture inspired by [20] to classify oral histopathology images into normal and OSCC and named it as ViT-14. Here, 14 represents the patch size. In this study, we also compared the effectiveness of the proposed approach using 8 pre-trained DL models named Xception [38], Resnet50 [11], InceptionV3 [39], InceptionResnetV2 [40], Densenet121 [41], Densenet169, [41], Densenet201 [41], EfficientNetB7 [13].
3.1 Dataset description
An oral cancer histopathological imaging dataset is publicly available in [42]. It has three directories, namely train, test, and val. We have utilized the train directory [43], as followed by the study [24]. There are two types of subjects in the considered oral histopathology dataset: the patients having oral squamous cell carcinoma and the healthy subjects. Table 3 shows number of images present in the dataset. Figures 1 and 2 shows samples from both dataset categories.
3.2 Preprocessing and data augmentation
All the images in the dataset have been reshaped to \(224 \times 224\) pixel resolution. Data augmentation procedures are employed to increase the image count as training the original dataset will result in overfitting the model. The Keras DL toolbox provides ImageDataGenerator function to generate images with appropriate data augmentation. The resized oral histopathology images undergoes different augmentation techniques such as normalization, randomly rotated, zoomed, horizontally flipped, varying height and width to enhance the generalizability of the model. The details of the augmentation techniques are shown in Table 4.
3.3 Description of the ViT-14 model used in our proposed work
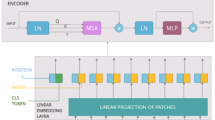
After pre-processing and data augmentation, the images are split into non-overlapping patches inspired by the [19] architecture before being fed to the encoder section. However, non-overlapping style partly breaks the internal framework of an image [44]. Multi-headed self attention (MSA) blocks alleviate this issue by integrating information from several patches. Additionally, when non-overlapping patches are fed into Transformer, computational redundancy does not exist. In our study, an input image of size 224 \(\times \) 224 \(\times \) 3 (H=224, W=224, C=3) is splitted into flattened patches of size 588 (\(P^{2}C\), where P=14,C=3). Thus, 256 patches (N=HW/\(P^{2}\)) are generated before heading into the Transformer encoder section. It is also noted that the sequence length of the Transformer and square of patch size are inversely related, hence, models having smaller patch size requires more computation.
The resulting flattened patches are used to create linear embeddings of a lower dimensional latent space (D) of size 64, known as patch embeddings. The size of the latent space remains constant through all layers of the encoder. ViT does not use convolution or recurrence in the multi-head self attention module in encoder section, hence to ensure that the images preserve their positional knowledge, position embeddings are then linearly added to the patch embeddings using Eq. 1.
where, \(y_{j}\) denotes the patch embedding of the jth patch and \(x_{j}\) denotes the position embedding of the jth patch and \(y_{j}\), \(x_{j}\) \(\in \) \(D_{y}\). \(D_{y}\) is the dimensionality of the jth patch embedding. An additional learnable (class) embedding is also added similar to BERT’s (class) token as shown in Eq. 2. The class of the input image is predicted using this class embedding.
where, \(y_\textrm{class}\) is the additional learnable class embedding and \(P_\textrm{o}^\textrm{o}\) is \(y_\textrm{class}\). The outcome of the Transformer encoder (TE) at the Lth layer (\(L=8\)) is denoted as \(P_{L}^\textrm{o}\). The series of patches are then passed to the TE layer. The TE module is composed of alternating layers of multi-head self attention (MSA) layer and feed forward network (FFN). The patch embedding passes through a number of layers in encoder section depicted by Eqs. 3 and 4.
where, LN is the layer normalization layer. The output of the encoder at the Lth layer, \(P_{L}^\textrm{o}\) is layer normalized and passed through a learnable classification network known as multi-layer perceptron (MLP) head as shown in Fig. 6.
In basic terms, a group of patches splitted from an input image is transformed into a latent vector with a specified size. Then position embeddings are added to the transformed patch embeddings and a class token is also prepended. Further, the modified input passes through a chain of encoder layers. The pictorial representation of the ViT-14 model is shown in Fig. 4.
