Abstract
Cancer is a lethal illness that requires an initial stage prognosis to enhance patient survival rate. Accurate brain tumor and their sub-structure segmentation through Magnetic Resonance Images (MRIs) is a tough endeavor. Owing to the heterogeneous tumor areas, automatically segmenting brain tumors has proved to be a critical task even for neural network-based algorithms, some tumor regions remain unidentified due to their small size and the variation in area occupancy among tumor sub-classes. Current progress in the area of neural networks has been employed to enhance the segmentation performance. This study designed an intelligent 3D U-Net encoder-decoder-based system for automatic detection and brain tumor sub-structure segmentation. Our proposed 3D model constitutes neural units (the basic building blocks) followed by transition layer blocks and skip connections. BraTS 2018 and private local datasets are used to evaluate the proposed model which segments the Whole Tumor (WT), Tumor Core (TC), and the Enhancing Tumor (ET). The training accuracy, validation accuracy, dice score, sensitivities, and specificities of WT, CT, and ET zones are computed. The experimental results demonstrate that dice scores are 0.913, 0.874, and 0.801 for the BraTS 2018 dataset. The developed models performance was further evaluated by utilizing the dataset from a local hospital containing 71 subjects. The dice scores of 0.891, 0.834, and 0.776 are achieved by the proposed model on the private dataset. The practicability of the proposed model was assessed by the comparative studies of our model with existing literature.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumors are a prevalent type of nervous system disorder that proved to be extremely harmful to human health. Gliomas are considered the most threatening forms of brain tumors [1]. These are often categorized as high-grade glioma (HGG) or low-grade glioma (LGG), with an average survival time of about two years for individuals who have progressed into HGG. LGG is less aggressive and infiltrative. Grade I gliomas are generally treatable with surgical resection; grade IV, Glioblastoma Multiforme (GBM), is the most dangerous due to the poorest survival rate.
Numerous imaging methods, including MRI, computed tomography (CT), positron emission tomography (PET), etc. have been used to examine brain malignancies. Among them, MRI has been considered the key imaging method for brain tumor diagnosis, owing to its benefits of strong tissue contrast, diverse directions, and being non-invasive in nature. Various MRI sequences emphasize different areas of brain picture detail and depict the features of tumors from several perspectives. Its very important to distinguish brain tumor tissues like enhancing core, non-enhancing core, necrosis, and edema from healthy tissues. However, correct segmentation of these tissues is a difficult task, owing to the following factors. Tumors can vary widely in location, appearance, and size from subject to subject [2]. Second, tumors normally infect nearby tissues, blurring cancer and non-cancerous tissues [3]. The third is the problem of image distortion and noise induced by imaging devices or acquisition procedures.
In medical imaging and brain tumor image segmentation, many conventional machine-learning approaches have been utilized [4,5,6,7,8,9]. For example, Yang et al. [4], tto identify Glioblastoma multiforme (GBMs) from isolated METs, a 3D morphometric analytic approach was presented. The shape index and curvedness morphometric characteristics were calculated for each tumor surface described by the diffusion tensor imaging (DTI) segmentation technique [5]. To categorize diverse mixtures of first and second-order statistical properties of an area of interest (ROIs), M. Soltaninejad et al. [6] employ support vector machines (SVM). Soltaninejad [7] offers a fully automatic technique for segmenting brain tumors, which uses superpixels to generate Gabor texton features as well as fractal examination, and statistical intensity characteristics. The superpixels are then classified into tumor or non-tumor brain tissue using extremely randomized trees (ERT). Information fusion from multimodal MRI scans is used to calculate super voxels. A variety of characteristics are retrieved from each super voxel and put into a well-known classifier named random forests [8]. Wu et al. [9] segmented brain tumors using superpixel features inside a conditional random field (CRF) system, although the result was low in LGG scans. Geometry-based techniques [10], including level sets and active contour techniques, are also useful since they are computationally efficient, but they necessitate user participation, expert knowledge, and feature engineering to be effective. At the moment, these methods are frequently employed as pre-or post-processing steps in deep learning-based systems.
Manual segmentation of tumors is considered the gold standard. But, it is a laborious process, and if an inexperienced radiologist delineates the ROI, the results are poor [11, 12]. Hence, automatic tumor segmentation is ideal, particularly when dealing with huge volumes of data and when regular tumor monitoring and adaptive patient management are necessary. Thus, researchers are interested in learning more about fully automatic or semi-automated segmentation methods [13]. However, due to the wide range of tumor sizes, shapes, and patterns, effective automatic tumor segmentation is generally a tough task.
Recently, deep learning-based segmentation algorithms have shown substantial benefits in medical image analysis [14] including breast cancer diagnosis, Skin lesion classification [15], Covid-19 [16], and brain tumor segmentation [17, 18]. One of the main research objectives is to improve the model using a fully convolutional network (FCN) [19] and the U-Net standard network [20, 21]. Zhao et al. [22] suggested a strategy for segmenting brain tumors by integrating FCN and CRF into a combined framework. Pereira et al. [23] ccollected image context information by segmentation of Squeeze Excitation (SE) blocks while retaining spatial information for the creation of highly complicated semantic features. Vijay et al. [24, 25] and Sajid et al. [26] designed various SegNet-based architectures, while FCN-based U-Net [20] networks were also proposed. Both modified SegNet and FCN showed improvement in segmentation performance. Dong et al. [27] eemployed a variety of data augmentation strategies to enhance brain tumor MRI segmentation performance and address the problem of class imbalance using the soft dice loss function. Zhou et al. [28] created a sequence of stacked, dense skip paths to connect the encoder and decoder models, therefore bridging the semantic divide between the encoder and decoder models feature maps. The original U-Net architecture is applied as the standard architecture in this work, and a novel modified, and intelligent 3D U-Net-based system is designed. The following is a summary of the significant contributions:
-
1.
