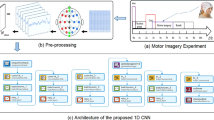
Abstract
Recently, deep learning-based methods have achieved meaningful results in the Motor imagery electroencephalogram (MI EEG) classification. However, because of the low signal-to-noise ratio and the various characteristics of brain activities among subjects, these methods lack a subject adaptive feature extraction mechanism. Another issue is that they neglect important spatial topological information and the global temporal variation trend of MI EEG signals. These issues limit the classification accuracy. Here, we propose an end-to-end 3D CNN to extract multiscale spatial and temporal dependent features for improving the accuracy performance of 4-class MI EEG classification. The proposed method adaptively assigns higher weights to motor-related spatial channels and temporal sampling cues than the motor-unrelated ones across all brain regions, which can prevent influences caused by biological and environmental artifacts. Experimental evaluation reveals that the proposed method achieved an average classification accuracy of 93.06% and 97.05% on two commonly used datasets, demonstrating excellent performance and robustness for different subjects compared to other state-of-the-art methods.In order to verify the real-time performance in actual applications, the proposed method is applied to control the robot based on MI EEG signals. The proposed approach effectively addresses the issues of existing methods, improves the classification accuracy and the performance of BCI system, and has great application prospects.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
A brain-computer interface (BCI) is a system that connects the human brain with computational devices by decoding neuronal activity (Lotte et al. 2018). EEG is among the most widely used BCI signals because of its non-invasive nature, high temporal resolution and low cost.Motor imagery electroencephalogram (MI EEG) is the only BCI paradigm that reflects a user’s voluntary conscious movement consciousness without any external stimuli. It is a promising technology owing to its very widespread domains in both medical applications and human augmentation technologies (Zhang et al. 2019). When a subject actively imagines a body movement, the power of \(\mu \) (8-12 Hz) and \(\beta \) (16-26 Hz) rhythms decrease or increase in their brain’s sensorimotor cortex area of the contralateral and ipsilateral hemispheres, which is denoted as event-related desynchronization (ERD) and event-related synchronization (ERS), respectively. The core problem of MI EEG classification is that of decoding the low signal-to-noise ratio (SNR) and significant individual variance of MI EEG signals into correct instructions effectively. In this study, our goal is to accurately analyze brain activity for helping people, such as post-stroke and paralyzed patients, to solve the problem of communication with the outside world.
Numerous studies have reported on the classification of MI EEG signals. These studies can be divided into two categories, including machine learning- and deep learning-based methods. Here, we only provide a general summary; more details can be found in Sect. 2.
Conventional machine learning (ML)-based MI EEG classification methods consist of hand-crafted feature extraction and subsequent different objective feature classifiers. The most popular method is the common spatial pattern (CSP) and its variants (Ang et al. 2012; Kwon et al. 2019; Dong et al. 2020). However, the effectiveness of CSP is highly affected by the frequency band and time window of the EEG segments’ range for each subject, which influences the final performance.
In addition to CSP-based methods, other ML-based methods combined with other feature extraction algorithms can also extract potentially valuable components of EEG signals and show satisfactory results on MI classification tasks (Xie et al. 2016; Miao et al. 2021). Unfortunately, because they generally depends on manually designed features based on human knowledge and experience to extract features at a fixed time period or frequency band, the existing ML-based methods are not capable of achieving a high-performance MI EEG classification (Gaur et al. 2021).
In contrast, deep learning (DL) integrates the feature extraction phase and the feature classification phase into a single end-to-end architecture to jointly learn all parameters, and has achieved excellent performance in medical image processing (Zhang et al. 2020), computer-aided diagnosis (Zhang et al. 2019), and computer vision (Pang et al. 2020). In contrast to machine learning-based algorithms, deep learning-based methods are empowered to learn distinct high-level representations from raw brain signals without the limitation of human handcrafted features, and have been proven to be more suitable for EEG single processing (Penaloza and Nishio 2018). Therefore, researchers have paid more attention to the DL-based methods for multi-class MI EEG classification (Schirrmeister et al. 2017; Dai et al. 2020). In addition, with the development of graphics processing units, the real-time performance of MI EEG classification has also been enhanced (Zhao et al. 2019; Li et al. 2019; Zhang et al. 2021). According to the input format definition, two main branches of research have been developed in DL-based multiclass MI EEG classification methods. One is to take the feature maps extracted from the original MI EEG signals as input, and the other directly focuses on the input format as the original MI EEG signals.
The first case represents MI EEG signals into a series of two-dimensional feature maps as the input format for reducing noise and enhancing low SNR signals by manually selected feature extraction methods (Lei et al. 2019; Ma et al. 2020; Sun et al. 2021). However, the key potential problem of the feature-based input case is that the extracted features must be manually designed by human experts. More importantly, MI EEG signals are non-stationary and easily corrupted by various biological fluctuations and events (e.g., eye blinks, muscle artifacts, fatigue, and concentration levels), which results in different subjects exhibiting activities in different time periods, and the most optimal features being subject-specific. Therefore, it is difficult to manually choose a suitable feature extraction method across subjects, which usually leads to poor generalization ability and low decoding performance for multi-class classification tasks.
Another input format of the network is to represent the original MI signal as a two-dimensional array, which takes the number of time sampling points as the array width and the number of electrodes as the array height (Schirrmeister et al. 2017; Hong et al. 2021; Liu et al. 2021). However, when representing the raw MI EEG as a 2D array input in the abovementioned manner, deep learning-based methods typically omit the spatial dependencies of MI EEG data, which has been proven to be important for improving the classification performance (Bashivan et al. 2015). Nor can the correlation among nearby sampling electrodes cannot be fully reflected the 2D array. Consequently, the final performance of the MI classification model is affected. To address these issues, some studies have attempted to incorporate a topological structure into deep learning architectures (Zhao et al. 2019; Zhang et al. 2019).
However, MI EEG data are recorded from many electrodes (typically 32, 64, 128, and even more) placed at different locations across the brain. Each electrode channel EEG signal is a time sequence whose features also vary over time and exhibit significant variations between different individuals and different tasks. Some researchers have used feature selection algorithms after applying the channel selection algorithms to further improve the system performance (Li et al. 2019; Zhang et al. 2021; Li et al. 2020). Therefore, how to determine the best optimal ones remains a changeling question (Baig et al. 2020).
Based on the above discussion, MI EEG classification task still has the following problems.
-
The traditional 2D representation of MI EEG does not consider spatial topological dependencies among electrode channels. Furthermore, the correlation among nearby electrode channels cannot be fully reflected using a traditional 2D convolution kernel. As a result, the performance of MI EEG classification systems is affected.
-
The purpose of EEG electrode channel selection is to determine the most discriminative EEG nodes. The motor regions (\(C_3\), \(C_4\), and \(C_Z\)) are three commonly hand-selected channels located in the brain motor regions, which may have certain effects on different subjects or MI tasks.
