Abstract
Brain-Computer Interface (BCI) enables human beings to interact with the outside world through brain intention. Human-computer interaction (HCI) based on electroencephalogram (EEG) has become the main research direction in the field of BCI. Though many achievements have been made in EEG research recently, the lack of sample data and individual differences, effective motor imagery (MI) classification based on EEG signals is still a challenge. Compared with the 2D and 3D CNN models that are widely used, however, there are few researches on extracting EEG sequence features using 1D CNN model. To this end, considering the temporal structure of multi-channel EEG signals, we propose a EEG-Based temporal one-dimensional convolution neural network (ETIODCNN) to classify MI. First, we extract temporal correlation from EEG signals by introducing the core blocks. Then, we use the global average pooling (GAP) layer and the fully connected (F) layer to fuse the temporal series features, and realize classification task. Our model can automatically learn effective features from EEG signals. We trained and tested the proposed method on BCI competition IV datasets (BCICID). The experiments were conducted on two open-source EEG datasets. The comparison results show that this method has good performance in real-time and accuracy.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain-computer interface (BCI) can enable disabled people to interact with the outside world through brain signals [48]. Moreover, BCI plays a great role in virtual reality and meta-universe [23].
BCI is a novel approach that directly controls external equipment by bypassing the typical peripheral nervous system. Thanks to the merits of the lower hardware cost, the higher temporal resolution, and portability, EEG-based BCIs are widely used. Many recent studies on BCI have obtained achievements in the field of health, such as emotion recognition [49]. The EEG signal is easy to obtain in a non-invasive way. It not only has high time resolution, but also can be obtained in real-time. Therefore, at present, EEG is regarded as the most practical method for BCI.
To control external devices, such as ground vehicle [51], wheelchairs [1, 26], robot arms [44], through imagination, MI can be interpreted as human imaginary movement without actual movement. We can get the corresponding EEG signal from some active brain areas. The BCI system is still frequently used in the field of rehabilitation engineering today. According to certain researchers, MI-BCI has a high practicability in the rehabilitation of limb illnesses, which has prompted many researchers to invest in MI-BCI research [47]. The results of these practical applications have attracted more attention from experts in the field.
However, the current EEG-based BCI system is still immature and faces many challenges. Firstly, EEG signals usually contain a lot of noise. In addition to the system noise caused by circuit system and power line interference, EEG signals also have some unique inherent noise. In addition, studies have shown that subjects’ emotions will also bring uncertain noise to EEG signals [29]. It is difficult to ensure that participants focus on the task throughout experiments. Secondly, compared with image or video analysis tasks, EEG-based analysis tasks usually involve only 8-128 signal channels, leading to limited signal resolution. Thirdly, the correlation between EEG signals and the corresponding brain activities is fuzzy. For example, in other pattern recognition tasks, the label of the sample can be given by directly observing the picture or video, but it is not easy to infer the intention by directly observing the EEG signal. Finally, most current EEG-based MI recognition methods often rely heavily on manual processing in the raw data preprocessing stage [16].
The proposed method is end-to-end trainable, which can be extended to real-world applications. our model is compared with some baseline models and the most advanced models. The results show that the proposed method can identify different types of human intentions in different BCI systems. Notably, we seek to improve the robustness and adaptability for various persons in the two-class and four-class MI classification tasks. The proposed method is evaluated on two publicly available EEG datasets, involving cross-subject and multi-scene motion intention detection. Experimental results show that our model has better classification accuracy than the baseline and advanced methods.
The rest of this paper is organized as follows: the relevant work is described in Section 2, Section 3 introduces our method, Section 4 gives experiments and results, and the conclusions are presented in Section 5.
2 Related works
Existing EEG-based MI recognition methods mainly include two stages: feature processing and feature recognition. The first stage often requires a complex and error-prone manual design [14]. Therefore, the whole feature processing process is time-consuming, and the obtained features usually have limitations, such as specific testers and specific acquisition environment, which is not easy to be extended to other subjects and testing environments. Besides, the current research on EEG classification mainly focuses on the subjects, i.e., the test data and train data from the same subject or binary EEG signal classification [45]. There are few EEG classification studies considering different scenes and subjects, which cannot meet the requirements of real-world BCI applications, and the accuracy of many existing methods is only about 75%. For example, Grosse-Wentrup et al. [15] proposes an algorithm that needs to extract common spatial pattern (CSP) features before classification. Hence, new methodologies must be considered that can complete the feature extraction and classification process directly from the EEG time series.
Deep learning does not require manual features and domain knowledge and can directly use the original EEG data to extract features without preprocessing. Researchers have recently used the deep learning (DL) method for EEG classification tasks because it helps improve the accuracy and decrease the EEG channel number. Kumar et al. [22] presents a DL approach for extracting features as the network’s input using CSP. A novel DL approach for classification of EEG MI signals, using SAE over 1D CNN features, a convolutional neural network (CNN)-based model [42] achieved the desired accuracy.