This study developed an intelligent deep learning-based 3D U-Net system for a brain tumor and sub-regions segmentation (i.e. WT, TC, and ET).
-
2.
The suggested method can effectively segment and detect brain tumors from MRI images under the existence of blurring, noise, and bias field-effect fluctuations in input images.
-
3.
This research also design several skip connections to support the transmission of high-resolution features from shallow layers to deeper layers. These skip connections also reinforce the synthesis of various scales by associates at several network layers.
-
4.
By efficiently generalizing distinct local patterns, the suggested model uses the transition layer block which helps us in reducing feature maps. Furthermore, the dice loss function aids in the appropriate segmentation of minor abnormalities that are likely to be misclassified.
-
5.
The locally collated new dataset first time was employed to check the efficiency and effectiveness of the 3D U-Net system.
-
6.
Lastly, the experimental implementation illustrates that our 3D U-Net model achieves robust performance regarding accuracy, dice score, sensitivity, and specificity which outperform existing systems.
The remaining paper is structured as follows: Section 2 represents the literature studies on brain tumor segmentation. Section 3 introduces the material and method. Section 4 illustrates the results and discussion. Finally, a conclusion is made in Section 5.
2 Related work
Brain tumor segmentation techniques can be divided into conventional segmentation techniques and deep learning-based segmentation techniques. Based on features including gradients and in-context which were extracted through handcrafted methods [29], machine learning-based approaches such as random forests, fuzzy C-means, and SVM [30,31,32] hhave been implemented to segment brain tumors from MRIs. Traditionally, threshold-based segmentation algorithms separated voxels into distinct tumor or normal tissues. These approaches begin by calculating a threshold based on the intensity of MRIs and then classifying each voxel based on the comparative study of the grey value with the threshold. Clustering-based segmentation approaches include the Markov random field (MRF), k-means clustering, and fuzzy c-means clustering, these approaches use unsupervised methods for training to differentiate the tissues of unseen data. However, these approaches continue to confront difficulties, such as low segmentation accuracy for brain tumor segmentation and classification.
In contrast, deep learning-based applications have express a high level of competitiveness in medical image detection [18], semantic segmentation [33] and also other classification and segmentation systems. For instance, it has been extensively employed in medical image analysis [34], and tissue segmentation [35]. When it comes to brain tumor segmentation, convolutional neural networks (CNNs) are by far the most popular deep learning technique to use. It can learn from real images by identifying promising hierarchies of features.
Havaei, et al. [36] proposed a two-path CNN, which was able to incorporate both local and global context information. A cascaded model was utilized to obtain more data, and then a two-phase training scheme was applied to reduce the issue of class imbalance. To accomplish coarse segmentation, Hu, Gan, et al. [37] developed a multi-cascaded model that considers both the voxel-level and multi-scale properties simultaneously. CNN segmentation was improved using a fully linked conditional random field (CRF). Pereira et al. [38] proposed two CNNs for two-grade brain tumor segmentation using 2D patches. Additionally, the eliminated discrete sections were post-processed using a morphological filter. This segmentation algorithm learns the characteristics of two grades to obtain pure features of various brain tumors. Nevertheless, their architectures need huge storage capability, as well as continual spatial loss was observed which results in poor segmentation performance. To avoid overfitting, a multiple-scale of the input image using CNN was utilized [39, 40]. Around the center pixel, this can fully hold the input patchs texture. However, a general drawback of this technique is that lacks the semantic knowledge of the lesion region in contrast with the image-based approach.
Kamnitsas et al. [41] suggested a three-dimensional network with dual pathways capable of collecting context information about the local position and connecting pixels inside a specified scope of the lesion region. However, it did not generate global features. The researchers in [42] proposed multiclass segmentation using a cascaded 3D-FCN technique by splitting it into three segmentation assignments. While the binary classification approach can simply be utilized to solve the four-class problem, however, three additional models significantly increase the computational cost.
Albishri et al. [43] demonstrate the effectiveness of the AM-UNet approach through comprehensive experiments and validation on brain MRI datasets, highlighting its potential for enhancing neuroimaging analysis and clinical applications in neuroscience research. Punn et al. [44] proposed a method that integrates 3D Inception U-Net and decoder models to effectively combine information from different imaging modalities, such as MRI and CT scans. By using advantages of multi-modality encoding and fusion techniques, the model achieves accurate segmentation of brain tumors with enhanced spatial and contextual information. Raza et al. [45] introduce a novel approach for segmenting brain tumors using a 3D deep residual U-Net architecture. The proposed method leverages multimodal MRI data to enhance the segmentation accuracy and robustness. Li et al. [46] present a robust approach for brain tumor segmentation using cascaded 3D U-Net and 3D U-Net++ architectures. Through the integration of multiparametric MRI data, the proposed method achieves accurate and comprehensive segmentation of brain tumors. The cascaded architecture allows for a more refined segmentation process, effectively capturing intricate tumor features and boundaries.