-
A subject usually concentrates at some time but is distracted at the other times, and different subjects pay attention at different times within a trial. Therefore, emphasizing the EEG temporal slices when a subject concentrates in the trial while neglecting the other slices is necessary for successful EEG analysis. Therefore, adequately extracting time-invariant high-level temporal features from a temporal slice to encode temporal information for a higher and more robust classification of different subjects is problematic.
To address these issues, we propose a end-to-end 3D CNN to extract multiscale spatial and temporal dependent features (MST-3DCNN) for the 4-class motor imagery classification tasks. MST-3DCNN is specifically composed of a 3D representation, a 3D spatial attention module (3D-SAM), multiscale temporal attention module (MS-TAM), and dense fused classification module.
The goal of 3D-SAM and MS-TAM is to adaptively assign higher weights to motor-related spatial channels and temporal sampling cues than motor-unrelated ones across all brain regions. They can define a new compact feature representation of MI EEG in space and time domains and prevent influences caused by biological and environmental artifacts to improve classification performance.
The major contributions of this study are summarized as follows.
-
1.
An end-to-end 3D convolution neural network with multiscale spatial and temporal cues was proposed for 4-class MI EEG classification tasks. It can further improve the robustness and accuracy of subject-dependent and subject-independent data with limited annotation data.
-
2.
To validate the robustness and accuracy, we carried out comprehensive experiments on two public benchmark datasets. The results demonstrate the superior generalization performance of the proposed methods, which exhibited significantly higher classification accuracy than the state-of-the-art methods.
-
3.
We provide insight into the intrinsic patterns inherent in MI EEG signals to explain the reason that the proposed approach enhanced the feature representation capability and obtaining more distinct feature representations for high-level applications.
-
4.
To validate the real-time capability of the proposed method, we design a 4-class trial to acquire an MI EEG dataset. Then, a BCI-based NAO robot system was developed through the online decoding of our acquired MI EEG signals.
The remainder of this study is organized as follows. “Previous works” section provides a description of the challenge. “Method” section discusses the details of the proposed method. The experimental results and discussion are presented in “Experiment results and discussion” section. Finally, “Conclusion” section provides our final conclusions and suggests some potential avenues for further research.
Previous works
Studies on MI EEG signal classification are divided into two categories: machine learning and deep learning-based methods.
Among the ML-based methods, the filter bank common spatial pattern (FBCSP) algorithm is a representative method using the common spatial pattern (CSP). It improved the classification accuracy by performing autonomous selection of the discriminative subject-specific frequency range for bandpass filtering of the EEG measurements. Ang et al. (2012). A sparse filter band common spatial pattern (SFBCSP) algorithm (Zhang et al. 2015) was proposed to automatically estimate significant CSP features on multiple signals from raw EEG data at a set of overlapping bands, which is considered as a potential method for improving the performance of MI EEG. However, the effectiveness of CSP is highly affected by the frequency band and time window of the EEG segment range for each subject, which influences the final performance.
The DL-based multiclass MI EEG classification methods consist of two categories, including those of the feature map-based input case and those of the original signal-based input case.
The first is to use the feature maps from MI EEG signals as input to reduce the effect of noise and enhance low SNR signals. Sun et al. (2021) proposed a new framework called sparse spectro-temporal decomposition and a CNN for MI-EEG classification tasks. Instead of using conventional time-frequency analysis methods for feature extraction, a sparse spectro-temporal decomposition method was proposed to enforce sparsity of EEG signals on the time-frequency plane, overcoming the drawbacks of the conventional model and enhancing the ERS and ERD phenomena. Xu et al. (2020) proposed a unified time-frequency energy calculation architecture to learn a topographical representation data structure of brain activities from EEG data with frequency band handle selection. They constructed an appropriate data structure by indicating the intrinsic connections of brain activity status in EEG to reduce computational complexity.
In contrast, the other format directly takes the original MI EEG as input. Schirrmeister et al. (2017) verified the feasibility of decoding MI EEG data using a convolutional neural network (CNN). They described architectural choices and training strategies for CNNs on EEG data, including how to represent the EEG input formations. Three CNN architectures were used in their study, and several specific design choices were evaluated for these architectures. Lawhern et al. (2018) used depthwise and separable convolutions to construct a compact EEG-specific network for the precise classification of EEG signals. Hong et al. (2021) proposed a dynamic joint domain adaptation network based on an adversarial learning strategy to learn domain-invariant feature representation and improve MI EEG classification performance in the target domain by leveraging useful information from the original EEG signals.
However, most DL-based classification methods use a single receptive kernel size in a limited number of convolutional layers, which has to change the parameters of the network for different subjects and cannot extract high-level features to improve the classification accuracy. Therefore, Amin et al. (2019) proposed a multi-layer CNN architecture for fusing different features. It contained different convolutional features with different depths and filter sizes to capture spatial and temporal features from raw EEG data to improve the accuracy of MI EEG classification. Dai et al. (2020) introduced a hybrid-scale CNN classification architecture to enhance the information in different domains (time, frequency, and space) with different convolution scales, and exploited an optimal convolution scale varying from subject to subject. They also generated artificial training data based on real training data to improve classification accuracy when the available training data were limited. Liu et al. (2021) attempted to fuse different models using their complementary characteristics to develop a multiscale space-time-frequency feature-guided multitask learning CNN architecture. Their method includes a multitask learning framework in which four modules are trained simultaneously and jointly optimized in an end-to-end manner.
However, representing the raw MI EEG as a 2D array usually omits the spatial information of MI EEG data, which is important for the final classification performance (Bashivan et al. 2015). Some studies have attempted to incorporate a topological structure into a deep learning architecture. The 1D-vector EEG format with the location information of electrodes was mapped to a 2D mesh-like EEG signal, which was further employed to form 3D format data that was fed to the designed 3DCNN (Zhang et al. 2019). Zhao et al. (2019) proposed a 3D representation by transforming the original EEG signals into a sequence of 2D arrays that contain the spatial structure of all electrodes. A multi-branch 3D CNN with different receptive field sizes is designed to extract high-level MI EEG-related features.
In addition, MI EEG data were recorded from many electrodes placed at different locations across the brain. Gong et al. (2018) selected signal channels in motor regions, such as (\(C_3\), \(C_4\), and \(C_Z\)). Furthermore, Li et al. (2019) employed raw multi-channel EEG as inputs to capture the multi-scale temporal features of EEG signals using a channel-projection mixed-scale convolutional neural network. However, because the strength of the MI EEG signals varies between subjects, it is impossible to determine exactly which brain regions are most associated with MI for different people (Ma et al. 2019). All brain functional areas may have certain effects on different MI tasks, instead of only the motor regions.However, all electrodes were selected by a priori knowledge, which might lead to missing some information available in other channels.