To learn the dynamic correlation in the MI signal, based on multi-layer CNN feature fusion, [2, 20] provides an improved CNN model for EEG MI classification. Hou et al. [17] developed a unique method for decoding EEG four-class MI problems using scout ESI and CNN, which can obtain competitive results. A method of generating spatial spectral feature representation is proposed, which can preserve multiple variable information in EEG data [7]. Zhao et al. [50] presents a system where the serial module can extract rough features in the time-frequency-space domain and the parallel module is used to fine feature learning in different scales. A stacked random forest model is used to enhance feature extraction and classification [40].
An end-to-end model [13] is constructed to reduce dimension and learn generalized features without specific approach to get the feature. CNN has been widely used in EEG signal sequence analysis [10, 30]. In [41], a combined structure of CNN and LSTM is used to obtain spatio-temporal features and perform high-precision classification of EEG data. Some works achieve similar classification accuracy in different MI tasks even for shorter length MI data [34]. Liu et al. [25] proposed to select the MI signal with a duration of 2s for comparative experiments. The results show that the appropriate selection of the length of MI signal for experiments has a great impact on the classification accuracy.
As mentioned above, our motivation is as follows: (1) Compared with the 2D and 3D CNN models that are widely used, however, there are few researches on extracting EEG sequence features using 1D CNN model. To improve the real-time performance of MI classification, we designed a 1D CNN architecture with core blocks. It is efficient for EEG processing and it saves computing resources and reduces the complexity of the model. (2) We use the sliding window technique and select the appropriate window length to generate more training EEG data. (3) We validate the performance of our model on two datasets for different classification tasks.
This paper proposes the method based on special 1D-CNN architecture and EEG data augmentation technology for EEG classification. In this method, the sliding window segmentation method are used to extend the training data, and special 1D-CNN framework is used to extract the temporal optimal EEG features. Based on this, our method can obtain a high EEG recognition accuracy. In the experiment, two EEG datasets are used to test the method performance, and satisfactory results are obtained.
3 Method
For the EEG classification task, we first consider the time-dependent that multi-channel EEG signals have. Secondly, reasonable information in different dimensions can make the model much more expressive. For this purpose, we designed a suitable network. Its specific model structure and implementation details of data enhancement will be described in detail. Lastly, we present the learning procedures.
3.1 Core block
In different neural network structures, such as convolutional and recursive layers are used to extract the dynamic properties of EEG signals and thus learn valid information about partial sequence segments. The temporal convolution layer is computationally efficient in time series classification tasks, so temporal convolution can be used as a feature extraction module to extract features of time series. Inspired by this, we propose the core blocks for four reasons. First, the convolutional layer in the core block can extract high-level features of multidimensional channel EEG signals. Second, in the core block, the BN [19] layer normalizes small batches of data for each channel independently in all observations.
After the convolutional operation, the BN layer is used to accelerate CNN training and reduce sensitivity to network initialization, which is very effective for fitting models and improving accuracy. Obviously, BN can normalize the data distribution to bring the data back to the unsaturated region. Third, in the core block, it is followed by a ReLU layer [28] after BN layer. The ReLU layer applies a threshold operation to each element, converting any input value less than zero to zero, which can control the saturation of the activation. BN layers and ReLU layers are included in the composition of each core block. Last, each FC layer in the core block integrates all the features learned in the previous layers across the sequence, especially in the case of large differences between the source and target domains. The FC layer can maintain a large model capacity thus ensuring the migration of the model representation capability. It helps to extract high-level information across time and space, and at the same time it can mitigate over-fitting.
At the feature level, the core blocks can be interpreted as follows. Each convolutional layer in the core block conducts a weighted feature combination with nonlinear activation. Then, the extracted features are repeatedly convolved across channels in the next convolutional layer. Here, we use this convolutional structure as the core block to extract multidimensional channel EEG signal features, allowing us to directly understand the feature extraction process in the temporal dimension.
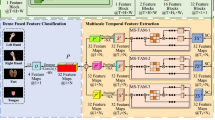
Schematic diagram of the MI classification system. Figure 1 (a) is an experimental table for EEG data collection, where the duration of the motion imagination task is taken as 4 s. Figure 1 (b) shows some preprocessing operations that have been performed on EEG data, including channel selection, data segmentation, and artifact removal. In addition, we standardize data processing by converting multi-channel EEG data into a two-dimensional matrix format. We enhance the EEG data by DA operation. Figure 1 (c) shows our proposed 1D CNN model. Three 1D convolutional layers were used for feature extraction of EEG signals, which can capture some local patterns or temporal features in the input signal
3.2 The framework of MI classification system
Figure 1 shows the proposed framework of EEG-based MI classification system. The framework can learn valid information through a combination of core blocks. Let the matrix \(\mathcal {X}\in \mathbb {R}^{N \cdot T}\) serve as the input samples. Where N denotes the number of recorded electrode channels, T denotes the number of signal sample points.