In TE block, the embedded patches pass through MSA layer and feed forward network. A residual connection [11] is added prior to layer normalization [45] layer around MSA layer and FFN. The TE module is shown in Fig. 5. Muti-headed attention boosts the performance of the model by performing multiple self-attention mechanism simultaneously. Each self attention operation serves as a head in multi-headed attention mechanism, and each head tries to learn something unique, thus improving the representation power of the encoder module. Therefore, the model is able to capture intricate correlations of different patches present at distinct locations in a histopathology image. It focuses on local and global features encompassed within an image in contrast to conventional CNN models which emphasizes on local attention. The parameters of the adopted ViT-14 model is tabulated in Table 5. Details of the layers, output shape and number of parameters are shown in Table 6.
3.4 Pre-trained deep learning models for comparison
This subsection gives a brief description of the various DL models used in our work for comparative analysis. Architecture of the DL models used in our study is shown in Fig. 7. Table 7 lists few details of the DL models.
3.4.1 Xception
The elementary theory of Inception has been pushed to an extreme in Xception architecture [38]. In Inception, 1x1 convolutions were used to extract features from the initial input, and filters of varying sizes were employed at every depth space. The reverse occurs in Xception, it uses filters at every depth space independently prior to compressing the input image at once using 1x1 convolution. The feature extraction backbone in the Xception architecture is composed of 36 convolutional layers. The Xception architecture can be summed up as a linear stack of residually connected depthwise separable convolution layers. As a result, developing and altering the architecture is relatively simple.
3.4.2 Resnet50
The Resnet50 utilizes a bottleneck framework for its building block. The residual block consists of 1\(\times \)1 convolutions termed as bottleneck, which minimizes the matrix multiplications and parameter count. This makes training each layer considerably faster. Instead of using a stack of two layers, it leverages three layers [11]. It is widely known that increasing the depth of the model for deeper feature extraction reduces model performance due to exploding or vanishing gradient issue. To resolve this issue and enable the training of deeper networks, residual blocks were introduced.
3.4.3 InceptionV3
InceptionV3 is an image recognition model that achieved an accuracy higher than 77.9% on the ImageNet dataset. It is an optimized and upgraded adaptation of InceptionV1 model. Factorized convolutions, smaller convolutions, asymmetric convolutions, auxillary classifiers, and grid size reduction forms the architecture of InceptionV3 [39].
3.4.4 InceptionResnetV2
A convolutional neural network known as InceptionResNetV2 expands on the Inception group of architectures while incorporating residual connections. It replaces the filter concatenation step of the Inception model [40].
3.4.5 Densenet121/169/201
Densenets are deep CNNs that enhance the training of deeper networks by connecting the feature map of one layer with all the layers preceding it [41]. This increase the effectiveness with regard to memory utilization and computation. It can extract minute features of the input images with few channels. DenseNet further improves feature propagation, increases feature reuse, and significantly lowers the number of parameters, obviate the vanishing gradient issue and mitigates its impacts.
3.4.6 EfficientNetB7
EfficientNetB7 is a non-repetitive, nonlinear neural network search that optimizes floating point operations per second (FLOPS) and accuracy by balancing resolution, network depth and breadth. Seven flipped residual blocks, each with its own parameters, are used in the architecture. These blocks employ swish activation, squeeze, and excitation blocks [13].
4 Experiments and analysis
All studies related to this work were carried out using Python 3.7.6, TensorFlow 2.7.0, and Keras 2.7.0 on a PC with 2.40 GHz Intel(R) Core(TM) i5-1135G7 processor, Intel(R) Iris(R) Xe graphics and 16.0 GB of RAM.
4.1 Evaluation indicators
The overall performance of our proposed approach is evaluated on the basis of the contents of the confusion matrix. There are four terms namely true positive (TP), false positive (FP), false negative (FN), and true negative (TN) included in this evaluation matrix. TP means a person has OSCC and the model accurately predicts it. TN means a person has healthy oral mucosa and the model accurately predicts it. FP means a healthy oral mucosa is inaccurately predicted as OSCC. FN means an OSCC is inaccurately predicted as healthy oral mucosa. Evaluation indicators, namely specificity, sensitivity, F1-score, precision, cohen kappa score (CKS), matthews correlation coefficient (MCC), error rate, false omission rate (FOR), false discovery rate (FDR), negative predictive value (NPV), false negative rate (FNR), and false positive rate (FPR) were evaluated to assess the performance of our proposed approach. These evaluation indicators can be calculated utilizing the formulas given as follows:
-
1.
Precision: It represents the proportion of accurately predicted positive instances out of the total instances predicted as positive.
$$\begin{aligned} {\text {Precision}}=\frac{{\text {TP}}}{{\text {TP}}+{\text {FP}}} \end{aligned}$$(5) -
2.