Li et al. (2020) proved the feasibility of improving the classification performance with the attention mechanism. They introduced the attention mechanism to a multi-scale fusion convolutional neural network. Their network extracted multi-scale features from multi-brain regions representation signals and was supplemented by a dense fusion strategy to retain the maximum information flow.
As may be noted from the discussion above, previous algorithms have usually resulted in methods of extracting general representations involving significant limitations, as well as a low accuracy for subject-specific and subject-independent classification
Schematic architecture of the proposed method. The color cuboids are extracted features in different phases, and their corresponding sizes are indicated around cuboids. The convolution and pooling operations are listed in the arrow line. a The 3D spatial feature extraction phase and multiscale temporal feature extraction phase are shown in the green and pink rectangles, respectively. b Dense fused Feature classification phase. \(M_j^i\) is the 3D representation of the \(i-th\) trial of \(j-th\) subject
Method
In this section, the detailed configurations of the MI classification architecture are presented. As shown in Fig. 1 and Table 1. The proposed architecture is an end-to-end framework that can be trained using standard back-propagation.
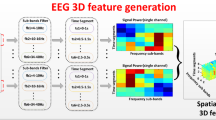
First, we explain the definition of 3D representations. Let \(X_j^i\) be the original MI EEG signal, where \(X_j^i\in \mathbb {R}^{C\times T}\) represents the \(i-th\) trial of \(j-th\) subject as a traditional 2D matrix in which the width of the matrix is the number of discretized time sampling points (T) and the height of the matrix is the number of electrode channels (C). However, as discussed above, traditional 2D representation \(X_j^i\) cannot fully reflect the correlation among nearby electrode channels without considering the space topological dependencies among them. Therefore, as shown in Fig. 2, we expanded the traditional 2D matrix \(X_j^i\) to a 3D tensor (\(M_j^i\)) by using the locations of the EEG electrode channels. For example, the color number k in \(M_j^i\) (see Fig. 2c) indicates that it has the same relative location as the electrode channel k in Fig. 2a, whose temporal sampling values are formed as sequential data in \(M_j^i\). The blue number 0 in \(M_j^i\) indicates that there was no electrode channel, and its temporal sampling values were zero. The purpose of adding zero to \(M_j^i\) is to retain \(M_j^i\) as a 3D cube tensor, to support the use of 3D convolution without introducing any noise.
This 3D representation not only uses the electrode distribution to explicitly preserve the relative space topological information between electrode channels, but also uses the sequential form of temporal sampling values to preserve the temporal information. Therefore, it is easily used to extract spatiotemporal features using 3D convolution.
Spatial feature extraction
As shown by the green rectangle in Fig. 1, based on the 3D representation, the purpose of this section is to automatically explore most motor-related discriminative spatial dependent features and the corresponding hierarchical correlation between any two electrode channels. It is independent of subjects, MI tasks, and manual-selected parameters, which can eliminate artifacts caused by manually selected channels and adaptively improve the accuracy on different subjects or MI tasks.
-
1.
3D spatial feature attention module (3D-SAM) learns a new 3D spatial representation of \(M_j^i\), which automatically assigns higher weights to the most motor-related channels and lower weights to the motor-unrelated channels (see Fig. 2 ).
-
As shown in Fig. 3 and Table 2, \(M_j^i\) is first fed into three separable 3D convolutions (\(Conv3d\_1\), \(Conv3d\_2\), \(Conv3d\_3\)) to generate different 3D spatial feature blocks \(L_1\), \(L_2\) and \(L_3\), respectively. \(L_1\), \(L_2\) and \(L_3\) belong to \(\mathbb {R}^{C_1\times T\times H\times W}\), and \(C_1\) is 4, which indicates the number of feature blocks. Furthermore, \(L_1\), \(L_2\) and \(L_3\) are reshaped and transposed (\(\mathrm {RT}_1\), \(\mathrm {RT}_2\) and \(\mathrm {RT}_3\)) to different sizes as \(\mathbb {R}^{(H\times W)\times (C_1\times T)}\), \(\mathbb {R}^{(C_1\times T)\times (H\times W)}\) and \(\mathbb {R}^{(C_1\times T)\times (H\times W)}\), for performing a matrix multiplication (Mat) between them. Then, a softmax function is applied to the matrix multiplication result of \(L_4\) and \(L_5\) to obtain the space attention weight matrix (\(L_7\in \mathbb {R}^{D\times D}\)).
$$\begin{aligned} L^{ij}_{7}=\frac{Mat(L^{i}_{4},L^{j}_{5})}{\sum ^{D}_{j=1}Mat(L^{i}_{4},L^{j}_{5})} \end{aligned}$$(1)where D is (\(H\times W\)) which is equal to the number of electrode channels in \(M_j^i\). \(L_7^{ij}\in L_7\) ranges from 0 to 1, which represents the similarity weights between the \(i-th\) and the \(j-th\) electrode channels of \(M_j^i\). The more similar the motor-related characteristics, the larger the weight \(L_7^{ij}\).
-
Another matrix multiplication between \(L_7\) and \(L_6\) was performed to obtain \(L_8\). \(L_8\) is the new attention-based spatial representation of \(M_j^i\) that updates each electrode channel by adaptively aggregating 3D spatial features of other electrode channels according to the space attention weight matrix (\(L_7\)).
-
A \(1\times 1\) convolutional kernel (Conv3d_4) is implemented because of its powerful ability to decrease the dimension of feature blocks, while the \(1\times 1\) convolution operation only focuses on the feature block dimension, and the number of input data is constant. Therefore, it is suitable for handling a large number of input channels efficiently.
-
Finally, by multiplying a learnable parameter \(\gamma _1\) to \(L_9\) and performing an element-wise sum operation with the original EEG 3D representation \(M_j^i\), the attention-based adaptive spatial feature was obtained as follows.
$$\begin{aligned} L=M_j^i+L_9\times \gamma _1,L\in \mathbb {R}^{(T\times H\times W)} \end{aligned}$$(2)\(\gamma _1\) is a real number that automatically learns the optimistic value when training the proposed architecture. In contrast to other spatial feature extractions such as 2D convolution, 3D-SAM focuses on the location of electrode channels in the real world. It enhances the valuable motor-related features and suppresses useless motor-unrelated features based on 3D space information, which is consistent with our hypothesis that different brain functional regions may have certain effects on different MI tasks for different subjects. Therefore, by incorporating the self-attention mechanism into 3D convolution for the first time, 3D-SAM can be seen as a complement to the existing 3D neural networks.
-
-
2.
The attention-based spatial adaptive feature and the original EEG 3D representation \(M_j^i\) are combined via concatenation as
$$\begin{aligned} Y_1=\{M_j^i,L\} \end{aligned}$$(3) -
3.