CNN has also been recognized as an effective DL method in recent years. It has a hierarchy in the form of different layers, each of which uses specific operations to extract high-level features from the input data. In our experiments, we proposed 1D CNN architecture consists of 20 layers. It includes five core blocks. Each of the first three core blocks is composed of \(CONV+BN+ReLU\), and the rest two core blocks is composed of \(FC+BN+ReLU\). The GAP layer facilitates structurally regularizing the entire network, preventing over-fitting and preserving global feature information, directly giving each channel an actual special meaning. For classification problems, the softmax function is the output unit activation function. A softmax layer and a classification layer usually follow the final FC layer. The classification layer calculates the cross entropy loss of weighted classification tasks, which have mutually exclusive categories. The network analysis results and configuration parameters are shown in Table 1.
In this system, the recorded EEG signals were split into MI-EEG data with length of 4s. Then, data augmentation (DA) was performed on the splitting data. Through the sliding window technology, the MI-EEG signal with a length of 2s is extracted from the MI-EEG signal with length of 4s, and finally the samples are enhanced. The classification result of the 2s MI-EEG signal will be the output of the 1D CNN.
3.3 EEG preprocessing and data augmentation
In our works, first, 22 channels were selected from the raw EEG signals. The sampling frequency is 250Hz. Then we remove artifacts and extract four categories MI-EEG data from the EEG signal of each channel automatically. The length of each MI-EEG data is 4s. Finally, we implement data augmentation for MI-EEG data.
Those EEG signals have been preprocessed before feeding the convolutional layer [25]. Suppose we have a series of MI-EEG sequence signals as inputs, which we denote by \(\mathcal {X}\). We normalize each channel signal of input data as follows:
where \(\sigma _{i}\), j, i and \(N_{len}\) refer to the standard deviation of channel i, the position in the signal, the channel and the number of sample sequences in a batch of inputs. The signal \(S_{i,j}\) is then divided into batches. After preprocessing, these signals will be fed into convolution layer and it is then written in the form of matrix \(N_{i} \times N_{len}\), where \(N_{i}\) and \(N_{len}\) refer to 22 and 500, representing 2 seconds.
Here, we have a sequence of MI-EEG signals S with n sample points, assuming the following three parameters are defined: \(L_{non-overlaping}\) is the length of sample non-overlapping region for two neighbor segments. \(L_{seg}\) is the length of each segment. N is the number of segments. Their relationships are defined in (2).
In this paper, n is the sample points, \(L_{non-overlaping}\) and \(L_{seg}\), are specified as 50 and 500, respectively. Then, the MI signal with 1000 sample points can obtain 11 training samples with 500 sample points. If we specify the three parameters (n, \(L_{non-overlaping}\), \(L_{seg}\)), the MI-EEG signal S can be split into N shorter MI-EEG signals \(\{S_1[n]\), \(S_2[n]\), ..., \(S_N[n]\}\) with an overlap of 90%, the schematic diagram of data augmentation as shown in Fig. 2.
3.4 Model Learning
The proposed network consists of five core blocks combined with other layers to form a 20-layer network, as shown in Fig. 3. First, the input multidimensional signal is fed to the first convolutional layer. In our proposed ETIODCNN method, the size of the convolution kernel used in the convolution layer of all core blocks is 5. The first convolutional layer uses a convolutional kernel in the shape of 5\(\times \)22. It was mentioned earlier that the signal of the input layer is in the form of a 22\(\times \)500 matrix. In the convolution process, the padding mode is set to ‘same’ and stride is set to 1, so we get the feature map as an array of 500\(\times \)1 (1D). The number of convolution kernels in the first convolution layer is specified as 16, so the size of the output is 500\(\times \)16. After convolution, the feature map is fed to the BN layer and then to the ReLU layer for nonlinear activation. For the first convolution layer in the core block, we have the mathematical expression:
where \(\varrho _{m}^{l}\left( u \right) \) stands for neuron u in layer l of map m. It also refers to the scalar product between the input neuron and the weighted value. \(\mathcal {W}_v(l, m, i)\) is the filter used in the neural network, l, m, i, and v represent the layer, map, kernel/channel and position in the kernel, respectively. \(b^l\left( u\right) \) refers to the bias of neuron u in layer l, here l is 1.
The input to the second convolutional layer is a 500\(\times \)32 matrix. The shape of the convolution kernel used in it is 5\(\times \)32. Its structure and properties are similar to those of the previous layer. It just has a different feature map size. For the second convolution layer in the core block, the mathematical expression as follows:
where \(N_{kernel}^{l}\) is the number of convolutional kernels used in the \(l_{th}\) convolutional layer. m refers to the elements i, j of the feature map produced after the convolution of a specific layer. The input to the third convolutional layer of the core block is a 500×48 matrix. The shape of the convolution kernel used in it is 5\(\times \)48. Similarly, for the third convolution layer in the core block, the feed-forward processing can be seen in (4).