Sensitivity: It denotes the proportion of accurately predicted positive instances relative to all instances in the actual positive class.
$$\begin{aligned} {\text {Sensitivity}}=\frac{{\text {TP}}}{{\text {TP}}+{\text {FN}}} \end{aligned}$$(6) -
3.
Specificity: It measures the ability of the model to correctly identify true negatives.
$$\begin{aligned} {\text {Specificity}}=\frac{{\text {TN}}}{{\text {TN}}+{\text {FP}}} \end{aligned}$$(7) -
4.
Accuracy: It measures the ratio of accurately identified images to the total number of test images.
$$\begin{aligned} {\text {Accuracy}}=\frac{{\text {TP}}+{\text {TN}}}{{\text {TP}}+{\text {TN}}+{\text {FP}}+{\text {FN}}} \end{aligned}$$(8) -
5.
F1-Score: It is the harmonic mean of precision and recall, serves as a means to optimize the model either for recall or precision.
$$\begin{aligned} F1 \; {\text {Score}} =\frac{2 * {\text {Precision}} * {\text {Recall}}}{{\text {Precision}}+{\text {Recall}}} \end{aligned}$$(9) -
6.
Cohen Kappa score: It is a metric used to measure the agreement between predicted and actual classifications while accounting for the possibility of random agreement.
$$\begin{aligned} {\text {CKS}} = \frac{P_\textrm{o}-P_\textrm{e}}{1-P_\textrm{e}} \end{aligned}$$(10)where \(P_\textrm{o}\) is the observed agreement and \(P_\textrm{e}\) is the expected agreement.
-
7.
MCC: It is a correlation coefficient between the observed and predicted binary classifications.
 (11)
(11) -
8.
Error rate: It provides a measure of misclassification.
$$\begin{aligned} {\text {Error \; rate}}=1-{\text {Accuracy}} \end{aligned}$$(12) -
9.
False omission rate (FOR): It is the proportion of false negatives out of the total actual negative instances.
$$\begin{aligned} {\text {FOR}} = \frac{{\text {FN}}}{{{\text {FN}} + {\text {TN}}}} \end{aligned}$$(13) -
10.
False discovery rate (FDR): It is the proportion of false positives out of the total predicted positive instances.
$$\begin{aligned} {\text {FDR}} = \frac{{\text {FP}}}{{{\text {FP}} + {\text {TP}}}} \end{aligned}$$(14) -
11.
Negative predictive value (NPV): It is the proportion of correctly predicted negative instances out of the total predicted negative instances.
$$\begin{aligned} {\text {NPV}} = \frac{{\text {TN}}}{{{\text {TN}} + {\text {FN}}}} \end{aligned}$$(15) -
12.
False negative rate (FNR): It is the proportion of false negatives out of the total actual positive instances.
$$\begin{aligned} {\text {FNR}} = \frac{{\text {FN}}}{{{\text {FN}} + {\text {TP}}}} \end{aligned}$$(16) -
13.
False positive rate (FPR): It is the proportion of false positives out of the total actual negative instances.
$$\begin{aligned} {\text {FPR}} = \frac{{\text {FP}}}{{{\text {FP}} + {\text {TN}}}} \end{aligned}$$(17)
4.2 Model parameters
We selected sparse categorical crossentropy as a loss function for our binary classification task. The training is done over 100 epochs with the AdamW optimizer. We have used a patch size of \(14 \times 14 \times 3\) with each image having 256 patches. In our Transformer encoder architecture, we employed a configuration with 4 heads and opted for 8 layers in the Transformer encoder. Batch size of 32, learning rate of 0.001 and weight decay of 0.0001 are chosen for model training. Table 8 lists the optimal hyperparameters used in our study.
4.3 Ablation study on model parameters
We perform an ablation study to analyze how different components and hyperparameters in our proposed model contribute to the overall performance of the model.
4.3.1 Impacts of different parameters
In the initial experimentation phase with the ViT-14 model, default hyperparameters were initially assumed. This included a learning rate of 0.001, a batch size of 8, weight decay of 0.0001, a patch size of \(14 \times 14 \times 3\), a latent dimension of 64, the number of Transformer encoder layers set to 6, and the number of heads set to 4. Subsequent exploration involved varying batch sizes to 8 and 16 while keeping other parameters constant, and it was found that a batch size of 32 yielded the highest accuracy, as outlined in Table 9. Further experiments focused on patch size variations while maintaining other parameters constant, confirming that the initial dimensions of \(14 \times 14 \times 3\) for the patch achieved highest accuracy. Likewise, alternative latent dimensions of 16 and 32 were explored while keeping other parameters constant, with the initial choice of 64 demonstrating the highest accuracy, as illustrated in Table 9. Once optimized values for batch size, patch size, and latent dimension were obtained, experiments on the number of layers indicated that 8 layers outperformed 6 and 10, as detailed in Table 9. Finally, experiments on the number of heads, exploring values of 6 and 8, validated the initial choice of 4 as yielding the highest accuracy, as indicated in Table 9.