Conv3d_A. is a function that transforms the \(Y_1\) into feature blocks \(Y_2\) using a 3D convolution, which, along with the width and height of \(M_j^i\) resulting a more compact structure.
-
4.
Conv3d_B+BN+NA. \(Conv3d\_B\), whose aim is to extract the spatial feature and reducing dimensions, is used to reshape the size as \(32\ \times \ T\ \times \ 1\ \times \ 1\). More compact features along the height and width of \(Y_2\) are obtained, but the distribution of input features may change, and the shift of the data distribution affects the training of the network (Santurkar et al. 2018). BN and NA are the batch normalization (Bjorck et al. 2018) and square nonlinear active function (Schirrmeister et al. 2017), which is applied to extract the final output spatial feature \(Y_3\) of this extraction phase.
Multiscale temporal feature extraction
In order to enhance the robustness of subject-specific and subject-independent classification, as shown by the yellow rectangle in Fig. 1 and Table 3, the extracted spatial features (\(Y_3\)) from an EEG 3D representation were cut into three time slices (\(TS^i\in \mathbb {R}^{32\times T_i\times N_i},i=\{1,2,3\}\), \(T_i\) is the length of each time slice, and \(N_i\) is the number of time slices with scale i) along the time dimension and fed into a designed multiscale temporal attention modular (MS-TAM)-based neural network (\(MA-TAM-i,i=\{1,2,3\}\)).
-
1.
Multiscale temporal attention module (MS-TAM). A subject usually concentrates on the trial some of the time, but is distracted at other times, and different subjects pay attention at different times within a trial, emphasizing on the EEG temporal slices when a subject concentrates on the trial while neglecting the other slices is necessary for successful EEG analysis. To better extract the time-invariant high-level features within each time slice, we assigned adaptive weights to different time slices by utilizing the attention mechanism to meet the requirement of EEG analysis, where different subjects concentrate on different temporal periods. In contrast to previous methods, the MS-TAM does not rely on subjects or tasks, but is also more robust to new subjects or tasks. Taking the MS-TAM of the \(i-th\) scale as an example, as shown in Fig. 4, \(TS^i\) is fed into two separate 2D convolutions (Conv2d_i1 and Conv2d_i2) to generate initial temporal features \(S_1^i\) and \(S_2^i\), the sizes of all of which were \(\mathbb {R}^{C_2\times T_i\times N_i}\). Then \(S_1^i\), \(S_2^i\) and \(TS^i\) are reshaped and transposed (\(\mathrm{RT}_1^i\), \(\mathrm{RT}_2^i\) and \(\mathrm{RT}_3^i\)) to generate \(S_3^i\), \(S_4^i\) and \(S_5^i\), whose sizes are \(\mathbb {R}^{N_i\times (C_2\times T_i)}\), \(\mathbb {R}^{(C_2\times T_i)\times N_i}\) and \(\mathbb {R}^{(C_2\times T_i)\times N_i}\). Matrix multiplication (Mat) between \(S_3^i\) and \(S_4^i\) with a softmax function is applied to obtain the temporal attention weight matrix (\(S_6^i\in \mathbb {R}^{N_i\times N_i}\)).
$$\begin{aligned} S^{i}_{6}(k,l)=\frac{Mat(S^{i}_{3}(k),S^{l}_{4})}{\sum ^{D}_{j=1}Mat(S^{i}_{3}(k),S^{l}_{4})} \end{aligned}$$(4)where \(N_i\) is the number of time slices under scale i. \(S_6^i(k,l)\) is the similarity weight between the \(k-th\) and the \(l-th\) time slices in a trial. \(S_6^i\) focuses on specific motor-related temporal slices that are more distinguishable than other motor-unrelated slices. The larger \(S_6^i(k,l)\) is, the more similar the \(k-th\) and the \(l-th\) time slices are to each other. Then, a weighted sum of all EEG temporal slices (\(S_5^i\)) is computed to learn an attention-based temporal representation (\(S_7^i\)). Finally, another learnable parameter \(\gamma _2^i\) to \(S_7^i\) and perform an element-wise sum operation with \(TS^i\) to obtain the attention-based adaptive temporal feature under scale i as follows.
$$\begin{aligned} S^i=TS^i+S_7^i\times \gamma _2^i,S^i\in \mathbb {R}^{\left( 32\times T_i\times N_i\right) }. \end{aligned}$$(5) -
2.
Poolingi + NA. Next, the \(i-th\) attention-based temporal adaptive representation (\(S^i\)) is fed into an Avg-Pooling layer (Poolingi) with a log nonlinear active function (NA) to aggregate the features of the temporal dimension in parallel. It further reduces the temporal dimension and transforms the low-level context features to high-level abstract features (\(P^1\), \(P^2\) and \(P^3\)) , which are concatenated as the multiscale spatiotemporal feature \(P=\{P^1,P^2,P^3\}\),\(P\in \mathbb {R}^{(32\times 1\times N)}\).
Dense fused classification
The dense fused classification phase consists of a dropout layer (Li et al. 2019), a convolutional layer (\(Conv2d\_C\)),a batch normalization layer and a LogSoftmax classification layer, as shown by the gray rectangle in Fig. 1. To reduce the computation and increase the robustness of the model, a dropout layer is employed before Conv2d_C. It randomly selects a portion of the input features with a certain Bernoulli probability distribution (p = 0.5) to reduce the risk of overfitting. Batch normalization was applied to high-level abstract features extracted by Conv2d_C. Finally, the typically LogSoftmax function is used for multi-classification by converting O to the conditional probability of four labels.
Implementation strategy
For the classification task, we used the negative log-likelihood loss function (NLLoss) to evaluate the proposed architecture. Let \(\theta \) be all parameters of our architecture, and the total loss function is defined as follows.
where \(\Omega \) denotes the number of trials, \(O_j^i\) represents the ground truth label of the \(i-th\) trial of \(j-th\) subject. \({{\overline{O}}}_j^i\) is the predicted result of the dense fused classification phase. \(\gamma \) is a hyperparameter whose value is between 0 and 1 (\(\gamma =0.01\)). \(\Vert \theta \Vert ^2\) is denoted as a regularization term to alleviate overfitting. Our goal is to find an optimal parameters \(\theta \) so as to obtain the minimum loss Loss(\(\theta \)).
The training configuration of the network was as follows.
-
1.
The Adam (2017) algorithm is employed as an optimizer.
-
2.
Convolution layer parameters are initialized by the Xavier algorithm (Glorot and Bengio 2010).
-
3.
The initial learning rate is 0.0001, and the decay weight is 0.01.
-
4.
The batch size is 32.
-
5.
80% and 20% trials in each dataset were selected randomly for training and cross-validation, respectively, while all test data were selected for the testing. The proposed architecture is optimized by the backpropagation of the total loss function in (6) on the training and validation sets.
-
6.