In each core block, a BN layer and an ReLU layer are included. After being computed by convolutional or fully connected layers, the obtained feature information is first input to the BN layer and then processed by a ReLU function. The reconstructed feature information can be calculated by the (5)-(9):
where \(o_i^{'}\) is the feature information obtained from the previous layer and i represents the feature value of \(o^{'}\). \(\mu \) is the mean, \(\sigma ^2\) is the variance, \(\gamma \) and \(\beta \) are the parameter learned during training. Mini-batch training is used, which divides the entire training sample into smaller parts and updates all parameters after learning a mini-batch instead of a single sample. Where \(\epsilon \) is a constant, when the variance is small, numerical stability can be improved.
A 1D global average pooling (1D-GAP) layer implements down-sampling by calculating the average of the feature map. Through this operation, the global information is preserved and the local-global information features are extracted by representing the original channel feature vectors in one value after weighted average calculation.
The fully connected (FC) layer flattens the output. FC layer is included in core block4 and core block5, respectively, and the number of hidden units is set to 128. To prevent loss of desired feature information, the FC layer merges all previous feature representations and often contains many consecutive layers. For MI-EEG classification problems, the last fully connected layer combines all the features before using the softmax operation. For example, on the task of MI-EEG four-classification, the softmax operation can map the outputs of multiple neurons to the interval (0,1), and gives four values ranging between 0 and 1, which of the four values (four indexes in total) has the largest value represents the class corresponding to that position. Therefore, the softmax value of the \(i^{th}\) class can be calculated. So, for multi-class classification issues, the softmax function is the output unit activation function after the last fully connected layer, we have:
where \(0 \le P(r_i|x,\beta ) \le 1\) and \(\sum _{j=1}^K{P(r_i|x,\beta )} =1\), \(c_i = \ln (P(x,\beta |r_i)P(r_i))\), \(P(x,\beta |r_i)\) is the conditional of probability of the sample given class i, \(\beta \) is a system parameter in the multi-classification task, and \(P(r_i)\) is the class prior probability. For classification and weighted classification problems with mutually exclusive classes, a classification layer computes the cross-entropy loss. The best parameters can be found by minimizing the cost function, and it can be specified as:
where D is the number of samples, K is the number of classes, \(w_i\) is the weight for class i, \(t_{di}\) is the indicator that the \(d^{th}\) sample belongs to the \(i^{th}\) class, and \(y_{di}\) is the output for sample d for class i, which in this case, is the value from the softmax function. In other words, \(y_{di}\) is the probability that the network associates the \(d^{th}\) input with class i.
4 Experiment and results
4.1 Description of EEG datasets
BCI competition IV dataset 2a (BCICID 2a)
[3]. The dataset contains 22 channel EEG signals from 9 subjects (A01-A09). The sampling rate of the signal is 250 Hz. The data correspond to four different MI tasks, including left hand, right hand, tongue, and foot. For each subject, the data contain training data and testing data. There are 288 trials in the training data (72 trials per MI task) and 288 trials in the testing data. Note that a period of [2, 6] seconds was considered in our experiment. Herein, the training and testing sets of the BCICID 2a can be separated into two categories. To train and test our model, we used data from 9 subjects with around 28000 samples.
BCI competition IV dataset 2b (BCICID 2b)
[43]. This dataset records three bipolar channel EEG signals of nine subjects (B01-B09), i.e., those involving left-hand and right-hand MI activities. The sampling rate is 250 Hz. For each subject, five times of data collection were performed. The first three times were used for training and the rest for testing. The period of [3, 7] seconds was used in the experiment.
4.2 Experimental setup
Implementation details
In this experiment, we implement the proposed 1D-CNN network framework using the Matlab 2021b software. As for the hardware system configuration, the processor is an Intel (R) Xeon (R) silver 4110 CPU @ 2.10 GHz, the RAM has a capacity of 32.0 GB, and the graphics card is NVIDIA RTX 2080 Ti. Network training and testing are performed on a workstation with the Microsoft Windows 10 operating system. The network hyper-parameter of our model mainly includes the following aspects: the size of the mini-batch used for each training iteration, the number of hidden node, the kernel size, the initial learning rate and the max-epoch. Meanwhile, the adaptive learning rate algorithm chooses adaptive moment estimation (Adam), which is a method for stochastic optimization.
We trained our model with non enhanced data samples and enhanced data samples respectively, and finally verified and analyzed the performance of our model in different classification tasks.
Optimizing parameters and training network
To find the optimal network hyper-parameters and choose the best combination of network hyper-parameters, we employed the experiment over the training data of the BCICID 2a. In this study, we use the deep learning toolbox to construct and execute experiments. The purpose is to train the deep learning network and find the optimal parameter configuration of the network.
Based on the experimental design and statistical analysis, as shown in Table 2. In this paper, the initial learn rate is set to 0.001, the filter size is set to 5, the hidden node is set to 128, the epoch is set to 30, and the min-batch size is set to 128. Therefore, we choose this best combination of four network hyper-parameters to train our model. For example, the network training performance of trial 41 and trial 80 are shown in Fig. 4. The accuracy and error diagrams of two trials are shown in Fig. 4 (a) and (b). The results of experiment verify that the model has a excellent performance.