4.4 Results
After obtaining the optimal model hyperparameters, we have evaluated the performance of the model. The dataset is divided into two subsets: the training set, comprising 90% of the data, with 10% of this subset allocated for validation; and the testing set, which constitutes the remaining 10% as shown in Table 10.
It is extremely important that the model does not exhibit significant overfitting to ensure the overall effectiveness of the proposed method. Figure 8a shows the training and validation accuracy and loss curves plotted over 100 epochs. It is observed that the model exhibits no major overfitting, and robustness is maintained. The confusion matrices (CM) generated have been displayed in Fig. 8b which further helps in understanding the results. It is inferred from CM that FP and FN are very less in number as compared to TN and TP, thus showing correct predictions of images into normal and OSCC classes. Table 11 shows the evaluation metrics of the proposed model. As shown in Table 11, ViT-14 model achieved an accuracy, specificity, and sensitivity of 97.78%, 96.72%, and 98.80%, respectively.
In our study, we implemented a fivefold cross-validation methodology, running the model five times to ensure a comprehensive and robust evaluation of its generalization to unseen data. For each iteration, the dataset was shuffled to create unique training and test sets, with the training set comprising 90% of the data and the test set representing the remaining 10%. Evaluation metrics were computed in each iteration on the assigned test set, offering a thorough assessment of performance of the model across diverse data splits. The CM generated from each of the five folds have been displayed in Fig. 9 which further helps in understanding the results. Evaluation metrics for ViT-14 model using fivefold cross validation are listed in Tables 12 and 13.
4.4.1 Impacts of split ratio
The model underwent evaluation with different combinations of training and testing ratios. This evaluation retained consistency with the same set of hyperparameters, as detailed in Table 8. This approach allowed for an assessment of its performance under different data split scenarios while keeping the experimental conditions uniform.
Case 1 (90:10) Training images constitute 90%, and testing images make up 10% of the entire dataset. For the ViT-14 model, 4451 images were used for training, and 495 for testing. The model underwent five runs to assess generalizability, and the resulting average accuracy values are detailed in Table 14.
Case 2 (80:20) Training images constitute 80%, and testing images make up 20% of the entire dataset. For the ViT-14 model, 3956 images were used for training, and 990 for testing. The model underwent five runs to assess generalizability, and the resulting average accuracy values are detailed in Table 14.
Case 3 (70:30) Training images account for 70%, with testing images at 30% of the overall dataset. The ViT-14 model was trained on 3462 images and tested on 1484. Similar to Case 1, the model underwent five runs to evaluate generalizability, and the corresponding average accuracy values are presented in Table 14.
It is observed from Table 14 that the accuracy appears to decrease as the proportion of training data decreases relative to testing data. Thus, a higher proportion of training data (90:10 ratio) contributes to better model performance.
4.5 Comparative analysis of model performance across different datasets
After the ablation study, the optimal model configuration is used for further analysis on two publicly available oral cancer histopathological datasets.
Dataset 1 [46] was collected from a histopathological image repository of the normal epithelium of the oral cavity and OSCC images. The repository consists of 1224 total images. They are divided into two sets in two different resolutions, 100x magnification and 400x magnification. In total, there are 290 normal epithelium images and 934 OSCC images.
Dataset 2 is an oral cancer histopathological image dataset available in [42]. It comprises three directories: train, test, and val, containing a total of 5192 images. There are 2,494 normal images and 2,698 images with OSCC.
We employed a fivefold cross-validation approach, executing the model five times to ensure a thorough and robust evaluation of its ability to generalize to new data. In each iteration, the dataset was shuffled, creating distinct training and test sets. The training set constituted 90% of the data, while the test set comprised the remaining 10%. Evaluation metrics were calculated in each iteration on the assigned test set, providing a comprehensive assessment of the performance of the model across a variety of data partitions. Tables 15 and 16 present the evaluation metrics for dataset 1 and dataset 2, respectively, respectively, utilizing the fivefold cross-validation technique.