In addition, an early stopping strategy (Schirrmeister et al. 2017) was applied in this architecture.
The EEG signal preprocessing was carried out using MNE-Python (Gramfort et al. 2013) in the Ubuntu 16.04, 64bit system with an Intel(R) Core i9-9900X 3.50Ghz. For deep learning, we used one NVIDIA 2080Ti GPU with 12 GB. The Braindecode framework (Schirrmeister et al. 2017) was implemented using the PyTorch deep learning framework (Paszke et al. 2019).
Experiment results and discussion
To verify the accuracy and robustness of the proposed method, a series of experiments with two changeling public datasets (“Overall quantitative and qualitative evaluations for subject-specific classification using the IV-2a and HGD datasets” section).We provide a detailed insight into the overall performance of our method with those of other state-of-the-art methods using five ten-fold cross-validation. The confusion matrix is used for the quantitative evaluations. T-distributed stochastic neighbor embedding (T-SNE) (Maaten and Hinton 2008) was used for the qualitative evaluations.
The accuracy (7), which is the most widely used indicator for MI classification, was used as the evaluation metric.
where TP is the true positive, TN is the true negative, FP is the false positive, and FN is the false negative.
Public benchmark datasets
Two public MI EEG datasets were used to evaluate the proposed method. The first dataset was the BCI Competition IV-2a (IV-2a) (Ang et al. 2012). IV2a is a 25-channel (22 EEG and 3 EOG) with 4-class MI tasks (left hand, right hand, foot, and tongue, "http://www.bbci.de/competition/iv/#schedule") dataset from nine healthy subjects. It contained 72 trials per subject. The total number of trials was 5184. IV2a was separately recorded at 250 Hz and bandpass-filtered between 0.5 Hz and 100 Hz. Each MI EEG signal was divided into several labeled time fragments ([0.5s, 4s]), which were called trials. Thus, the input MI EEG signals were 22 electrode channel time series with 1125 sampling points.
In contrast to IV-2a, the high gamma dataset (HGD) (Schirrmeister et al. 2017) RECORDED the 4-class EEG signal of executed movement (left hand, right hand, rest, and both feet, "https://web.gin.g-node.org/robintibor/high-gamma-dataset"). All EEG signals of HGD were obtained from 44 electrode channels of 14 subjects. For each subject, approximately 880 trials and 160 trials were included in the training and testing datasets, respectively. The Tortola trial number was 14560. For a fair comparison with IV2a, HGD used the same time fragment ([0.5s, 4s]) and sampling rate (250 Hz), which contained 44 channel time series with 1125 sampling points per trial.
Overall quantitative and qualitative evaluations for subject-specific classification using the IV-2a and HGD datasets
Evaluations with the IV-2a dataset
Evaluations of the IV-2a dataset (Ang et al. 2012) (“Public benchmark datasets” section) were conducted to validate the performance and advancement of the subject-specific classification between the proposed method and other state-of-the-art methods in the past three years with various model structures and feature extraction strategies, such as deep learning-based methods (M3DCNN (Zhao et al. 2019), HSCNN (Dai et al. 2020), MS-AMF (Li et al. 2020), ETRCNN (Xu et al. 2020), TCNet-Fusion (Musallam et al. 2021), DJDAN (Hong et al. 2021), SSD-SE-CNN (Sun et al. 2021)), and machine-learning-based methods (PSCP (Dong et al. 2020) and TSGSP (Zhang et al. 2018).
As listed in Table 4, the proposed method had the best performance, with an average accuracy of 93.06% for the subject-specific classification. We first compared two recently published ML-based methods. PSCSP (Dong et al. 2020) proposed a new hybrid kernel function to fit both local (Gaussian) and global (polynomial) kernel functions for relevance vector machines. With the ‘one versus one’ CSP feature extraction strategy, it constructed six sets of spatial filters to extracted the phase space CSP features, which were classified by the hybrid kernel function. TSGSP (Zhang et al. 2018) used a joint sparse optimization of filter bands and time windows with temporal smoothness constraints to extract robust CSP features under a multitask learning framework. Although TSGSP achieved the best average accuracy of ML-based methods on the IV-2a dataset, reaching 84.00%, it was also nearly 10% lower than ours. It is limited by human expert knowledge and experience, and usually results in the extracted features may not be the most suitable for classification when different subjects exhibit significantly dynamic characteristics of EEG in different MI tasks.
Then, the proposed method was compared with SSD-SE-CNN Sun et al. (2021) and ETRCNN Xu et al. (2020), which are representative deep learning-based methods using feature maps as input formations and the time-frequency information in EEG data is fully used. The accuracy of these two methods is 79.30% and 84.57% respectively. However, the selection of the most optimal feature-extraction method for different subjects or different tasks is a changeling task and mainly depends on human experience. In contrast, we used the original signals as inputs to satisfy the relay time requirement. Furthermore, the proposed 3D-SAM and MS-TAM modules can eliminate the artifacts caused by the manually selected electrode channels and various biological, even adaptively improving the robustness and accuracy of different subjects or MI tasks for subject-specific and subject-independent. Thus, our average accuracies are 13.76% and 8.49% higher than those of SSD-SE-CNN and ETRCNN, respectively.
A further comparison with the most advanced representative original signal input case-based deep learning methods since 2019 was performed. Their average accuracy values range from 75.01% to 91.57%.
M3DCNN (Zhao et al. 2019) first introduced a 3D data structure into the EEG signal process. Its greatest contribution is to indicate that a deeper and more complex representation of the original MI EEG can help improve the performance. Our model demonstrated a better performance than M3DCNN (18.05% higher average accuracy).
MS-AMF (Li et al. 2020) proposed a multi-scale fusion CNN based on the attention mechanism, which extracts spatiotemporal multi-scale features from multi-brain regions representation signals and improves the network’s expression ability. However, when the channel is manually selected, the characteristic information of the MI signal will be lost. Compared with the average accuracy of 79.90% for MS-AMF, ours was 13.16% higher.
DJDAN (Hong et al. 2021) proposed a dynamic joint domain adaptation CNN to learn discriminative features for MI classification, simultaneously reducing the marginal and conditional distribution discrepancies across domains via global and local discriminators. However, a single receptive kernel size in a limited convolutional layer restricts DJDAN to extract high-level features for improving classification performance. Our average accuracy was 11.54% higher than that of DJDAN (81.52%).
TCNet-Fusion (Musallam et al. 2021) was proposed with the EEG-TCNet (Ingolfsson et al. 2020) architecture to improve accuracy by 83.61%. It extends the receptive field while increasing the number of parameters linearly, as opposed to traditional CNNs. In addition, based on the backbone of the inception-time network (Fawaz et al. 2020), EEG-inception (Zhang et al. 2021) attempted to explore the role of different depths and filter sizes on capturing space and time features from raw EEG data. The average accuracy reached 88.39%. However, the best convolution scale differs from subject to subject, which limits classification accuracy.