Isensee et al. [47] designed a network for dealing with the issue of generalization that performed well across a variety of datasets. The outcomes of the BraTS 2018 competition showed that architectures like the U-shape with skip connection have a great ability to learn representations of features. Isensee et al. [48] applied a typical U-Net algorithm with small adjustments in the encoder and decoder. McKinley, Meier, and Wiest [49] designed a new framework with dense blocks through dilated convolutions inserted in U-Net architecture. Authors have also proposed a loss function shared focal loss by standard binary cross-entropy (BCE) loss to decrease uncertainty. To improve information flow, Li et al. [50] and Kayalibay et al. [51] proposed novel U-Net models. These techniques, however, unavoidably produced results in a huge number of false positives. Furthermore, to attain great segmentation accuracy, the most advanced models require huge computer resources.
In comparison to the conventional CNN models, U-Net possesses the following advantages: (1) U-Net utilizes both local and global features; (2) owing to the great value of the segmentation labels, this approach requires fewer training images to achieve optimal results, and (3) end-to-end input and output generate comprehensive segmentation performance. Inspired by previous studies and to overcome the problems in the literature, this study developed a modified and improved 3D U-Net architecture for brain tumor sub-regions whole, core, and enhanced tumor segmentation using MRI brain images. The suggested system used a deeper encoder path and decoder path followed by transition and batch normalization layers that achieved high performance using different evaluation matrices.
3 Material and methods
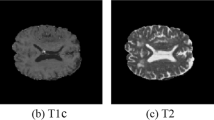
To evaluate the 3D U-Net model this study utilized two MRI datasets. The BraT- S 2018 dataset [52] and locally collected dataset are termed DATASET1 and DATASET2, respectively. DATASET1 contains 210 HGG and 75 LGG training subjects’ MRIs. Each subject in the dataset has four MRI scanning volumes including T1-weighted imaging (T1), T1-weighted imaging with enhancing contrast (T1c), T2-weighted imaging (T2), and FLAIR with an image size of \(240\times 40\times 55\). DATASET2 consists of 71 subjects. The whole data was in DICOM format which was converted into .nifti format before being given to the proposed model. Figure 1 illustrates a few classical MRI images of a patient case in the BraTS dataset [53].
The included pictures in the BraTS dataset depict classical MRI brain scans, arranged from left to right as follows: T1, T1ce, T2, Flair, and Ground Truth. Each color corresponds to a specific tumor class, namely, yellow indicates necrotic and non-enhancing regions, red indicates enhancing tumors, and green indicates areas of edema
3.1 Data pre-processing
For computer vision, a pre-processing step is important. In this research, we employed data cropping, data normalization, and data augmentation as pre-processing approaches. The following are the details of the data processing method.
Firstly, crop MRI images to fit the system memory, all of the brain MRI scans are taken randomly with a size of \(128\times 128\times 128\). This method preserves the majority of the image content within the crop region without affecting the images, however, it does reduce image size and computing complexity. Secondly, normalization is the process of transforming data into more usable MRI scans which are typically acquired using a variety of scanners and acquisition procedures. Thus, image intensity value normalization is critical for compensating for image heterogeneity. Entire input MRI data are normalized to set a mean of zero and a variance of one. Thirdly, data augmentation techniques are often used in deep learning to increase the number of samples without providing new data. This procedure aids in the generalization of neural networks. For example, they can generate additional data from a smaller dataset. This is beneficial for those datasets where categorizing new data is difficult and laborious, such as in medical imaging. Another benefit is that they assist in avoiding overfitting. One issue with the deep learning model is that they are capable of remembering vast volumes of data, which can lead to overfitting. Rather than learning the core concepts of the input volume, deep learning-based networks learn all of the inputs, reducing system competence. To enhance the number of images, we applied data augmentation approaches for example rotation, flipping, etc. to the local dataset.
3.2 Proposed 3D U-Net architecture
This study utilizes U-Net as a basic network. The proposed scheme comprises three main steps performing pre-processing, feature extraction and selection, and segmentation of brain tumor sub-regions represented in Fig. 2.
The proposed system expanded the U-Net framework by substituting all 2D processes with their 3D counterparts. We have applied a modified U-Net algorithm based on the several number of convolutional blocks. In our modified U-Net architecture a combination of three convolutional blocks followed by one transitional layer for each block is employed. This makes the model deeper. The depth of the network is quite important for the model accuracy; the deeper the model, the superior the performance. On the other hand, a deeper model means the use of more parameters, which leads to an overfitting problem, particularly in the medical imaging domain. Moreover, deeper architectures need extra computational time and resources. For this reason, a size of \(128\times 128\times 128\) for each scan is used to reduce the computational time and resources. Since the transition layers are missing among the encoding and decoding stages of classical U-Net, this study builds them after every Conv-Net block to emphasize the role of map representations in the decoding stages.