4.6 Comparison with deep learning models
The proposed approach is compared with eight pre-trained DL models to demonstrate its effectiveness. The details of the hyperparameters are listed in Table 18. The dataset is divided into two subsets: the training set, comprising 90% of the data, with 10% of this subset allocated for validation; and the testing set, which constitutes the remaining 10% as shown in Table 17. The training-to-testing split ratio of 9:1 was maintained, consistent with the proposed ViT-14 method. We selected binary cross-entropy as a loss function for our binary classification task. Adam optimizer is used for training over a course of 100 epochs. The reduction of the generalization gap between training loss and validation loss was our main objective during model training. A batch size of 32 and a learning rate of 0.001 is used. Additionally, a dropout rate of 0.2 is used to address overfitting during training time [29]. Then, we saved the weights of the model having lowest validation loss for evaluation purposes. We adhered to the original architectural descriptions of convolutional filters, padding, pooling and strides in Xception, Resnet50, InceptionV3, InceptionResnetV2, Densenet121/169/201, EfficientNetB7 models.
We have utilized a model pre-trained on the ImageNet dataset for the analytical results of DL models on the oral histopathology dataset. The training and validation accuracy and loss curves are plotted over 100 epochs as displayed in Figs. 10 and 11. The confusion matrix (CM) for DL models were also calculated to help in understanding the results as shown in Fig. 12. It is inferred from the CM that FP and FN have increased in comparison to the ViT-14 model, thus showing lesser correct predictions of images into normal and OSCC classes. Table 19 lists the various evaluation measures of the compared DL models and ViT-14 model. Table 20 shows the superior performance of ViT-14 model in terms of accuracy, specificity and sensitivity in comparison to the DL models.
4.7 Comparison with previous works
Table 21 provides a comprehensive comparative analysis of diverse methods and models applied to various publicly available oral cancer datasets. In previous research [23], transfer learning methods using Resnet50, MobileNet, and InceptionV3 achieved accuracies ranging from 76.61% to 91.13% on a dataset containing 290 normal and 934 OSCC images [46]. A customized 10-layer CNN [25] attained a higher accuracy of 97.82% on the same dataset [46]. A hybrid approach involving both CNNs and SVM, and the integration of deep and texture-based features, the study [47] demonstrated an accuracy of 97.00% on 2698 OSCC images and 2494 healthy tissue images [42]. Additionally, Gabor filter combined with a Catboost classifier [48] achieved 94.92% accuracy on the same dataset [42]. A transformer with external attention [49] attained an accuracy of 96.97% on 2511 OSCC images and 2435 healthy tissue images [43]. Transfer learning using Alexnet [24] achieved 90.06% accuracy on same set [43]. While the proposed method demonstrated an accuracy of 95.12% on the dataset [46], it is noteworthy that the 10-layer CNN model [25] achieved a higher accuracy of 97.82%. However, it is important to highlight that the proposed method showcased competitive performance on other datasets, achieving accuracies of 97.69% and 97.78% on datasets [42] and [43] respectively. It is evident that a reduction in performance on the dataset [46] is significantly due to class imbalance, potentially impacting the model’s ability to effectively learn and generalize across both classes. While the presently employed data augmentation techniques, such as rotation, zoom, flip, height, and width variations, contribute to model resilience, addressing the class imbalance may require additional augmentation strategies. This could involve applying techniques like synthetic minority over-sampling technique (SMOTE) and employing generative adversial networks (GANs) for the creation of realistic synthetic samples, particularly for the minority class. By implementing such additional data augmentation techniques tailored to address class imbalances, the proposed model is likely to achieve improved generalization and classification accuracy across all datasets, ensuring consistent performance in the presence of varied class distributions.
5 Conclusion
Histopathological assessment by pathologists stands as the gold standard for detecting oral squamous cell carcinoma (OSCC). However, the intricate morphological variations in cancerous conditions pose a significant challenge for human evaluation. This study is a dedicated effort to aid clinicians in early OSCC identification. While deep learning (DL) models have advanced to enhance various applications for effective medical assessments, the incorporation of attention mechanisms into Vision Transformers (ViTs) introduces a level of precision that is essential in the medical industry, where inaccuracies could have profound consequences. The study introduces ViT-14, a fine-tuned ViT framework, specifically designed for classifying oral histopathology images into normal and OSCC categories across diverse publicly available datasets. The ViT-14 model demonstrates performance on par with or exceeding that of state-of-the-art models, emphasizing its effectiveness in early oral cancer detection using histopathological images. This study not only underscores the capabilities of ViTs in the field of medical imaging but also establishes ViT-14 as a promising instrument to assist clinicians in achieving more precise and timely diagnoses in cases of oral cancer.