4.3 Statistical analysis and performance evaluation
Statistical analysis. Several statistical characteristics were analyzed in this study to estimate and compare the performance of various approaches. TP, TN, FP, and FN stand for true positive, true negative, false positive, and false negative, respectively. Accuracy, confusion matrix, and Kappa (\(\kappa \)) are used to evaluate the performances based on these metrics.
Accuracy (AC) : describes the ratio of true positives and true negatives for all predictions.
Kappa (\(\kappa \)) : is a robust measure since it considers the possibility of the agreement obtained by chance. The \(\kappa \) value can be computed as below:
Where \({P_e}\) is the theoretical probability of agreement. The higher the \(\kappa \) value, the better the performance.
Performance evaluation
In addition, we set the optimal network hyper-parameters, we use the BCICID 2a and the BCICID 2b to train and test our model. Datasets were divided into training, validation, and testing sets in a 7:2:1 ratio.
The training results of our proposed method for the BCICID 2a are illustrated in Fig. 5. The confusion matrix with column and row summaries are shown in Fig. 5 (a)-(i), and the “True Class” is the true label, while the “Predicted Class” is the predicted label. The true positive rates and false positive rates are displayed in the row summary, while the positive predictive values and false discovery rates are displayed in the column summary.
To evaluate the classification accuracy of MI tasks, someone considered the effects of different numbers of conv1D layers and different input signal processing lengths [35]. Based on the consideration and inspiration, our experimental results show that different configurations of network parameters and the number of channels of EEG signals are also very important to improve MI classification performance, it can be seen in Table 2 and Fig. 6. According to the correlation coefficient [6] of EEG signals, the EEG signals of adjacent channels in the same brain functional area have strong correlation and similar characteristics. We generated three sub-datasets with 3 channels, 6 channels and 22 channels respectively from BCICID 2a.
At the same time, we also use the original two datasets (BCICID 2a and BCICID 2b) without data augmentation to train and test our model. We evaluated through experiments and found that: the MI task of the acquired EEG signal is closely related to the channels of various brain functional areas. The information of each channel is also highly correlated, and an MI task is generated by the combined action of EEG channel signals from different brain functional areas. The data enhancement operation improves the performance of the model for MI classification. And the result is shown in Table 3.
To adequately test the method’s performance and stability, the average of all subjects classification results for the different datasets after applying different 20 predictions. It indicates an improvement in the quality of the prediction findings and the confusion matrix with column and row summaries. With the increase of the number of channels, the classification performance is excellent, as shown in Fig. 7 and Table 3. The BCICID 2b with 3 channels is shown in Fig. 7(a) and the confusion matrix of 3, 6, 22 channels of BCICID 2a are shown in Fig. 7(b), (c), and (d), respectively. The results of experiment verify that our proposed method has a excellent performance.
10-fold cross-validation
We used the BCICID 2a data with DA operation to evaluate the performance of the proposed method. In addition, we divided them into ten subsets, respectively. Each time, one subset was used as the validation set, and the other nine were used as the training set. Ten cross-validation tests were conducted to obtain the average result. Similarly, we also conducted evaluations in the same way on each subject data. Our model achieved an average accuracy of 97.32(±0.17)% with a variance of 0.0176 on BCICID 2a, an average accuracy of 96.04 (±0.23)% with a variance of 0.0481 on BCICID 2b, and an average accuracy of 94.76 (±0.55)% with a variance of 0.0314 on PhysioNet [18], respectively.
4.4 Performance comparison
Some methods propose different network framework based on different convolution structures, and some combine the traditional feature extraction methods and DL methods to improve MI classification accuracy. For example, the accuracy is 92.9%, 84.3%, 76.2%, 73.6%, 71.9% in the studies of Sadiq et al. [33], Joadder et al. [21], Samek et al. [37], Devlaminck et al. [11], and Atyabi et al. [5], respectively. In order to verify the effectiveness of the proposed method, we also compared our method with PSD, ABS, VSBS, variations based PSD (VPSD) methods on the BCICID 2b. Table 4 gives the accuracy and kappa values of PSD, VPSD, ABS, VSBS and our methods. When compared to other approaches for extracting features, it can be demonstrated that our method produces superior results. Our approach has an average kappa of 0.955 and an average accuracy of 96.29%. When compared to VSBS, the proposed technique improves average accuracy by 13.61% and average kappa by 0.27.
The within-subject test is implemented, i.e. the training data set and test data set are from the same subject. More importantly, the comparison results of performance of different networks as shown in Table 5. Our method outperforms others in terms of parameter quantity, real-time performance and accuracy. Table 6 show the test accuracy of each subject under different networks and the test accuracy of corresponding networks. According to the experimental results, it displays that the accuracy of subject 2 is the lowest and the accuracy of subject 3 is the highest on BCICID 2a. However, a higher standard deviation in the model indicates that the model may fit extremely well for some subjects while being just fair for others. More subject data for model training may overcome this, thus decreasing the gap.
The CNN-based model [42] also used SAE over 1D CNN features and attained an EEG classification accuracy of 70%. The BCICID is used to implement and test this concept. Table 7 shows a comparison of the proposed method’s overall accuracy with that of other approaches. Our proposed strategy outperformed the baseline, which also used CNN, in terms of accuracy.
