Abstract
With deep learning playing a crucial role in biomedical image segmentation, multi-modality fusion-based techniques have gained rapid growth. For any radiologist, identification and segmentation of brain tumor (gliomas) via multi-sequence 3D volumetric MRI scan for diagnosis, monitoring, and treatment, are complex and time-consuming tasks. The brain tumor segmentation (BraTS) challenge offers 3D volumes of high-graded gliomas (HGG), and low-graded gliomas (LGG) MRI scans with four modalities: T1, T1c, T2 and FLAIR. In this article, the tumor segmentation is performed on the preprocessed multi-modalities by proposed 3D deep neural network components: multi-modalities fusion, tumor extractor, and tumor segmenter. The multi-modalities fusion component uses the deep inception based encoding to merge the multi-modalities. Tumor extractor component is passed with the fused images to recognise the tumor patterns using the 3D inception U-Net model. Finally, tumor segmenter utilises the progressive approach to decode the extracted feature maps into the tumor regions. The architecture segments each lesion region into the whole tumor (WT), core tumor (CT), and enhancing tumor (ET) using the five target classes: background, necrosis, edema, enhancing tumor and non-enhancing tumor. To tackle the class imbalance problem, the weighted segmentation loss function is proposed based on the dice coefficient and the Jaccard index. This article illustrates the significance of each component on the BraTS 2017 and 2018 datasets by achieving better segmentation results. The performance of the proposed approach is also compared with the other state-of-the-art methods.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Deep learning is bringing revolution in technological advancement for the welfare of living society, especially in the domain of biomedical image analysis [9]. Diagnosis and treatment of any severe health hazard is followed by critical observation from the medical scans such as magnetic resonance imaging (MRI), computed tomography (CT or CAT), X-Rays, etc. Analysing such scans require experienced neuro-radiologist, which however is a tedious and time consuming task. Deep learning image processing techniques [17] are utilised to mitigate the complication of such tasks and produce faster and efficient diagnoses.
1.1 Multi-modalities fusion
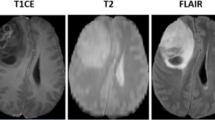
T1 weighted (T1), T1 contrast enhanced (T1c), T2 weighted (T2), and T2 fluid attenuated inversion recovery (FLAIR) are the most common sequences or modalities captured using MRI. Figure 1 shows a slice of each of these modalities that highlight certain tissues of the brain. Table 1 presents the comparison of each of these modalities in presenting the brain tissues [23]. Since tumors vary in shape, size, appearance, and location, thereby forms the complicated structure and makes it hard to fully identify and segment the lesion regions using unary-modality sequences. The fusion of multi-modalities offers the multi-information of the brain tissues for comprehensive analysis to improve upon the tumor segmentation results [18]. Fusion approaches are broadly classified into three different levels, input level, layer level, and decision level [11]. Zhe et al. [11] observed that input level fusion produced the best segmentation results that completely exploits the information from the multi-modalities.
1.2 Overview
Following from the brain tumor segmentation (BraTS) challenge, in this article, an efficient approach is proposed for the tumor segmentation via 3D deep neural network components, inspired by the success of inception U-Net architecture [24] in 2D segmentation of nucleus. Figure 2 illustrates the proposed framework, divided into the following components: data preprocessor, multi-modalities fusion, tumor extractor, and tumor segmenter. In data preprocessing, the input sequences are corrected using advanced normalisation tools (ANTs) [1] to remove the bias field distortion caused by the non-uniformity of the magnetic fields [15]. Later, the scans are cropped to maximum fit the brain structure, which reduces the number of features without the loss of information. The multi-modalities fusion component uses the deep inception based encoding to merge the input sequences T1, T1c, T2, and FLAIR with the objective to make it more informative. Then the tumor extractor component is passed with the encoded scans to extract the feature maps for gliomas using the 3D inception U-Net model. Finally, tumor segmenter decodes the obtained feature maps into the tumor regions as whole tumor (WT), core tumor (CT), and enhancing tumor (ET) from the five target classes as background, necrosis, edema, enhancing tumor and non-enhancing tumor.
For the training phase, the input is normalised using the instance normalisation technique [33], and a segmentation loss function is proposed based on the dice coefficient and intersection-over-union metrics to address the class imbalance problem. The proposed network is implemented using the tensorflow-gpu version 1.12.0 and trained on a high-performance computing (HPC) environment of two Nvidia Tesla V-100 16 GB GPUs. Mostly, the models proposed for the BraTS challenge utilise F1 score (dice coefficient) as an evaluator metric for performance measurement; hence, in this article, the dice coefficient is considered for comparative analysis. The experimental results are compared on the BraTS 2017 and 2018 datasets with the other pre-existing approaches along with the ablation study of the proposed components and impact of different loss functions.