The 3D U-Net model contains several blocks in the expanding and contracting path. Every block uses a stack of convolutional layers followed by transitional layers. Each block contains 3 convolutional layers with a \(3\times 3\) kernel size, subsequently a ReLU activation function, and a batch normalization layer. Each input is the concatenated output of all preceding layers in the block, which supports preserving certain information misplaced during convolutional operations. Since each block may have a different amount of feature maps as a result of channel-wise concatenation, a transitional layer is added after the convolutional block to guarantee that the resultant feature maps are similar to those in the desired output of each step. Additionally, this layer serves as a compressing layer, reducing the size and parameters required for each level. The convolutional blocks are applied in the both encoder and decoder paths in a similar fashion. In comparison to the encoder path of the U-Net, the max-pooling operation is employed with a stride value of 2 for downsampling, while the decoder path employs an up-convolutional operation with a stride value of 2 for upsampling. We have employed transition layers in the encoding-decoding path for the enhancement of feature maps. Moreover, each convolutional block contains batch normalization to overcome the over-fitting issue. Finally, we use dice loss to balance loss from several anomalies like WT, TC, and ET. The softmax function is implemented for the output labels. The architecture of the 3D U-Net implemented in this study is illustrated in Fig. 3.
This study utilized transition layers blocks between Conv-Net blocks. The blocks in question are responsible for carrying out convolution and pooling operations in both the encoding and decoding components of the introduced 3D U-Net framework. The blocks consist of a batch normalization layer, a \(1\times 1\) convolutional layer, and a \(2\times 2\) average pooling layer. The inclusion of a transition layer is necessary to reduce the dimensionality of the feature maps by half before progressing to the subsequent Conv-Net block. Additionally, this feature provides advantages in terms of network cohesiveness. Figure 4 represents the overview of transition layer block uses in this work.
3.3 Class imbalance issue
The segmentation is an inherent issue owing to variations in the physiological properties of glioma. The background is tremendously dominated by the foreground. As a result, selecting the appropriate loss functions is critical in resolving the imbalance issue. The following are three different loss functions. Focal Tversky loss may be useful for solving intra-class imbalance in binary classification issues. However, it is less effective in addressing inter-class disparities. Categorical cross-entropy necessitates extra caution when working with a dataset that has a strong class imbalance. Dice loss can effectively cope with the datasets class disparity issue and can be expressed by the following (1).
Where N belongs to the set of the whole dataset, C belongs to the set of all classes, \(P_jc\) represents the probability of the tumor class, \(g_jc\) is showing the true tumor class and \(\varepsilon \) is a constant to avert dividing with 0. In this study, we apply dice loss to reduce the problem of class imbalance.
3.4 Optimization
We utilize the Adam optimizer to start with a learning rate and gradually reduce it according to (2):
Where the variable e indicates the epoch counter, whereas Te is the aggregate amount of epochs.
3.5 Network implementation and training details
During the training phase, we employed the Pycharm software development environment and the TensorFlow library to carry out the suggested 3D U-Net framework. Additionally, we employed the NVIDIA Geforce GTX1660Ti 6GB GPU, along with a system equipped with 16 GB of RAM.
The algorithm is trained throughout 20 epochs, with each epoch representing a complete iteration of the dataset. The choice of 20 epochs for training was determined through iterative experimentation to balance training time and performance. This duration allows the model to sufficiently learn from the data while avoiding overfitting.
The input MRI data is of dimensions \(128\times 128\times 128\), indicating a three-dimensional structure with a height, width, and depth of 128, 128, and 128 units respectively. Additionally, the Adam optimizer is employed with an initial learning rate of \(10^{-4}\) and a learning rate decay of 10% after 5 epochs to aid in convergence.
Moreover, a comprehensive assessment was conducted to evaluate the influence of these features on the overall efficacy of the proposed framework. This evaluation involved rigorous experimentation and a comparison with U-Net baselines. A thorough examination was undertaken to evaluate the algorithm’s responsiveness to changes in parameters and to determine the most effective configurations that achieve the highest level of accuracy and robustness in segmentation. The hyperparameters employed for the training and testing are represented in Table 1.
3.6 Performance metrics
Based on BraTS databases protocol procedures [13], the various sub-regions in the brain tumor structures for each subject are given below:
-
i.
The whole tumor area (It contains all 4 intra-tumoral regions labeled as 1, 2, 3, and 4).
-
ii.
The core tumor area (It contains the whole tumor area except the edema region and is labeled as 1, 3, and 4).
-
iii.
The enhanced tumor area (only comprising the enhancing region, i.e., label 4).
In this study, we use dice score, sensitivity, specificity, and area under the curve (AUC) for the evaluation of each tumor sub-region. DSC offers a dimension criterion for overlapping regions among manually labeled and predicted segmentation outcomes. It can be calculated using the (3).
The variables TP, FP, FN, and TN represent the quantities of true positive, false positive, false negative, and true negative voxels, correspondingly. Sensitivity is a valuable matrix of the number of TP and FN and could be computed using (4). Specificity shows the ratio of the identified background voxels to the entire background voxels and can be calculated through (5). Moreover, the AUC can be calculated using the ratio between true-positive and false-positive examples. AUC is computed using (6).
4 Experimental results and analysis
4.1 Results of segmentation on MICCAI BraTS 2018 (DATASET1)
The segmentation outcomes on DATASET1 accomplish average dice scores of 0.913, 0.874, and 0.76 for WT, TC, and ET, respectively as illustrated in Table 2. The segmentation performance for DATASET1 on the proposed architecture is considerably superior to that in DATASET2. It could be due to an intrinsic lower signal-to-noise ratio (SNR) among 1.5T and 3T MRI images. The scanning process yields sliced scans with poor resolution, characterized by diffused features and low intensity in their projection output. The identification of tissue margins presents a challenge, leading to a disruption in the efficacy of deep learning algorithms. Another possibility could be that both 1.5T and 3T images were used in the training set, enabling neural networks to develop a representation specific to both magnetic fields. The WT still has top performance and ET is the supreme challenging region.