On the BCICID, the MCNN approach only achieved accuracy of 75.7% for subject-specific training and assessment. With cropped training, the shallow and deep CNN model [38] was constructed for MI classification and achieved 72.0% decoding accuracy. Before DL techniques were used to EEG data, the most successful conventional machine learning methodology was filter bank common spatial patterns (FBCSP), which generated the greatest results for EEG decoding. FBCSP and CNN are used to extract spatio-temporal information from BCICID data [36], on the BCICID, this approach was able to attain 74.4% test accuracy. Notably, we enhanced our training samples and our model got better classification accuracy. Our method improves 13.67%, 21.7%, 23% in accuracy than Musallam et al. [27], Amin et al. [2], Sakhavi et al. [36], respectively.
5 Discussion
The validity and generalizability of research results in small-sample studies on EEG classification and EEG pattern recognition may be influenced by several potential factors, such as low statistical power, overfitting risks, instability, and feasibility in practical applications. Small-sample studies are prone to low statistical power, making it difficult to detect true differences or correlations. Moreover, due to the limited sample size, the research results may not adequately represent the entire population, thus limiting the reliability and generalizability of the findings. Additionally, small samples can lead to overfitting of the training data and poor generalization to new data, causing the research results to deviate from expectations when tested on larger samples or other datasets. Furthermore, small samples can result in result instability, where the same data may yield different outcomes under different sampling or partitioning schemes, challenging the reliability and consistency of the research. It’s worth noting that due to the small sample size, the research results may not accurately reflect EEG classification issues in different populations or contexts. Therefore, when applying the findings of small-sample studies to real-world scenarios, more validation and empirical research support are needed. Recent EEG studies [9, 12, 31] have endeavored to address these limitations in small-sample research by using similar sample sizes across different contexts and successfully mitigating these issues.
In our work, to enhance the reliability and generalizability of the research on small-sample EEG classification, we have increased the training sample size and employed methods such as cross-validation, while emphasizing the stability of the proposed model’s EEG classification results. The MI task categories in BCICID 2a and PhysioNet are both four categories. We tested the training model on two datasets with the same hyper-parameters. We find that the MI samples in BCICID 2a are more than PhysioNet. The model accuracy and test variance obtained from BCICID 2a are superior to PhysioNet. Therefore, the specific MI task categories and the MI sample numbers of subject in different datasets will have a certain impact on our model.
In practical brain-machine interface applications, precise and effective decoding and encoding of EEG signals are crucial for controlling a robotic hand to perform actions such as finger grasping and releasing. First, our approach can achieve accurate classification of EEG signals, encode the classification results into control commands, and send these commands wirelessly or through a USB interface to an intelligent peripheral device (robotic hand) equipped with a Raspberry Pi system. Secondly, the main control system receives the command information and drives the motors of the robotic hand accordingly. Finally, the robotic hand performs actions based on the control of EEG signals. Applying generative techniques to EEG signals can increase the amount of data, data diversity, and data privacy protection, helping explore unknown areas and optimize EEG experiment design. EEG generation technology can contribute to improving the performance and applications of EEG signal classification and pattern recognition tasks. When implementing this technology in the future, there may be challenges in the following aspects:
1) Consideration of EEG signal quality and noise during the EEG acquisition process. We know that EEG signals are susceptible to muscle activity, electromagnetic interference, and other noise. Obtaining high-quality EEG signals is essential for accurately identifying intentions and executing corresponding actions by the robotic hand.
2) Training a universally stable and effective model is challenging. Each individual’s EEG signals have unique features and patterns, so personalized models need to be established for recognition and control. Additionally, EEG signals may change over time, due to emotions and physiological states, requiring models with good adaptability and robustness.
3) Real-time and latency issues in brain-machine communication. Controlling the robotic hand requires real-time and low-latency conditions. The acquisition, processing, and decoding of EEG signals need to be performed as quickly as possible to achieve real-time control of the robotic hand’s actions. Reducing signal processing and transmission latency poses a technological challenge.
4) Multi-action classification. Robotic hands typically have multiple actions and movements, such as grasping, lifting, and rotating. Accurately identifying and classifying different intentions is a challenge, especially when transitioning between actions smoothly. Ensuring the precision and accuracy of the robotic hand’s actions is also crucial. These challenges mainly focus on the decoding and encoding aspects of EEG signals.
6 Conclusion
In this paper, the method of EEG classification of motor imagery based on a special 1D CNN architecture and data augmentation is proposed. We use core blocks to build the network and choose the time sliding window with a length of 2s to enrich MI samples. BCICID 2a and 2b are used to verify the effectiveness of proposed method. The comparison with other state-of-the-art works shows the superiority of our ETIODCNN model. In conclusion, ETIODCNN has a good real-time classification performance. Our training model outperforms other models in terms of stability and robustness. Furthermore, our method can improve the performance of MI tasks without any complicated and time-consuming feature engineering. The experimental results help us better understand how to use DL methods to solve EEG-based classification problems. In the future work, we hope to apply EEG recognition and generation for actual control systems, such as the control of wheelchairs. On the other hand, we also try to embed algorithms into mobile devices to control multiple daily devices.