2 Related work
The progressive advancements in deep learning and computer vision methodologies are expanding the dimensions towards the applications in the healthcare sector [21]. With the evolving medical image acquisition systems, multi-modality based segmentation [2, 3, 20] approach is widely studied. Most of the approaches are being proposed based on the modifications in the architectural design, training strategy, fusion methodology, pre and post-processing of data. Table 2 highlights the recent contributions and achievements in the field of MRI segmentation. The U-Net topology [26] is the most widely used base architecture for biomedical image segmentation as observed from the findings of Isensee et al. [14]. The symmetrical contraction and expansion pathways of convolution and pooling layers in U-Net discovers the high-level features and localise the objects in the original feature space, respectively. Later, with the advent of 3D medical images, e.g. MRI, CT, various approaches employed 3D convolution networks [37, 39] to manoeuvre the spatial contextual information of the image; whereas the expense of its computation cost is reduced by GPU accelerated hardware resources.
Wang et al. [34] proposed the fusion based brain tumor segmentation approach using anisotropic convolution network to hierarchically segment the brain tumor regions into WT, CT, and ET one after the other. The segmentation results are further fused across multiple views (axial, sagittal, and coronal). Though the authors achieved the dice scores of 0.78 (WT), 0.87 (CT), and 0.77 (ET) on the BraTS 2017 dataset, but did not consider the inter class correlation. Later, Chen et al. [6] employed a 3D convolution neural network (CNN) based model with partial fusion in which a two-stage model is proposed. The first stage uses patches (38×38×38) of FLAIR and T2 to segment WT, whereas the second stage uses patches of T1, T1c and output of the first stage to segment CT and ET. This approach achieved the dice score of 0.72 (WT), 0.83 (CT), and 0.81 (ET) on BraTS 2017. Afterwards, Zhou et al. [38] proposed one pass multi-task convolution network (OM-Net) to improve the performance of the model [34], using curriculum learning that preserves the class correlation through shared backbone model. This model is first trained to segment the WT region from the extracted correlated features then the resulting image is utilised to segment the CT region, and finally ET region is segmented similarly. The authors achieved better segmentation results than previous work [34] on BraTS 2017 dataset, but the approach requires substantial resources to train 14M parameters. Since the segmentation of the regions of varying size requires different sizes of the receptive field, Qin et al. [25] proposed the autofocus convolution that uses multiple convolution layers of varying dilation rates. The dilated convolution [36] increases the receptive field of the network without increasing the number of parameters. The proposed method achieved promising results on multi-organ and brain tumor segmentation task. Later, due to the increasing popularity of generative adversarial networks (GANs) [10], Yang et al. [35] proposed the contour aware residual network and adversarial training framework, for brain tumor segmentation. The residual U-Net is utilised as a segmentation generator to output the identified tumor regions which are later compared with ground truth masks under the supervision of the discriminator neural network. The authors achieved competitive and robust results on BraTS 2018 dataset, but additional resources are needed to train discriminator. Later, Myronenko et al. [22] employed autoencoding regularisation to learn important features that are effective for 3D MRI image classification. Indeed, the approach was ranked first in BraTS 2018 challenge. In Hatamizadeh et al. [12] boundary aware convolutional neural network with end-to-end training is proposed to segment organ boundary information using the edge-aware loss terms. The network utilises the multi-scale contextual information while fusing the boundary level feature maps via tuning of weight parameters. The approach achieved more accurate segmentation results on BraTS 2018 as compared to state-of-the-art U-Net and V-Net architectures.
Following from these notions, in this article, an efficient framework is proposed based on the inception U-Net architectural design [24] with the idea to exploit multi-information of deep encoded fused modalities to extract tumor patterns and later segment into the complete tumor and its sub-regions using the task specific components.