Table 2 summarizes the results of sensitivity, dice, and specificity of ET, WT, and TC obtained on DATASET1. Training and validation accuracy and loss plots for both DATASET1 and DATASET2 are illustrated in Fig. 5(a) and (b). The training and validation loss decreases sharply between 0-5 epochs. However, the change in training and validation loss curve is very moderate, after the 10th epoch. This signifies that the model is well-trained. Some segmentation results of DATASET1 are also presented in Fig. 6.
Figure 7(a) shows the training and validation dice score plot of three regions (WT, TC, ET) differing with epoch. The validation dice score increases rapidly when the epoch is small. However, the change in training and validation dice score curve is moderate, when the epoch is greater than 10. Figure 7(a) also demonstrates that training and validation dice scores are in the order of WT>TC>ET. This might be attributed to the size of the region area. The region area of WT is greater than TC, similarly, the region area of TC is larger than ET. Therefore, it is easier to predict the region with a larger area. Thus, the Dice score is higher for the modality with a large tumor area. Figure 7(b) and (c) also represent the sensitivity and specificity in terms of line graphs.
4.2 Results of segmentation on the local dataset (DATASET2)
This research utilized a local dataset obtained from a local hospital containing 71 patients MRI data to analyze the outcomes of the developed system. The local dataset comprises 41 HGG and 30 LGG patients. We computed the average dice scores, sensitivity, and specificity to analyze the results of the suggested 3D U-Net model on DATASET2 for WT, TC, and ET, respectively. The average results of dice, sensitivity, and specificity obtained by our model for WT, TC, and ET are presented in Table 2. The whole tumor attains the highest results of segmentation by an average dice score of 0.891. Figure 5(b) shows a little low accuracy and higher loss for DATASET2. One main reason for these low results of DATASET2 is a scarcity of data. Even applying data augmentation on DATASET2, we dont have enough data which is normally required for deep learning systems. DL architectures need a huge number of data to produce high performance. Moreover, Fig. 8(a),(b) and (c) demonstrate that training and validation dice scores, sensitivity, and specificity are in the form of a line graph. Some segmentation results are presented using the proposed network on DATASET2 in Fig. 9.
4.3 Ablation study
The ablation studies are undertaken to evaluate the impact of each component in the proposed technique. To demonstrate the productivity of our system, we compared the performance of our system to that of a standard U-Net architecture on the BraTS 2018 and local private datasets. In our system, first, we changed the U-Net block with a conventional CNN convolutional layers network (namely Traditional CNN). Second, in the Standard U-Net, we have added transition layers and batch normalization after each ConvNet block. Tables 3 and 4 display the outcomes of the experiments of DATASET1 and DATASET2, respectively. After embedding transition and batch normalization layers, accordingly improves the performance of these sub-regions results, which determines the effects of transition and batch normalization layers on the brain tumor segmentation task. When compared to the Standard U-Net architecture, this work for brain tumor segmentation outperforms both datasets. It is assumed that it may be due to the upsampling layers in the U-Net module, which can filter out some of the noise data and collect various context features. Taking a look at how the standard U-Net performs in comparison to the conventional simple CNN architecture, we can see that the standard U-Net offers marginally superior results. However, ablation test results verify the usefulness of the proposed network for the brain tumor segmentation task. To evaluate our proposed approach the AUC was computed for both datasets, as shown in Fig. 10(a),(b). It is evident that the AUC curve of the designed model is higher than the Traditional CNN and Standard U-Net. A comprehensive evaluation of both datasets indicated that the proposed approach was vastly superior to Traditional CNN, and Standard U- Net. The experimental outcomes showed that the 3D U-Net model could effectively segment brain tumors from MRI scans, which might aid doctors and radiologists in early brain tumor detection.
4.4 Performance comparison with the state-of-the-art techniques
In this section, we have analyzed previously published state-of-the-art work on brain tumor segmentation and compared it with the proposed network.
Myronenko [54] won the BraTS 2018 challenge. They trained a model by applying big patches of \(160\times 192\times 128\). The variational auto-encoder branch of the model was used to reconstruct brain images. Big patches offer additional contextual evidence, allowing for a better understanding of the whole image, therefore is critical for optimizing 3D methods performance. As shown in Table 5, the suggested system outperforms other methods in the ET dice evaluation measure. Nevertheless, the disadvantage of big patches is that neural networks require extra time to learn local information. In comparison to this method, our method exceeds 0.62% and 1.38% in terms of WT and TC regions, respectively.
Isensee et al. [48] came in the second position in the challenge because they used a three-dimensional U-Net with supplementary information. They were able to demonstrate that a well-trained basic U-Net was sufficient for achieving the best possible results. To establish a reliable contrast, this study reports the results of their technique without including any additional information. As seen in Table 5, our system outperforms this method. It is worth mentioning that our strategy improves dice scores by 0.38% and 2.18% in the WT and TC regions, respectively. However, in the ET region, the Isensee et al.’s (2018) method shows slightly better results.