Data Availability
The data (1. Four class motor imagery) and (4. Two class motor imagery) used to support the findings of this study have been deposited in the repository [http://bnci-horizon-2020.eu/database/data-sets].
References
Al-Qaysi Z, Zaidan B, Suzani M (2018) A review of disability EEG based wheelchair control system: Coherent taxonomy, open challenges and recommendations. Comput Methods Programs Biomed 164:221–237
Amin SU, Alsulaiman M, Muhammad G et al (2019) Deep Learning for EEG motor imagery classification based on multi-layer CNNs feature fusion. Future Gener Comput Syst 101:542–554
Ang K, Chin Z, Wang C, Guan C, Zhang H (2012) Filter bank common spatial pattern algorithm on BCI competition IV datasets 2a and 2b. Frontiers Neurosci. 6:39
Ang KK, Chin ZY, Wang C et al (2012) Filter bank common spatial pattern algorithm on BCI competition IV datasets 2a and 2b. Front Neurosci 6:39
Atyabi A, Luerssen M, Fitzgibbon SP et al (2017) Reducing training requirements through evolutionary based dimension reduction and subject transfer. Neurocomputing 224:19–36
Bahador N, Erikson K, Laurila J et al (2020) A correlation-driven mapping for deep learning application in detecting artifacts within the EEG. J Neural Eng 17(5):056018
Bang J S, Lee M H, Fazli S et al (2021) Spatio-spectral feature representation for motor imagery classification using convolutional neural networks. IEEE Trans Neural Netw Learn Syst
Bishop CM, Nasrabadi NM (2006) Pattern recognition and machine learning. New York: Springer
Castiblanco Jimenez IA, Gomez Acevedo JS, Olivetti EC et al (2022) User Engagement Comparison between Advergames and Traditional Advertising Using EEG: Does the User’s Engagement Influence Purchase Intention? Electronics 12(1):122
Croce P, Zappasodi F, Marzetti L et al (2018) Deep Convolutional Neural Networks for feature-less automatic classification of Independent Components in multi-channel electrophysiological brain recordings. IEEE Trans Biomed Eng 66(8):2372–2380
Devlaminck D, Wyns B, Grosse-Wentrup M et al (2011) Multisubject learning for common spatial patterns in motor-imagery BCI. Comput Intell Neurosci 2011
Di Flumeri G, De Crescenzio F (2019) Berberian B et al Brain-computer interface-based adaptive automation to prevent out-of-the-loop phenomenon in air traffic controllers dealing with highly automated systems. Front Hum Neurosci 13:296
Dose H, Møller JS, Iversen HK et al (2018) An end-to-end deep learning approach to MI-EEG signal classification for BCIs. Expert Syst Appl 114:532–542
Feng JK, Jin J, Daly I et al (2019) An optimized channel selection method based on multifrequency CSP-rank for motor imagery-based BCI system. Comput Intell Neurosci
Grosse-Wentrup M, Buss M (2008) Multiclass common spatial patterns and information theoretic feature extraction. IEEE Trans Biomed Eng 55(8):1991–2000
Gupta A, Bhateja V, Mishra A (2019) Autoregressive modeling-based feature extraction of EEG/EOG signals. In Proc. Inf. Commun. Technol. Intell. Syst., pp. 731-739
Hou Y, Zhou L, Jia S et al (2020) A novel approach of decoding EEG four-class motor imagery tasks via scout ESI and CNN. J Neural Eng 17(1):016048
Hou Y, Zhou L, Jia S et al (2020) A novel approach of decoding EEG four-class motor imagery tasks via scout ESI and CNN. J Neural Eng 17(1):016048
Ioffe S, Szegedy C (2015) Batch normalization: Accelerating deep network training by reducing internal covariate shift. International conference on machine learning. PMLR: 448-456
Jiao Z, Gao X, Wang Y et al (2018) Deep convolutional neural networks for mental load classification based on EEG data. Pattern Recognit 76:582–595
Joadder MAM, Siuly S, Kabir E et al (2019) A new design of mental state classification for subject independent BCI systems. IRBM 40(5):297–305
Kumar S, Sharma A, Mamun K (2016) A deep learning approach for motor imagery EEG signal classification. et al (2016) 3rd Asia-Pacific World Congress on Computer Science and Engineering (APWC on CSE). IEEE, 34–39
Leeb R, Lancelle M, Kaiser V (2013) Thinking Penguin: Multimodal Brain Computer Interface Control of a VR Game. IEEE Trans Comput Intell AI Games 5(2):117–128
Liu X, Shen Y, Liu J et al (2020) Parallel spatial-temporal self-attention CNN-based motor imagery classification for BCI. Front Neurosci: 1157
Liu J, Ye F, Xiong H Recognition of multi-class motor imagery EEG signals based on convolutional neural network. J Zhejiang Univ (Eng Sci) 55(11): 2054-2066
Lopes AC, Pires G, Nunes U (2013) Assisted navigation for a brain-actuated intelligent wheelchair. Robot Auton Syst 61(3):245–258