HSCNN (Dai et al. 2020) performed convolution on a hybrid scale to improve the classification. Three kernel sizes distributed with distances were used to extract EEG information in time, space, and frequency domains to suit different subjects. However, these methods used raw MI EEG as a 2D array input, omitting the potential spatial information in 3D space. In contrast, we automatically resolved the above problems using the proposed 3D representation and the MS-TAM module. Compared with the average accuracy of 91.57% for HSCNN, ours was still 1.49% higher.
In addition to the average accuracy, we also achieved the best results in three out of nine subjects (1, 3, 7 and 9). The confusion matrix and the T-SNE distribution of subject 1 and subject 3 are shown in Figs. 5 and 6.
Evaluations with the HGD dataset
To further verify the effectiveness and robustness of our method, evaluations were conducted on the HGD dataset (Schirrmeister et al. 2017) for subject-specific classification. The corresponding results are presented in Table 5. This clearly shows that our method was even more robust for different datasets and achieves a significantly better accuracy of 97.05% to the discussion above, the proposed method has been demonstrated as more powerful than other state-of-the-art methods.
Influence of different modules
In this section, we analyze the learning process of the proposed architecture to show how EEG features are encoded by the 3D spatial attention module (3D-SAM) and the multiscale temporal attention module (MS-TAM) modules for 4-class MI classification. In each evaluation, one module was omitted, and the others remained. All evaluations were performed on the VI-2a dataset for subject-specific classification.
Firstly, one modular was omitted and the others remained. Two evaluations were performed on the VI-2a dataset for subject-specific classification.
Confusion matrices of IV-2a dataset using the different modulars over all subjects. a-b Confusion matrixes of Evaluation 1 and 2. c Confusion matrix of the proposed method for subject-specific classification. d Confusion matrix of the proposed method for subject-independent classification. e-f Confusion matrixes of Evaluation 1 and 2 for subject-independent classification
In Evaluation 1, we omitted the MS-TAM while keeping the others. Here, we extracted the motor-related potential spatial dependent features and the corresponding hierarchical correlation between any two channels, which reflects the intrinsic connection of brain activity status in EEG. However, because the MI EEG has a low SNR and non-stationary signal, the evaluation result is easily affected by various biological (e.g., eye blinks, muscle artifacts, fatigue, and mood of a subject) and environmental artifacts (e.g., external noises). The result sharply decreases from 93.06% to 80.90%, which is nearly 12.16%. Meanwhile, as shown in Fig. 7a and c, our method efficiently reduces the misclassification error between the right hand and feet.
In Evaluation 2, we simply omitted the 3D-SAM while keeping the others. In this case, we extracted implicit multiscale temporal features based on the 3D representation. However, the intensity of the MI EEG signals varies among subjects, and it was impossible to manually determine which channels were most associated with the MI task. Hence, it was necessary to automatically assign the most suitable weights to motor-related brain electrode channels. When omitting the 3D-SAM, the corresponding average accuracy sharply decreases from 93.06% to 78.55%, which is nearly 14.51%. Meanwhile, as shown in Fig. 7b and c, the mistakes of misclassifying the right hand to the left hand and misclassifying feet to tongue were increased. The purpose of designing the 3D-SAM module was to improve the accuracy of cross-subject adaptation and eliminate the artifacts caused by the manual selection of signal channels, the corresponding subject-independent classification average accuracy increases from 60.61% to 62.77%, which is nearly 2.16%. Meanwhile, as shown in Fig. 7e and f.
Furthermore, we visualize the learned space and time attention weights from 3D-SAM and MS-TAM to discuss how these modulars encode spatial and temporal similarities among different electrode channels and different time slices, respectively.
According to the space attention similarity matrix (\(L_7\) , (1)), Fig. 8 presents 9 representative topological scalp plot maps of extracted spatial attention weights between different electrode channels. We define the blue color as the similarity between different electrode channels when subject imagery left hand movement (Fig. 8a and c). More blue the color, the stronger the similar correlation. In contrast, the red color is used as the similarity between different electrode channels when subject imagery right hand movement (Fig. 8b and d). More red the color, the stronger the positive correlation. Taking the Fig. 8a as an example, when a subject #1 imagery the left hand movement, channels with similar motor-related characteristics can mutually promote, regardless of their location in the space domain, not just the electrode channel \(C_3\) that traditional choose in previous methods. We can conclude that the 3D-SAM can explore motor-related potential spatial dependent features and the corresponding hierarchical correlation between any two channels, which eliminates the artifacts caused by the manual-selected electrode channels, and adaptively improves the accuracy of different subjects or MI tasks.
Topological scalp plot maps of extracted spatial attention weights between different electrode channels. The blue color as the similarity between different electrode channels when subject imagery left hand movement (Fig. 9(a and c). The red color is used as the similarity between different electrode channels when subject imagery right hand movement (Fig. 9b and d). These electrodes (\(C_3\), \(C_4\), and \(C_Z\)) directly over the motor cortex areas. More blue and red the color, the stronger the positive correlation
On the contrary, in order to explain how the MS-TAM module learns time attention similar weights, we collect the attention matrices (\(S^{i}_{6}\), (4), i means the scale 1, 2, 3) of the correctly classified samples of the MS-TAM model and plot the statistical results in Figs. 9, 10, 11 and 12. The elements in the attention matrix indicate the similar weight values between time slices under different scales. Larger number on horizontal axis means later in time. It is obvious that the weight values of temporal slices are varied from subject to subject for the same MI task (Figs. 9 and 10), or from task to task for the same subject (Figs. 11 and 12). This trend shows that the MS-TAM module can focus on the most MI task-related time slices, satisfying the phenomenon that different subjects have different ways of thinking and would concentrate on different temporal periods.
Taking Fig. 9 as an example, when subject #2 imagery right movement, the peak and valley trends of the curve throughout the attention matrices are located in the same time range (the sampling rectangles location on horizontal axis of scales). In contrast, although subject #7 imagery the same movement, the weight value distribution of temporal slices are different from subject #2. Hence, the MS-TAM modular can automatically learn weights to different time slices under different scales to adequately extract time-invariant high-level temporal features for a higher and more robust classification.
Evaluation for subject-independent classification using the IV-2a dataset
The main reason for the low accuracy performance of subject-independence is that the MI EEG signals vary over time from subject to subject, or from time to time for the same subject. It is impossible to determine exactly which electrode channels or time periods are most associated with MI. Therefore, traditional methods have limited performance in subject-independent classification.