3 Proposed framework
The overall architectural design consists of inception module, data preprocessing, multi-modality encoded fusion, tumor extractor and tumor segmenter units with 10.5M trainable parameters. The preprocessed multi-modalities are progressively fused to gain multi-information that enables the tumor extractor component to analyse the deep patterns using the contraction and expansion pathways associated with the tumor regions. The contraction path extracts the features for each downsampled sequence that is carried through the max-pooling operation. Whereas, the expansion path attempts to retain the learned features while progressively restoring the dimension of the activations using the transposed convolutions. Later, the tumor regions are segmented into WT, ET, and CT by tumor segmenter component that merges the features learned at different hierarchical layers of tumor extractor component with the help of upsampling operation. All convolution operations are activated with the most widely used leaky rectified linear unit (LReLU) activation function [19], except that the final convolution operation at the output layer follows the sigmoid activation function [28] for voxel classification. Each convolution layer is also accompanied by the instance normalisation [33] to standardise the activations and accelerate the training process. The training phase is employed with the objective function defined using the dice coefficient and Jaccard index to assist in fine-tuning of weight parameters and improving the dice score of the tumor segmentation.
3.1 Inception module
It is quite unlikely to use a single type of convolution to detect objects of varying size and shape like brain tumor. Following the concept of Google’s inception neural network [30], the inception segment is composed of concatenated convolutions of 1×1, 3×3, and 5×5 kernels which is accompanied by 1×1 convolution to reduce the 3D channels. Each of these convolutions is followed by instance normalisation [33], LReLU activation function [19] and downsampled enabled max pooling. Figure 3 illustrates the architecture of the inception module for the proposed framework.
3.2 Data preprocessing
MRI multi-modality scans incur the problem of the bias field, which is caused by the intensity inhomogeneity that corrupts the MRI sequences [29], which therefore makes it difficult for the desired analysis like segmentation. In this article, ANT’s N4BiasFieldCorrection [32] is utilised to correct the MRI sequences and perform the bias correction. In order to have lossless dimensionality reduction (reducing the number of features), dark slices and voxels are removed for each modality resulting in reduced 3D dimensions, i.e. from (240×240×155) to (144×144×144) as shown in Fig. 4. Finally, the volumes are normalised as (x − μ)/(σ + ε) where μ is mean, and σ is the standard deviation of input intensities, and ε is a constant with a minimal value, added to avoid the division by zero cases.
3.3 Multi-modality fusion
Figure 5 illustrates the inception based encoded fusion approach for the T1, T1c, T2, and FLAIR modalities. Instead of performing direct fusion along the batch axis, each preprocessed modality is passed as an input to the multi-modality fusion component consisting of hierarchical inception blocks. Each MRI sequence is initially processed through inception module (IM) with a filter followed by concatenation of T1 with T1c and T2 with FLAIR resulting modalities, forming two sequences of size 2×144×144×144. Later, the process is repeated for the resulting sequences to form 4×144×144×144 dimension. The fused output of this component is created via the concatenation of 2×(2×144×144×144) and 1×(4×144×144×144), resulting in a sequence of dimension 8×144×144×144. Each inception block extracts features from the associated modality, which is progressively encoded and fused to incorporate weighted multi-information. The encoded fused output is then passed to the tumor extractor for deep patterns discovery of lesion regions. After extensive trials of manipulating the architectural design, the proposed fusion approach is claimed to be effective in improvement of the segmentation results.
3.4 Tumor extractor
The tumor extractor component follows from the results of the fusion component and aims to understand the deep patterns associated with the brain tumor. It follows contraction and expansion paths for the discovery of tumor patterns and reconstruction of identified tumor regions into the same dimension as that of input respectively. Figure 6 shows the residual building block of the tumor extractor component consisting of IM blocks and a spatial dropout [31] of 30% for regularisation. The dropout layer tends to prevent the overfitting by reducing the correlation among the learned features in the intermediate blocks [13]. The spatial dropout deactivates the entire feature map along the axis, which tends to improve the performance than a standard dropout.
The complete tumor extractor architecture is developed using the interconnected layers of residual inception blocks to form the U-Net structure [26], as shown in Fig. 7. The residual skip connection [8] shown as the horizontal arrows in Fig. 7, enables to preserve the features learned in the previous layers. It avoids the vanishing gradient problem and aids in the upsampling of MRI sequences using transposed convolutions in the reconstruction or expansion phase. The results of the intermediate RI-32, RI-16, RI-8, and RI-3 layers are utilised for final segmentation into WT, ET, and CT regions by tumor segmenter component.