Musallam YK, AlFassam NI, Muhammad G et al (2021) Electroencephalography-based motor imagery classification using temporal convolutional network fusion. Biomed Signal Process Control 69:102826
Nair V, Hinton G E (2010) Rectified linear units improve restricted boltzmann machines. Icml
Noureddin B, Lawrence P, Birch G (2012) Online removal of eye movement and blink EEG artifacts using a high-speed eye tracker. IEEE Trans. Biomed. Eng. 59(8):2103–2110
Peng D, Liu Z, Wang H et al (2018) A novel deeper one-dimensional CNN with residual learning for fault diagnosis of wheelset bearings in high-speed trains. IEEE Access 7:10278–10293
Perera D, Wang YK, Lin CT et al (2022) Improving EEG-Based Driver Distraction Classification Using Brain Connectivity Estimators. Sensors 22(16):6230
Pérez-Enciso M, Zingaretti LM (2019) A guide on deep learning for complex trait genomic prediction. Genes 10(7):553
Sadiq MT, Yu X, Yuan Z (2021) Exploiting dimensionality reduction and neural network techniques for the development of expert brain-computer interfaces. Expert Syst Appl 164:114031
Saini M, Satija U, Upadhayay MD (2022) One-dimensional convolutional neural network architecture for classification of mental tasks from electroencephalogram. Biomed Signal Process Control 74:103494
Saini M, Satija U, Upadhayay MD (2022) One-dimensional convolutional neural network architecture for classification of mental tasks from electroencephalogram. Biomed Signal Process Control 74:103494
Sakhavi S, Guan C, Yan S (2018) Learning temporal information for brain-computer interface using convolutional neural networks. IEEE Trans Neural Netw Learn Syst 29(11):5619–5629
Samek W, Meinecke FC, Müller KR (2013) Transferring subspaces between subjects in brain-computer interfacing. IEEE Trans Biomed Eng 60(8):2289–2298
Schirrmeister RT, Springenberg JT, Fiederer LDJ et al (2017) Deep learning with convolutional neural networks for EEG decoding and visualization. Hum Brain Mapp 38(11):5391–5420
Sharma G, Parashar A, Joshi AM (2021) DepHNN: A novel hybrid neural network for electroencephalogram (EEG)-based screening of depression. Biomed Signal Process Control 66:102393
Shen Y, Lu H, Jia J (2017) Classification of motor imagery EEG signals with deep learning models. International Conference on Intelligent Science and Big Data Engineering. Springer, Cham, 181–190
Sun Y, Lo FPW, Lo B (2019) EEG-based user identification system using 1D-convolutional long short-term memory neural networks. Expert Syst Appl 125:259–267
Tabar YR, Halici U (2016) A novel deep learning approach for classification of EEG motor imagery signals. J Neural Eng 14(1):016003
Tangermann M (2012) Review of the BCI competition IV. Front Neurosci 6:55
Wang T, Wu D J, Coates A et al (2012) End-to-end text recognition with convolutional neural networks. Proceedings of the 21st international conference on pattern recognition (ICPR2012). IEEE 3304-3308
Wu S (2017) Fuzzy integral with particle swarm optimization for a motor-imagery-based brain-computer interface. IEEE Trans Fuzzy Syst 25(1):21–28
Wu H, Niu Y, Li F et al (2019) A parallel multiscale filter bank convolutional neural networks for motor imagery EEG classification. Front Neurosci 13:1275
Zhang R, Li Y, Yan Y et al (2015) Control of a wheelchair in an indoor environment based on a brain-computer interface and automated navigation. IEEE Trans Neural Syst Rehabilitation Eng 24(1):128–139
Zhang Y, Nam CS, Zhou G (2019) Temporally constrained sparse group spatial patterns for motor imagery BCI. IEEE Trans Cybern 49(9):3322–3332
Zhang T, Zheng W, Cui Z, Zong Y, Li Y (2019) Spatial-temporal recurrent neural network for emotion recognition. IEEE Trans Cybern 49(3):839–847
Zhao X, Liu D, Ma L et al (2022) Deep CNN model based on serial-parallel structure optimization for four-class motor imagery EEG classification. Biomed Signal Process Control 72:103338
Zhuang J, Geng K, Yin G (2019) Ensemble Learning Based Brain-Computer Interface System for Ground Vehicle Control. IEEE Trans Syst Man Cybern: Syst 51(9):5392–5404
Acknowledgements
This work was supported by the Nature Science Foundation of China (Nos. 61671362 and 62071366).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interest
The authors report no declarations of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chu, C., Xiao, Q., Chang, L. et al. EEG temporal information-based 1-D convolutional neural network for motor imagery classification. Multimed Tools Appl 82, 45747–45767 (2023). https://doi.org/10.1007/s11042-023-16536-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-023-16536-x