The potential for enhancing oral cancer classification with fine-tuned ViT models is promising, but it is crucial to recognize certain limitations. Limited and imbalanced datasets may hinder generalization, and interpreting complex models like ViT remains difficult. Class imbalance and the "black-box" nature of these models can introduce bias and limit explainability. Computational demands pose challenges for resource-limited institutions, and integrating these models into clinical workflows requires addressing privacy and regulatory issues. Despite these challenges, the future outlook is promising, with ongoing efforts to overcome these limitations through the accumulation of more diverse and expansive datasets, advancements in model interpretability, and optimization of computational efficiency for broader applicability in clinical settings.
Data Availability
The obtained datasets for our analysis are from free public database.
Code Availability
The codes that support the findings of this study are available from the corresponding author upon reasonable request.
References
Scully, C., Bedi, R.: Ethnicity and oral cancer. Lancet Oncol. 1(1), 37–42 (2000)
Tsai, M.-T., Lee, H.-C., Lee, C.-K., Yu, C.-H., Chen, H.-M., Chiang, C.-P., Chang, C.-C., Wang, Y.-M., Yang, C.: Effective indicators for diagnosis of oral cancer using optical coherence tomography. Opt. Express 16(20), 15847–15862 (2008)
Montero, P.H., Patel, S.G.: Cancer of the oral cavity. Surg. Oncol. Clin. 24(3), 491–508 (2015)
Albrecht, M., Schnabel, C., Mueller, J., Golde, J., Koch, E., Walther, J.: In vivo endoscopic optical coherence tomography of the healthy human oral mucosa: qualitative and quantitative image analysis. Diagnostics 10(10), 827 (2020)
Chakraborty, D., Natarajan, C., Mukherjee, A.: Advances in oral cancer detection. Adv. Clin. Chem. 91, 181–200 (2019)
Eckert, A.W., Kappler, M., Große, I., Wickenhauser, C., Seliger, B.: Current understanding of the hif-1-dependent metabolism in oral squamous cell carcinoma. Int. J. Mol. Sci. 21(17), 6083 (2020)
Ghosh, A., Chaudhuri, D., Adhikary, S., Chatterjee, K., Roychowdhury, A., Das, A.K., Barui, A.: Deep reinforced neural network model for cyto-spectroscopic analysis of epigenetic markers for automated oral cancer risk prediction. Chemometrics Intell. Lab. Syst. 224, 104548 (2022)
Kong, J., Sertel, O., Shimada, H., Boyer, K.L., Saltz, J.H., Gurcan, M.N.: Computer-aided evaluation of neuroblastoma on whole-slide histology images: classifying grade of neuroblastic differentiation. Pattern Recognit. 42(6), 1080–1092 (2009)
Deif, M.A., Hammam, R.E.: Skin lesions classification based on deep learning approach. J. Clin. Eng. 45(3), 155–161 (2020)
Santana, M.F., Ferreira, L.C.L.: Diagnostic errors in surgical pathology. J. Brasi. Patol. Med. Lab. 53, 124–129 (2017)
He, K., Zhang, X., Ren, S., Sun, J.: Deep residual learning for image recognition. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 770–778 (2016)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. Commun. ACM 60(6), 84–90 (2017)
Tan, M., Le, Q.: Efficientnet: Rethinking model scaling for convolutional neural networks. In: International Conference on Machine Learning, pp. 6105–6114 (2019). PMLR
Ariji, Y., Kise, Y., Fukuda, M., Kuwada, C., Ariji, E.: Segmentation of metastatic cervical lymph nodes from ct images of oral cancers using deep-learning technology. Dentomaxillofac. Radiol. 51(4), 20210515 (2022)
Jubair, F., Al-karadsheh, O., Malamos, D., Al Mahdi, S., Saad, Y., Hassona, Y.: A novel lightweight deep convolutional neural network for early detection of oral cancer. Oral Dis. 28(4), 1123–1130 (2022)
Zhang, X., Liang, Y., Li, W., Liu, C., Gu, D., Sun, W., Miao, L.: Development and evaluation of deep learning for screening dental caries from oral photographs. Oral Dis. 28(1), 173–181 (2022)
Park, J., Kim, Y.: Styleformer: Transformer based generative adversarial networks with style vector. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 8983–8992 (2022)