Zhou et al. [55] won 3rd position in the competition. We illustrate the consequence of their post-challenge modification (Zhou et al., 2019). By segmenting the tumor into three sub-regions, namely the WT, the TC, and the ET, they modified the metrics. Since they employed very small MRI patches of \(32\times 32\times 32\) for segmentation, their approach excluded global information from input images. As illustrated in Table 5, our strategy surpasses theirs by a significant margin on several measures. For instance, the proposed technique improves dice scores by 0.78% and 2.05% in the WT and TC regions, respectively. However, in the ET region to some extent, their method performs better than our model.
4.5 Discussion
This paper describes a brain tumor segmentation network using BraTS 2018 and local datasets namely DATASET1 and DATASET2, respectively. This research employs a modified 3D U-Net as a framework for model development to improve segmentation results. 3D U-Net showed its generalizability by obtaining top segmentation performance. Overall, the proposed 3D U-Net architecture offers accurate, efficient, and scalable solutions for brain MRI image segmentation tasks.
Figure 5(a) and (b) also demonstrate that the proposed model is well-trained and generalized. It attained validation dice scores are 0.913, 0.874, and 0.801 for the DATASET1 dataset and 0.891, 0.834, and 0.776 for WT, TC, and ET, respectively. Figure 11 (a),(b) and (c) presents the comparison results of DATASET1 and DATASET2. DATASET1 achieves better results in all statistical evaluation measures including dice, sensitivity, and specificity as compared with DATASET2. The reason is the shortage of MRI scans in DATASET2. The results may be improved by adding more patient data or using more data augmentation techniques. Moreover, some other reasons for the low performance of DATASET2 are explained in Sections (4.1 and 4.2) of the experimental results.
In this research, the ET class is debatably the most challenging to segment. The way this class is evaluated when the source segmentation of an image does not include this class makes it difficult. The BraTS evaluation scheme inspires postprocessing because it benefits the expulsion of small ET lesions. This property is not generally preferred in a clinical setting where the correct diagnosis of a small ET region is essential, and we suggest neglecting the postprocessing introduced in this paper.
One major shortcoming of our model is that our network is struggling to enhance tumor results. It will be of interest to train this model using ROIs as input instead of the whole MRI image. The scarcity of labeled medical imaging data is a significant impediment to model generalizability. One strategy for achieving good generalization is to augment the given dataset with more data. Post-processing techniques like CRF are effective in improving segmentation performance. It would be interesting to see how potentially meaningful models affect segmentation results. Many additional observations, modifications, and experimentations have been left for future work due to restricted computational resources and limited time. There are several ideas given below that can be implemented to enhance the proposed process even better.
5 Conclusion
To summarize, we developed an efficient 3D U-Net-based network for brain tumor substructure segmentation from non-invasive multimodal MRIs. This paper designed an improved and intelligent three-dimensional U-Net system that uses convolution blocks on the encoder and decoder side followed by transitional layers blocks for the brain tumor segmentation assignment. BraTS 2018 and the locally acquired dataset were used to evaluate the performance of the proposed approach. This method showed superior results owing to (1) the implementation of a transitional layer after each convolutional block in the encoder and decoder network, whereas the original 3D U-Net employed a simple convolution block. (2) Implementation of several skip connections to support the transmission of high-resolution features from shallow layers to deeper layers which improves the efficiency of information flow, therefore avoiding the degradation difficulty. In this study, the 3D U-Net approach is applied for brain tumor segmentation. However, this approach bears a vast amount of parameters. Furthermore, one major limitation of the proposed architecture is that the network is struggling in the performance of enhancing tumor results. Cascaded U-Net approaches include two or more encoder-decoder networks to resolve the issue of medical image segmentation. However, to overcome this limitation we can employ a U-Net-based cascaded scheme to enhance the performance of enhancing tumor class and also other regions in the future. Moreover, some advanced deep learning models can be employed to obtain more robust performance.
Data availability
The local dataset collected and/or analyzed during this research is not publicly available as it is the proprietary property of the Advanced Diagnostic Center. Any queries regarding to dataset or any raw data of this study can be directed to the first author S.A. at the email: saqibsaleem788@hotmail.com. The BRATS 2018 dataset is publicly available at https://www.med.upenn.edu/sbia/brats2018/data.html.