In contrast, one of the main contributions of our method is to improve the classification accuracy for subject-independence with the 3D-SAM and MS-TAM modules, which automatically assigns weights to the most motor-related electrode channels and time periods. As shown in Fig. 13b, the ‘Leave-One-Subject-Out (LOSO)’ was used as the training and testing strategy for the conducted evaluations on the IV-2a dataset. For example, when computing the subject-independent accuracy of subject #1, the testing samples of subject #1 (green rectangle in Fig. 13b) were used as the testing samples for the performance evaluation. Meanwhile, all datasets of the other subjects (blue rectangles in Fig. 13b) were used as the training samples.
As listed in Table 7. Owing to the highly dynamic characteristics between subjects, the final average accuracy is lower than the results of the subject-specific classification listed in Table 7. As shown in Fig. 7d, the mistake of misclassifying the right hand to the left hand and misclassifying feet to the tongue are increased. However, the proposed method achieved an average accuracy of 63.27acceleration of 7.93and showed better results for eight out of nine subjects. It is concluded that the proposed method achieves a more robust classification performance in different backgrounds.
MI BCI-Based Robot System
Although the proposed method has achieved high accuracy performance, it is important to extend it to real-time, online MI-BCI applications. Therefore, we further tested and validated the real-time capability of the proposed model through the online decoding of MI movements from streamed EEG signals for NAO robot control. We used a Neuroscan 32-channel EEG amplifier to collect the MI EEG data, the system flow chart is shown in Fig. 14. As shown in Fig. 14a, each trial lasted for 7 s. Each test block consisted of 40 trials (10 for the right hand,10 for the left hand, 10 for the tongue, and 10 for both feet) and lasted 280 s. In a trial, a white fixation cross first appeared in the center of the screen to indicate that the trial was about to begin. One second later, one of four visual cases (the left hand, right hand, tongue, and both feet) appeared randomly, and the subjects were asked to imagine the corresponding action immediately without feedback (see Fig. 14b) and avoid eye and muscle movement artifacts. After 4 s, the visual case disappeared and the white letters indicating ‘Break’ appeared in the center of the screen, lasting 1 s for resting. The total number of trials was set at 400.
According to the international standard 10/20 system, the EEG data were acquired using the Neuroscan system with 24 Ag/AgCl scalp electrodes to record the signal (see Fig. 14c). The sampling frequency used in the experiment was 250 Hz. Before the data collection process, we used conductive glue to adjust the skin impedance of the EEG electrode to less than 5 kilo-ohms. Of the 24 electrodes, all but two reference electrodes were used for the data analysis. All subjects were asked to concentrate during the experiment to prevent other actions and brain activities from affecting the data collection (see Fig. 14d).
Figure. 15 describes the entire scheme of how we used the collected data to control the movement of robots by decoding the original MI EEG signals. When the subject imagined the movement of the left hand, right hand, tongue, and feet, the robot moved left, right, forward, and backward, respectively. Another live demonstration video is provided as a supplementary material.
Conclusion
In this paper, we have proposed an end-to-end 3D CNN to extract multiscale spatial and temporal dependent features to improve the accuracy performance of 4-class EEG MI classification tasks. It consists of a 3D representation, a 3D spatial attention module, a multiscale temporal attention module, and a dense fused classification module. With the definition of a compact feature representation of MI EEG in space and time domains, the proposed method adaptively assigns higher weights to motor-related spatial channels and temporal sampling cues than the motor-unrelated ones across all brain regions, which can prevent influences caused by biological and environmental artifacts. This indicates that the 3D EEG topological representation with the attention mechanism improves the overall accuracy performance more significantly.
Quantitative and qualitative evaluations were conducted using two public challenge datasets (IV-2a and HGD) to validate the robustness and accuracy of our method against various characteristics from subject to subject, or from time to time for the same subject. For the IV-2a dataset, the proposed MI EEG classification method achieved an average classification accuracy of 93.06dataset, the proposed method achieved an average classification accuracy of 97.05extraction and the impact of different modules are demonstrated using statistical significance tests. Although our method has been applied as a part of an advanced MI BCI-based robot system, we plan to refine it to explore further applications on wearable devices in the future.
References
Amin SU, Alsulaiman M, Muhammad G, Mekhtiche MA, Hossain MS (2019) Deep learning for eeg motor imagery classification based on multi-layer cnns feature fusion. Futur Gener Comput Syst 101:542–554
Ang KK, Chin ZY, Wang C, Guan C, Zhang H (2012) Filter bank common spatial pattern algorithm on bci competition iv datasets 2a and 2b. Front Neurosci 6:39
Baig MZ, Aslam N, Shum HP (2020) Filtering techniques for channel selection in motor imagery eeg applications: a survey. Artif Intell Rev 53(2):1207–1232
Bashivan P, Rish I, Yeasin M, Codella N (2015) Learning representations from eeg with deep recurrent-convolutional neural networks. arXiv preprint arXiv:1511.06448
Bjorck N, Gomes CP, Selman B, Weinberger KQ (2018) In Advances in Neural Information Processing Systems, pp. 7694–7705
Dai G, Zhou J, Huang J, Wang N (2020) Hs-cnn: a cnn with hybrid convolution scale for eeg motor imagery classification. J Neural Eng 17(1):016025
Dong E, Zhou K, Tong J, Du S (2020) A novel hybrid kernel function relevance vector machine for multi-task motor imagery eeg classification. Biomed Signal Process Control 60:101991
Fawaz HI, Lucas B, Forestier G, Pelletier C, Schmidt DF, Weber J, Webb GI, Idoumghar L, Muller PA, Petitjean F (2020) Inceptiontime: finding alexnet for time series classification. Data Min Knowl Disc 34(6):1936–1962
Gaur P, Gupta H, Chowdhury A, McCreadie K, Pachori RB, Wang H (2021) A sliding window common spatial pattern for enhancing motor imagery classification in eeg-bci. IEEE Trans Instrum Meas 70:1–9
Glorot X, Bengio Y (2010) In: Proceedings of the thirteenth international conference on artificial intelligence and statistics, pp. 249–256