Dai, Y., Gao, Y., Liu, F.: Transmed: Transformers advance multi-modal medical image classification. Diagnostics 11(8), 1384 (2021)
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A.N., Kaiser, Ł., Polosukhin, I.: Attention is all you need. Advances in neural information processing systems, vol. 30 (2017)
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., Dehghani, M., Minderer, M., Heigold, G., Gelly, S. et al.: An image is worth 16x16 words: Transformers for image recognition at scale. arXiv preprint arXiv:2010.11929 (2020)
Parvaiz, A., Khalid, M.A., Zafar, R., Ameer, H., Ali, M., Fraz, M.M.: Vision transformers in medical computer vision-a contemplative retrospection. Eng. Appl. Artif. Intell. 122, 106126 (2023)
Xu, H., Xu, Q., Cong, F., Kang, J., Han, C., Liu, Z., Madabhushi, A., Lu, C.: Vision transformers for computational histopathology. IEEE Rev. Biomed. Eng. (2023)
Palaskar, R., Vyas, R., Khedekar, V., Palaskar, S., Sahu, P.: Transfer learning for oral cancer detection using microscopic images. arXiv preprint arXiv:2011.11610 (2020)
Rahman, A.-U., Alqahtani, A., Aldhafferi, N., Nasir, M.U., Khan, M.F., Khan, M.A., Mosavi, A.: Histopathologic oral cancer prediction using oral squamous cell carcinoma biopsy empowered with transfer learning. Sensors 22(10), 3833 (2022)
Das, M., Dash, R., Mishra, S.K.: Automatic detection of oral squamous cell carcinoma from histopathological images of oral mucosa using deep convolutional neural network. Int. J. Environ. Res. Public Health 20(3), 2131 (2023)
Myriam, H., Abdelhamid, A.A., El-Kenawy, E.-S.M., Ibrahim, A., Eid, M.M., Jamjoom, M.M., Khafaga, D.S.: Advanced meta-heuristic algorithm based on particle swarm and al-biruni earth radius optimization methods for oral cancer detection. IEEE Access 11, 23681–23700 (2023)
Muthu Rama Krishnan, M., Shah, P., Chakraborty, C., Ray, A.K.: Statistical analysis of textural features for improved classification of oral histopathological images. J. Med. Syst. 36, 865–881 (2012)
Jeyaraj, P.R., Samuel Nadar, E.R.: Computer-assisted medical image classification for early diagnosis of oral cancer employing deep learning algorithm. J. Cancer Res. Clin. Oncol. 145(4), 829–837 (2019)
Nanditha, B., Geetha, A., Chandrashekar, H., Dinesh, M., Murali, S.: An ensemble deep neural network approach for oral cancer screening (2021)
Flügge, T., Gaudin, R., Sabatakakis, A., Tröltzsch, D., Heiland, M., Nistelrooij, N., Vinayahalingam, S.: Detection of oral squamous cell carcinoma in clinical photographs using a vision transformer. Sci. Rep. 13(1), 2296 (2023)
Zeid, M.A.-E., El-Bahnasy, K., Abo-Youssef, S.: Multiclass colorectal cancer histology images classification using vision transformers. In: 2021 Tenth International Conference on Intelligent Computing and Information Systems (ICICIS), pp. 224–230 (2021). IEEE
Chen, H., Li, C., Li, X., Rahaman, M.M., Hu, W., Li, Y., Liu, W., Sun, C., Sun, H., Huang, X., et al.: Il-mcam: An interactive learning and multi-channel attention mechanism-based weakly supervised colorectal histopathology image classification approach. Comput. Biol. Med. 143, 105265 (2022)
Wu, Y., Qi, S., Sun, Y., Xia, S., Yao, Y., Qian, W.: A vision transformer for emphysema classification using ct images. Phys. Med. Biol. 66(24), 245016 (2021)
Ambita, A.A.E., Boquio, E.N.V., Naval, P.C.: Covit-gan: vision transformer forcovid-19 detection in ct scan imageswith self-attention gan fordataaugmentation. In: Artificial Neural Networks and Machine Learning–ICANN 2021: 30th International Conference on Artificial Neural Networks, Bratislava, Slovakia, September 14–17, 2021, Proceedings, Part II 30, pp. 587–598 (2021). Springer
Tyagi, K., Pathak, G., Nijhawan, R., Mittal, A.: Detecting pneumonia using vision transformer and comparing with other techniques. In: 2021 5th International Conference on Electronics, Communication and Aerospace Technology (ICECA), pp. 12–16 (2021). IEEE