References
Bauer S, Wiest R, N olte L-P, Reyes M, (2013) A survey of mri-based medical image analysis for brain tumor studies. Phys Med Biol 58(13):R97
Işın A, Direkoğlu C, Şah M (2016) Review of mri-based brain tumor image segmentation using deep learning methods. Procedia Comput Sci 102:317–324
Goetz M, Weber C, Binczyk F, Polanska J, Tarnawski R, Bobek-Billewicz B, Koethe U, Kleesiek J, Stieltjes B, Maier-Hein KH (2015) Dalsa: domain adaptation for supervised learning from sparsely annotated mr images. IEEE Trans Med Imaging 35(1):184–196
Yang G, Jones TL, Barrick TR, Howe FA (2014) Discrimination between glioblastoma multiforme and solitary metastasis using morphological features derived from the p: q tensor decomposition of diffusion tensor imaging. NMR Biomed 27(9):1103–1111
Yang G, Jones TL, Howe FA, Barrick TR (2016) Morphometric model for discrimination between glioblastoma multiforme and solitary metastasis using three-dimensional shape analysis. Magn Reson Med 75(6):2505–2516
Soltaninejad M, Ye X, Yang G, Allinson N, Lambrou T et al (2014) Brain tumour grading in different mri protocols using svm on statistical features
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X (2017) Automated brain tumour detection and segmentation using superpixel-based extremely randomized trees in flair mri. Int J Comput Assist Radiol Surg 12(2):183–203
Soltaninejad M, Yang G, Lambrou T, Allinson N, Jones TL, Barrick TR, Howe FA, Ye X (2018) Supervised learning based multimodal mri brain tumour segmentation using texture features from supervoxels. Comput Methods Programs biomed 157:69–84
Wu W, Chen AYC, Zhao L, Corso JJ (2014) Brain tumor detection and segmentation in a crf (conditional random fields) framework with pixel-pairwise affinity and superpixel-level features. Int J Comput Assist Radiol Surg 9(2):241–253
Amarapur B et al (2019) Cognition-based mri brain tumor segmentation technique using modified level set method. Cogn Technol Work 21(3):357–369
Olabarriaga SD, Smeulders AWM (2001) Interaction in the segmentation of medical images: A survey. Med Image Anal 5(2):127–142
Yao J (2006) Image processing in tumor imaging. New techniques in oncologic imaging, pp 79–102
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom Wiest R et al (2014) The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans Med Imaging 34(10):1993–2024
Iqbal S, Qureshi AN, Aurangzeb K, Alhussein M, Haider SI, Rida I (2023) Amiac: adaptive medical image analyzes and classification, a robust self-learning framework. Neural Comput Appl pp 1–29
Bozkurt F (2023) Skin lesion classification on dermatoscopic images using effective data augmentation and pre-trained deep learning approach. Multimed Tools Appl 82(12):18985–19003
Bozkurt F (2022) A deep and handcrafted features-based framework for diagnosis of covid-19 from chest x-ray images. Concurr Comput Pract Experience 34(5):e6725
Ali S, Li J, Pei Y, Rehman KU (2022) A multi-module 3d u-net learning architecture for brain tumor segmentation. In: International conference on data mining and big data, Springer, pp 57–69
Ali S, Li J, Pei Y, Khurram R, Rehman Ku, Rasool AB (2021) State-of-the-art challenges and perspectives in multi-organ cancer diagnosis via deep learning-based methods. Cancers 13(21):5546
Shelhamer E, Long J, Darrell T (2016) Fully convolutional networks for semantic segmentation. IEEE Trans Pattern Anal Mach Intell 39(4):640–651
Ronneberger O, Fischer P, Brox T (2015) U-net: Convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 234–241
Qin C, Yujie W, Liao W, Zeng J, Liang S, Zhang X (2022) Improved u-net3+ with stage residual for brain tumor segmentation. BMC Med Imaging 22(1):1–15
Zhao X, Yihong W, Song G, Li Z, Zhang Y, Fan Y (2018) A deep learning model integrating fcnns and crfs for brain tumor segmentation. Med Image Anal 43:98–111
Pereira S, Alves V, Silva CA (2018) Adaptive feature recombination and recalibration for semantic segmentation: application to brain tumor segmentation in mri. In: International conference on medical image computing and computer-assisted intervention, Springer, pp 706–714
Badrinarayanan V, Kendall A, Cipolla R (2017) Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans Pattern Anal Mach Intell 39(12):2481–2495
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Iqbal S, Ghani MU, Saba T, Rehman A (2018) Brain tumor segmentation in multi-spectral mri using convolutional neural networks (cnn). Microscopy Res Tech 81(4):419–427
Dong H, Yang G, Liu F, Mo Y, Guo Y (2017) Automatic brain tumor detection and segmentation using u-net based fully convolutional networks. In: Annual conference on medical image understanding and analysis, pp 506–517. Springer
Zhou Z, Rahman Siddiquee MM, Tajbakhsh N, Liang J (2018) Unet++: A nested u-net architecture for medical image segmentation. In: Deep learning in medical image analysis and multimodal learning for clinical decision support, pp 3–11. Springer
Tu Z, Bai X (2009) Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE transactions on pattern analysis and machine intelligence 32(10):1744–1757
Meier R, Bauer S, Slotboom J, Wiest R, Reyes M (2013) A hybrid model for multimodal brain tumor segmentation. Multimodal Brain Tumor Segmentation 31:31–37
Meier R, Bauer S, Slotboom J, Wiest R, Reyes M (2014) Appearance-and context-sensitive features for brain tumor segmentation. Proceedings of MICCAI BRATS Challenge, pp 020–026
Bauer S, Nolte LP, Reyes M (2011) Fully automatic segmentation of brain tumor images using support vector machine classification in combination with hierarchical conditional random field regularization. In: International conference on medical image computing and computer-assisted intervention, pp 354–361. Springer
Sikka K, Sinha N, Singh PK, Mishra AK (2009) A fully automated algorithm under modified fcm framework for improved brain mr image segmentation. Magn Reson Imaging 27(7):994–1004