3.5 Tumor segmenter
The tumor segmenter decodes the feature maps from RI-32, RI-16, RI-8, and RI-3 layers of tumor extractor into WT, CT, and ET tumor regions, as illustrated in Fig. 8. In order to set the number of channels equivalent to the number of classes, RI layers are passed through 1×1 convolution with three filters followed by spatial dropout [31] to form a volume of size (3×H ×W ×D). The convoluted volumes are upsampled by mapping a voxel value of (1×1×1) to (2×2×2) dimension [7], in order to match the dimensions with higher layers such that it becomes compatible for the addition operation with the higher layers along the batch axis which lastly converges at (3×144×144×144). The final activation is applied as sigmoid to get intensities of each voxel within the range [0,1] and compute the segmentation loss for predicted WT, CT and ET regions.
4 Experimentation and results
4.1 Dataset setup
BraTS dataset is comprised of 3D volumes of MRI scans taken in four modalities of T1, T1c, T2, and FLAIR with 155 sequences of size 244×244 forming complete dimension as 244×244×155. Each of these modalities highlight distinct features of the tumor regions. The BraTS 2017 and 2018 challenges offer dataset consisting of MRI scans of 285 patients along with the ground truth. The datasets include five classes as background, necrosis, edema, enhancing tumor and non-enhancing tumor which forms three tumor regions as whole tumor (WT), core tumor (CT), and enhancing tumor (ET). Each dataset comprises of approximately 61%, 24%, and 15% distribution of WT, CT, and ET regions, respectively, resulting in highly imbalanced dataset. Each sequence is preprocessed with N4BiasFieldCorrection [32] and resampled to remove the dark voxels forming the cuboid structure of dimension 144×144×144. Each dataset is randomly split into 80% of the training set and 20% of the validation set. The training and validation sets are kept same throughout the experimental trials for comparative analysis. Due to the limited availability of the validation data, the results are compared on the validation split set of BraTS 2017 and 2018 datasets.
4.2 Training and testing
The proposed framework is trained and tested on BraTS 2017 and 2018 datasets. The training phase is assisted with the 5-fold cross validation, early-stopping to avoid the overfitting problem [4], and Adam as a learning rate optimiser [27]. The most widely used metrics for model training and performance analysis in segmentation are the dice coefficient (F1-score) and intersection-over-union (IoU / Jaccard index). In this article, trials were made with the dice loss, IoU loss and weighted average of IoU-dice loss as shown in the ablation study under results and discussion section. With deep insight into the literature, it is observed that generally, the authors use dice metric as an objective function (1), but the IoU metric as given in (2) is comparatively more effective in punishing the false predictions, thereby making it more suitable for imbalanced datasets. For example, consider that a single voxel is available in an image, but the model detects one more voxel along with the genuine one. The model’s dice and IoU score would be 2/3 and 1/2, respectively. Compared to dice, the lower IoU score penalises the training weights to improve upon the false prediction.
where TP indicates true positive predictions (correctly classified), FP indicates false positive (wrongly classified), and FN indicates false negatives (missed objects) of each voxel.
Equations (3) and (4) represent the dice loss (DCL), and IoU loss (IoUL) metrics as the loss functions and their weighted average (DIUL) is defined in (5). With several experiments, it is found that the proposed loss function DIUL offers a significant improvement over the segmentation results in the proposed framework than using dice coefficient loss and IoU loss individually, but with the cost of slow convergence, and hence increases the training time.
where y and p(y) indicates the ground truth and predicted values.
The backpropagation process fine tunes the training weights or parameters with the concern to minimise the DIUL value (L) by computing the loss gradient with respect to the prediction as shown in (6).
4.3 Results and discussion
The proposed model outputs the 3D volume of the segmented mask consisting of tumor regions WT, ET, and CT. Figure 9 displays the qualitative results for the MRI sequences viewed over the axial plane with randomly chosen slices. The visual representation and dice score confirm that results are quite close to the ground truth values of WT, CT, and ET.
Table 3 presents the result of the proposed framework in the form of dice coefficient values for WT, CT and ET, along with the comparative analysis with other popular approaches proposed for the BraTS 2017 and 2018 challenges. For BraTS 2017, the proposed framework outperforms the other approaches in segmenting the CT and ET regions with dice score of 0.88 and 0.83, respectively. In contrast, Zhou et al. [38] approach produce slightly better dice score to identify WT regions. Similarly, in BraTS 2018 challenge, the model proposed by Yang et al. [35] works better to identify the whole tumor and shows similar results to the proposed approach to segment CT and ET regions. We further tried to improve the results with post-processing technique, conditional random fields (CRF) [16] but resulted in degrading the performance of the model. In contrast to other methods, the main reason for the success of the proposed approach follows from the organised architectural design, comprised of task specific 3D deep inception based components and optimised loss function that tackles the class imbalance problem.