Kamran, S.A., Hossain, K.F., Tavakkoli, A., Zuckerbrod, S.L., Baker, S.A.: Vtgan: Semi-supervised retinal image synthesis and disease prediction using vision transformers. In: Proceedings of the IEEE/CVF International Conference on Computer Vision, pp. 3235–3245 (2021)
Zhang, J., Li, C., Liu, G., Min, M., Wang, C., Li, J., Wang, Y., Yan, H., Zuo, Z., Huang, W., et al.: A cnn-transformer hybrid approach for decoding visual neural activity into text. Comput. Methods Programs Biomed. 214, 106586 (2022)
Chollet, F.: Xception: deep learning with depthwise separable convolutions In: The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) (2017)
Szegedy, C., Vanhoucke, V., Ioffe, S., Shlens, J., Wojna, Z.: Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2818–2826 (2016)
Szegedy, C., Ioffe, S., Vanhoucke, V., Alemi, A.A.: Inception-v4, inception-resnet and the impact of residual connections on learning. In: Thirty-first AAAI Conference on Artificial Intelligence (2017)
Huang, G., Liu, Z., Van Der Maaten, L., Weinberger, K.Q.: Densely connected convolutional networks. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 4700–4708 (2017)
Oral cancer histopathology dataset. https://www.kaggle.com/datasets/ashenafifasilkebede/dataset. Accessed 17 Nov 2023
Oral cancer histopathology dataset. https://www.kaggle.com/datasets/ashenafifasilkebede/dataset?select=train. Accessed 17 Nov 2023
Han, K., Xiao, A., Wu, E., Guo, J., Xu, C., Wang, Y.: Transformer in transformer. Adv. Neural. Inf. Process. Syst. 34, 15908–15919 (2021)
Ba, J.L., Kiros, J.R., Hinton, G.E.: Layer normalization. arXiv preprint arXiv:1607.06450 (2016)
Rahman, T.Y., Mahanta, L.B., Das, A.K., Sarma, J.D.: Histopathological imaging database for oral cancer analysis. Data Brief 29, 105114 (2020)
Ahmad, M., Irfan, M.A., Sadique, U., Haq, I., Jan, A., Khattak, M.I., Ghadi, Y.Y., Aljuaid, H.: Multi-method analysis of histopathological image for early diagnosis of oral squamous cell carcinoma using deep learning and hybrid techniques. Cancers 15(21), 5247 (2023)
Haq, I.U., Ahmad, M., Assam, M., Ghadi, Y.Y., Algarni, A.: Unveiling the future of oral squamous cell carcinoma diagnosis: an innovative hybrid ai approach for accurate histopathological image analysis. IEEE Access (2023)
Deo, B.S., Pal, M., Pradhan, A.: External-attention-based deep neural network model for reliable detection of oral cancer from histopathological images. In: Women in Optics and Photonics in India 2022, vol. 12638, pp. 25–28. SPIE (2023)
Acknowledgements
Bhaswati Singha Deo is thankful to IIT Kanpur for the Institute fellowship. The author MPal wishes to thank ABB Ability Innovation Centre, India, for their support in this work.
Funding
This research received no specific support from governmental, private, or non-profit funding agencies.
Author information
Authors and Affiliations
Contributions
Bhaswati Singha Deo: Developed the model from the concept, Developed the code, Generated the results, Wrote the manuscript. Mayukha Pal: Conceived the idea and conceptualized it, Developed the methodology from the concept, Reviewed the manuscript and mentored the work. Prasanta K. Panigrahi: Reviewed the manuscript and mentored the work. Asima Pradhan: Developed the analysis methodology, Reviewed the manuscript and mentored the work.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethics approval
Datasets are publicly available for research purpose, hence not applicable for this paper.
Consent to participate
Datasets are publicly available for research purpose, hence not applicable for this paper.
Consent for publication
Datasets are publicly available for research purpose, hence not applicable for this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Deo, B.S., Pal, M., Panigrahi, P.K. et al. Supremacy of attention-based transformer in oral cancer classification using histopathology images. Int J Data Sci Anal (2024). https://doi.org/10.1007/s41060-023-00502-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s41060-023-00502-9