Pinto A, Pereira S, Correia H, Oliveira J, DMLD Rasteiro, Silva CA (2015) Brain tumour segmentation based on extremely randomized forest with high-level features. In: 2015 37th annual international conference of the IEEE engineering in medicine and biology society (EMBC), pp 3037–3040. IEEE
Hussain S, Anwar SM, Majid M (2017) Brain tumor segmentation using cascaded deep convolutional neural network. In 2017 39th annual International Conference of the IEEE engineering in medicine and biology Society (EMBC), pp 1998–2001. IEEE
Havaei M, Davy A, Farley DW, Biard A, Courville A, Bengio Y, Pal C, Jodoin PM, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
Hu K, Gan Q, Zhang Y, Deng S, Xiao F, Huang W, Cao C, Gao X (2019) Brain tumor segmentation using multi-cascaded convolutional neural networks and conditional random field. IEEE Access 7:92615–92629
Pereira S, Pinto A, Alves V, Silva CA (2015) Deep convolutional neural networks for the segmentation of gliomas in multi-sequence mri. In: BrainLes 2015, pp 131–143. Springer
Hussain S, Anwar SM, Majid M (2018) Segmentation of glioma tumors in brain using deep convolutional neural network. Neurocomputing 282:248–261
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2818–2826
Kamnitsas K, Ferrante E, Parisot S, Ledig C, Nori AV, Criminisi A, Rueckert D, Glocker B (2016) Deepmedic for brain tumor segmentation. In: International workshop on Brainlesion: Glioma, multiple sclerosis, stroke and traumatic brain injuries, Springer, pp 138–149
Wang G, Li W, Ourselin S, Vercauteren T (2017) Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: International MICCAI brainlesion workshop, pp 178–190. Springer
Albishri AA, Shah SJH, Kang SS, Lee Y (2022) Am-unet: automated mini 3d end-to-end u-net based network for brain claustrum segmentation. Multimed Tools Appl 81(25):36171–36194
Punn NS, Agarwal S (2021) Multi-modality encoded fusion with 3d inception u-net and decoder model for brain tumor segmentation. Multimed Tools Appl 80(20):30305–30320
Raza R, Bajwa UI, Mehmood Y, Anwar MW, Jamal MH (2023) dresu-net: 3d deep residual u-net based brain tumor segmentation from multimodal mri. Biomed Signal Proc Control 79:103861
Li P, Wu W, Liu L, Serry FM, Wang J, Han H (2022) Automatic brain tumor segmentation from multiparametric mri based on cascaded 3d u-net and 3d u-net++. Biomed Signal Proc Control 78:103979
Isensee F, Jäger PF, Kohl SAA, Petersen J, Maier-Hein KH (2019) Automated design of deep learning methods for biomedical image segmentation. arXiv preprint arXiv:1904.08128
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) No new-net. In: International MICCAI Brainlesion Workshop, pp 234–244. Springer
McKinley R, Meier R, Wiest R (2018) Ensembles of densely-connected cnns with label-uncertainty for brain tumor segmentation. In: International MICCAI Brainlesion Workshop, pp 456–465. Springer
Li H, Li A, Wang M (2019) A novel end-to-end brain tumor segmentation method using improved fully convolutional networks. Comput Biol Med 108:150–160
Kayalibay B, Jensen G, van der Smagt P (2017) Cnn-based segmentation of medical imaging data. arXiv preprint arXiv:1701.03056
Battalapalli D, Rao BP, Yogeeswari P, Kesavadas C, Rajagopalan V (2022) An optimal brain tumor segmentation algorithm for clinical mri dataset with low resolution and non-contiguous slices. BMC Med Imaging 22(1):1–12
Ngo DK, Tran MT, Kim SH, Yang HJ, Lee GS (2020) Multi-task learning for small brain tumor segmentation from mri. Appl Sci 10(21):7790
Myronenko A (2018) 3d mri brain tumor segmentation using autoencoder regularization. In: International MICCAI Brainlesion Workshop, pp 311–320. Springer
Zhou C, Ding C, Wang X, Lu Z, Tao D (2020) One-pass multi-task networks with cross-task guided attention for brain tumor segmentation. IEEE Trans Image Proc 29:4516–4529
Hu Y, Liu X, Wen X, Niu C, Xia Y (2018) Brain tumor segmentation on multimodal mr imaging using multi-level upsampling in decoder. In: International MICCAI Brainlesion Workshop, pp 168–177. Springer
Carver E, Liu C, Zong W, Dai Z, Snyder JM, Lee J, Wen N (2018) Automatic brain tumor segmentation and overall survival prediction using machine learning algorithms. In: International MICCAI Brainlesion Workshop, pp 406–418. Springer
Islam M, Jose V, Ren H (2018) Glioma prognosis: Segmentation of the tumor and survival prediction using shape, geometric and clinical information. In: International MICCAI Brainlesion Workshop, pp 142–153. Springer
Funding
No external funding was received for this study.
Author information
Authors and Affiliations
Contributions
Each author took part in the present works conception and/or design. Saqib Ali and Rooha Khurram were responsible for carrying out the tasks of material preparation, and original draft writing. Ghulam Mujtaba helps in data collection and data analysis. Khalil ur Rehman and Zareen Sakhawat helped in reviewing and editing the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Informed consent statement
Institutional Review Board (IBR) approval was obtained. From, Advanced Diagnostic Center (Pvt) Ltd, and Advanced International Hospital Institutional Review Board (Approval number: MR-20-2543). All procedures performed in this study were by the ethical standards of the institutional and/or local research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This study has an IRB approval of an informed consent waiver.
Conflict of interest
The authors declare that there is no conict of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Saqib Ali and Rooha Khurram contributed equally to this study.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ali, S., Khurram, R., Rehman, K.u. et al. An improved 3D U-Net-based deep learning system for brain tumor segmentation using multi-modal MRI. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-19406-2
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-19406-2