The improvement with reference to each of the proposed components (multi-modalities fusion, tumor extractor, tumor segmenter) and the objective function is shown in Table 4. Figure 10 shows the training loss per epoch and box plot of dice score on the validation set for all combinations of the model mode and loss function, as shown in Table 4.
In order to develop a base platform for volumetric segmentation of brain tumor, 3D U-Net model is utilised based on the standard U-Net model [26], where convolution, pooling, and other operations are extended to the third dimension. As shown in Table 4, the plain 3D U-Net model has displayed the worst performance. The non-inception model is developed by replacing the inception convolutions with the 3D convolutions while retaining other proposed components of the framework with the aim to identify the effectiveness of the inception convolution [30] module in the proposed approach. The dice value for non-inception model mode outperforms the plain 3D U-Net mode, especially in identifying the ET regions. Similarly, for non-encoded fusion mode, the multi-modality fusion component is replaced with the simple input level concatenation of the multi-modalities, while retaining other components of the framework. This model mode has some improvements in dice score over the other model modes, but still not close to the results of the proposed model. Despite bad results from the other model modes, it can be observed that in most of the cases, the combination of dice loss and IoU loss tends to improve the performance and achieves better dice score.
5 Conclusion
In the present article, the complex task of brain tumor segmentation is divided into multiple components: data-preprocessing, multi-modalities fusion, tumor extractor, and tumor segmenter. The proposed approach achieved significant improvements on BraTS 2017 and BraTS 2018 datasets by exploiting the advantages of inception convolutions, 3D U-Net architecture, and segmentation loss function that is based on Jaccard index and dice coefficient. With exhaustive experiments, it was found that each segment of the proposed framework serves its desired purpose in understanding the deep patterns associated with the tumor regions and contributing towards improvement in the segmentation results. As an extension, the work of the present article could further be improved by employing hybrid, cascaded, ensembled or other learning approaches. It is believed that this work can also be extended to wide variety of biomedical image analyses like image registration, disease quantification, etc. and other application areas.
References
Avants BB, Tustison N, Song G (2009) Advanced normalization tools (ants). Insight J 2(365):1–35
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby JS, Freymann JB, Farahani K, Davatzikos C (2017) Advancing the cancer genome atlas glioma mri collections with expert segmentation labels and radiomic features. Sci Data 4:170117
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M, et al. (2018) Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the brats challenge. arXiv:1811.02629
Brownlee J (2018) A gentle introduction to early stopping to avoid overtraining neural networks. https://machinelearningmastery.com/early-stopping-to-avoid-overtraining-neural-network-models/. [Online; accessed March 02, 2020]
Chen C, Liu X, Ding M, Zheng J, Li J (2019) 3d dilated multi-fiber network for real-time brain tumor segmentation in mri. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 184–192
Chen L, Wu Y, DSouza AM, Abidin AZ, Wismüller A, Xu C (2018) Mri tumor segmentation with densely connected 3d cnn. In: Medical imaging 2018: image processing. International society for optics and photonics, vol 10574, p 105741F
Chollet F (2015) Keras: deep learning for humans. https://github.com/fchollet/keras. [Online; accessed March 02, 2020]
Drozdzal M, Vorontsov E, Chartrand G, Kadoury S, Pal C (2016) The importance of skip connections in biomedical image segmentation. In: Deep learning and data labeling for medical applications. Springer, Berlin, pp 179–187
Esteva A, Robicquet A, Ramsundar B, Kuleshov V, DePristo M, Chou K, Cui C, Corrado G, Thrun S, Dean J (2019) A guide to deep learning in healthcare. Nat Med 25(1):24–29
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. In: Advances in neural information processing systems, pp 2672–2680
Guo Z, Li X, Huang H, Guo N, Li Q (2019) Deep learning-based image segmentation on multimodal medical imaging. IEEE Trans Radiat Plasma Med Sci 3(2):162–169
Hatamizadeh A, Terzopoulos D, Myronenko A (2019) End-to-end boundary aware networks for medical image segmentation. In: International workshop on machine learning in medical imaging. Springer, Berlin, pp 187–194
Hinton GE, Srivastava N, Krizhevsky A, Sutskever I, Salakhutdinov RR (2012) Improving neural networks by preventing co-adaptation of feature detectors. arXiv:1207.0580