Gong A, Liu J, Chen S, Fu Y (2018) Time-frequency cross mutual information analysis of the brain functional networks underlying multiclass motor imagery. J Mot Behav 50(3):254–267
Gramfort A, Luessi M, Larson E, Engemann DA, Strohmeier D, Brodbeck C, Goj R, Jas M, Brooks T, Parkkonen L et al (2013) Meg and eeg data analysis with mne-python. Front Neurosci 7:267
Hong X, Zheng Q, Liu L, Chen P, Ma K, Gao Z, Zheng Y (2021) Dynamic joint domain adaptation network for motor imagery classification. IEEE Trans Neural Syst Rehabil Eng 29:556–565
Ingolfsson TM, Hersche M, Wang X, Kobayashi N, Cavigelli L, Benini L (2020) In: 2020 IEEE International Conference on Systems, Man, and Cybernetics (SMC) (IEEE), pp. 2958–2965
Kwon OY, Lee MH, Guan C, Lee SW (2019) Subject-independent brain-computer interfaces based on deep convolutional neural networks. IEEE Trans Neural Netw Learn Syst 31(10):3839–3852
Lawhern VJ, Solon AJ, Waytowich NR, Gordon SM, Hung CP, Lance BJ (2018) Eegnet: a compact convolutional neural network for eeg-based brain-computer interfaces. J Neural Eng 15(5):056013
Lei B, Liu X, Liang S, Hang W, Wang Q, Choi KS, Qin J (2019) Walking imagery evaluation in brain computer interfaces via a multi-view multi-level deep polynomial network. IEEE Trans Neural Syst Rehabil Eng 27(3):497–506
Li Y, Zhang XR, Zhang B, Lei MY, Cui WG, Guo YZ (2019) A channel-projection mixed-scale convolutional neural network for motor imagery eeg decoding. IEEE Trans Neural Syst Rehabil Eng 27(6):1170–1180
Li D, Xu J, Wang J, Fang X, Ying J (2020) A multi-scale fusion convolutional neural network based on attention mechanism for the visualization analysis of eeg signals decoding. IEEE Trans Neural Syst Rehabil Eng 28(12):2615–2626
Li X, Chen S, Hu X, Yang J (2019) In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 2682–2690
Liu X, Lv L, Shen Y, Xiong P, Yang J, Liu J (2021) Multiscale space-time-frequency feature-guided multitask learning cnn for motor imagery eeg classification. J Neural Eng 18(2):026003
Lotte F, Bougrain L, Cichocki A, Clerc M, Congedo M, Rakotomamonjy A, Yger F (2018) A review of classification algorithms for eeg-based brain-computer interfaces: a 10 year update. J Neural Eng 15(3):031005
Ma X, Qiu S, Wei W, Wang S, He H (2019) Deep channel-correlation network for motor imagery decoding from same limb. IEEE Trans Neural Syst Rehabil Eng 28(1):297–306
Ma X, Wang D, Liu D, Yang J (2020) Dwt and cnn based multi-class motor imagery electroencephalographic signal recognition. J Neural Eng 17(1):016073
Maaten Lvd, Hinton G (2008) Visualizing data using t-sne. J Mach Learn Res 9(Nov):2579–2605
Miao Y, Jin J, Daly I, Zuo C, Wang X, Cichocki A, Jung TP (2021) Learning common time-frequency-spatial patterns for motor imagery classification. IEEE Trans Neural Syst Rehabil Eng 29:699–707
Musallam YK, AlFassam NI, Muhammad G, Amin SU, Alsulaiman M, Abdul W, Altaheri H, Bencherif MA, Algabri M (2021) Electroencephalography-based motor imagery classification using temporal convolutional network fusion. Biomed Signal Process Control 69:102826
Pang Y, Zhao X, Zhang L, Lu H (2020) In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9413–9422
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L et al. (2019) In: Advances in neural information processing systems, pp. 8026–8037
Penaloza CI, Nishio S (2018) Bmi control of a third arm for multitasking. Sci Robot 3(20):eaat1228
Sakhavi S, Guan C, Yan S (2018) Learning temporal information for brain-computer interface using convolutional neural networks. IEEE Trans Neural Netw Learn Syst 29(11):5619–5629
Santurkar S, Tsipras D, Ilyas A, Madry A (2018) In Advances in Neural Information Processing Systems, pp. 2483–2493
Schirrmeister RT, Springenberg JT, Fiederer LDJ, Glasstetter M, Eggensperger K, Tangermann M, Hutter F, Burgard W, Ball T (2017) Deep learning with convolutional neural networks for eeg decoding and visualization. Hum Brain Mapp 38(11):5391–5420
Sharma M, Pachori R, Rajendra A (2017) Adam: a method for stochastic optimization. Pattern Recogn Lett 94:172–179
Sun B, Zhao X, Zhang H, Bai R, Li T (2021) Eeg motor imagery classification with sparse spectrotemporal decomposition and deep learning. IEEE Trans Autom Sci Eng 18(2):541–551
Wu H, Li F, Li Y, Fu B, Shi G, Dong M, Niu Y (2019) A parallel multiscale filter bank convolutional neural networks for motor imagery eeg classification. Front Neurosci 13:1275
Xie X, Yu ZL, Lu H, Gu Z, Li Y (2016) Motor imagery classification based on bilinear sub-manifold learning of symmetric positive-definite matrices. IEEE Trans Neural Syst Rehabil Eng 25(6):504–516
Xu M, Yao J, Zhang Z, Li R, Yang B, Li C, Li J, Zhang J (2020) Learning eeg topographical representation for classification via convolutional neural network. Pattern Recognit 105:107390
Zhang Y, Zhou G, Jin J, Wang X, Cichocki A (2015) Optimizing spatial patterns with sparse filter bands for motor-imagery based brain-computer interface. J Neurosci Methods 255:85–91
Zhang Y, Nam CS, Zhou G, Jin J, Wang X, Cichocki A (2018) Temporally constrained sparse group spatial patterns for motor imagery bci. IEEE Trans Cybern 49(9):3322–3332
Zhang J, Xie Y, Wu Q, Xia Y (2019) Medical image classification using synergic deep learning. Med Image Anal 54:10–19
Zhang D, Yao L, Chen K, Wang S, Chang X, Liu Y (2019) Making sense of spatio-temporal preserving representations for eeg-based human intention recognition. IEEE Trans Cybern 50(7):3033–3044
Zhang L, Wang X, Yang D, Sanford T, Harmon S, Turkbey B, Wood BJ, Roth H, Myronenko A, Xu D et al (2020) Generalizing deep learning for medical image segmentation to unseen domains via deep stacked transformation. IEEE Trans Med Imaging 39(7):2531–2540
Zhang H, Zhao X, Wu Z, Sun B, Li T (2021) Motor imagery recognition with automatic eeg channel selection and deep learning. J Neural Eng 18(1):016004
Zhang C, Kim YK, Eskandarian A (2021) Eeg-inception: an accurate and robust end-to-end neural network for eeg-based motor imagery classification. J Neural Eng 18(4):046014
Zhang X, Yao L, Wang X, Monaghan J, Mcalpine D, Zhang Y (2019) A survey on deep learning based brain computer interface: Recent advances and new frontiers. arXiv preprint arXiv:1905.04149
Zhao X, Zhang H, Zhu G, You F, Kuang S, Sun L (2019) A multi-branch 3d convolutional neural network for eeg-based motor imagery classification. IEEE Trans Neural Syst Rehabil Eng 27(10):2164–2177
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this research study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, X., Wang, K., Liu, F. et al. 3D Convolution neural network with multiscale spatial and temporal cues for motor imagery EEG classification. Cogn Neurodyn 17, 1357–1380 (2023). https://doi.org/10.1007/s11571-022-09906-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11571-022-09906-y