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) No new-net. In: International MICCAI brainlesion workshop. Springer, Berlin, pp 234–244
Juntu J, Sijbers J, Van Dyck D, Gielen J (2005) Bias field correction for mri images. In: Computer recognition systems. Springer, Berlin, pp 543–551
Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3d cnn with fully connected crf for accurate brain lesion segmentation. Med Image Anal 36:61–78
Ker J, Wang L, Rao J, Lim T (2017) Deep learning applications in medical image analysis. IEEE Access 6:9375–9389
Lahat D, Adali T, Jutten C (2015) Multimodal data fusion: an overview of methods, challenges, and prospects. Proc IEEE 103(9):1449–1477
Maas AL, Hannun AY, Ng AY (2013) Rectifier nonlinearities improve neural network acoustic models. In: Proc icml, vol 30, p 3
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, et al. (2014) The multimodal brain tumor image segmentation benchmark (brats). IEEE Trans Med Imaging 34(10):1993–2024
Miotto R, Wang F, Wang S, Jiang X, Dudley JT (2018) Deep learning for healthcare: review, opportunities and challenges. Brief Bioinform 19 (6):1236–1246
Myronenko A (2018) 3d mri brain tumor segmentation using autoencoder regularization. In: International MICCAI brainlesion workshop. Springer, Berlin, pp 311–320
Preston DC (2006) Magnetic resonance imaging (mri) of the brain and spine: basics. MRI Basics, Case Med 30
Punn NS, Agarwal S (2020) Inception u-net architecture for semantic segmentation to identify nuclei in microscopy cell images. ACM Trans Multimed Comput Commun Appl (TOMM) 16(1):1–15
Qin Y, Kamnitsas K, Ancha S, Nanavati J, Cottrell G, Criminisi A, Nori A (2018) Autofocus layer for semantic segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 603–611
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 234–241
Ruder S (2016) An overview of gradient descent optimization algorithms. arXiv:1609.04747
Sibi P, Jones SA, Siddarth P (2013) Analysis of different activation functions using back propagation neural networks. J Theor Appl Inf Technol 47 (3):1264–1268
Song J, Zhang Z (2019) Brain tissue segmentation and bias field correction of mr image based on spatially coherent fcm with nonlocal constraints. Computational and mathematical methods in medicine 2019
Szegedy C, Liu W, Jia Y, Sermanet P, Reed S, Anguelov D, Erhan D, Vanhoucke V, Rabinovich A (2015) Going deeper with convolutions. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 1–9
Tompson J, Goroshin R, Jain A, LeCun Y, Bregler C (2015) Efficient object localization using convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 648–656
Tustison NJ, Shrinidhi K, Wintermark M, Durst CR, Kandel BM, Gee JC, Grossman MC, Avants BB (2015) Optimal symmetric multimodal templates and concatenated random forests for supervised brain tumor segmentation (simplified) with antsr. Neuroinformatics 13(2):209–225
Ulyanov D, Vedaldi A, Lempitsky V (2016) Instance normalization: the missing ingredient for fast stylization. arXiv:1607.08022
Wang G, Li W, Ourselin S, Vercauteren T (2017) Automatic brain tumor segmentation using cascaded anisotropic convolutional neural networks. In: International MICCAI brainlesion workshop. Springer, Berlin, pp 178–190
Yang HY, Yang J (2018) Automatic brain tumor segmentation with contour aware residual network and adversarial training. In: International MICCAI brainlesion workshop. Springer, Berlin, pp 267–278
Yu F, Koltun V (2015) Multi-scale context aggregation by dilated convolutions. arXiv:1511.07122
Yu L, Yang X, Chen H, Qin J, Heng PA (2017) Volumetric convnets with mixed residual connections for automated prostate segmentation from 3d mr images. In: Thirty-first AAAI conference on artificial intelligence
Zhou C, Ding C, Lu Z, Wang X, Tao D (2018) One-pass multi-task convolutional neural networks for efficient brain tumor segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, Berlin, pp 637–645
Zhou X, Takayama R, Wang S, Hara T, Fujita H (2017) Deep learning of the sectional appearances of 3d ct images for anatomical structure segmentation based on an fcn voting method. Med Phys 44(10):5221–5233
Acknowledgements
We thank our institute, Indian Institute of Information Technology Allahabad (IIITA), India and Big Data Analytics (BDA) lab for allocating the centralised computing facility and other necessary resources to perform this research. We extend our thanks to our colleagues for their valuable guidance and suggestions.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Punn, N.S., Agarwal, S. Multi-modality encoded fusion with 3D inception U-net and decoder model for brain tumor segmentation. Multimed Tools Appl 80, 30305–30320 (2021). https://doi.org/10.1007/s11042-020-09271-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09271-0