Abstract
The multi-organ contouring (MOC) task is required when a radiotherapy is performed to eradicate the cancerous tissue while minimizing the dosage delivered to the surrounding healthy organs. Currently most of the task is done manually with enormous labor and time cost. To reduce the scheduling waiting time from increasing cancer population, it is beneficial to both the patients and therapeutists to have an automatic contouring tool. In this work an Attention-LSTM fused U-Net model is proposed to perform the multiple organ segmentation from a CT image series. The organs to be delineated include lung, liver, stomach, esophagus, heart, and kidneys. To train and evaluate our model, the CT image series of 146 patients was acquired from a local hospital with IRB approval. The segmentation accuracy of the six organs in terms of Dice Similarity Coefficient (DSC) were 99.27%, 95.48%, 88.53%, 80.81%, 93.8%, and 93.46%, respectively. To make the AI-embedded MOC system readily applicable in clinical environments, a data processing workflow and the corresponding GUI were also implemented and published on Github. The doctors can download the CT image data from the PACS server, use our system to perform MOC tasks, and output the contouring results in DICOM-RT format so they can be uploaded back to the treatment planning system for further fine-tuning and dosage/path calculation. To our best knowledge the work might be the first non-commercial model-integrated system compatible with the commercial treatment planning systems and ready to be used by the doctors.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Radiotherapy plays an indispensable role in current tumor treatment. Starting from the linear accelerator, a high-energy radiation is generated to eradicate cancerous tissue or tumor. Even with a programmable and rotational emission source and a six degree-of-freedom couch, the radiation still inevitably has impact to the normal tissue around the target. To make sure the tumor receives the maximum dosage of radiation while the surrounding normal tissue receives the minimum, the computerized tomography (CT) scan should be taken to locate the tumor region and organ at risk (OAR) separately. With their contours the commercial treatment planning system can calculate the entry path of the radiation beam and the sufficient radiation dosage for tumor and OAR.
Using CT images to locate tumors and OAR is not difficult for a professional therapist, but manually delineating their contours for an image series that could contain hundreds of images is tedious and time-consuming. It usually takes an hour to finish the contouring task over a CT image series of a patient. Therefore an automatic contouring tool is highly desirable. Although there are some commercial solutions embedded in the treatment planning and radiotherapy systems, such as Eclipse (Smart Segmentation) from Varian, Monaco (Smart Sequencing) from Elekta, Pinnacle (Model-Based Segmentation) from Philips, iPlan from BrainLab, their contouring accuracy may not be satisfactory [5,6,7, 17, 20]. Sometimes it might need more manpower and time to refine the results and cannot reach clinically practical effectiveness. In addition, since these contouring modules are part of their treatment planning systems, the users have to pay a high price but cannot select a good contouring tool that is suitable enough for their existing clinical workflow in their hospital.
Although several multi-organ segmentation networks have been proposed, which will be reviewed later in the next section, there is still a barrier for the medical doctors. They do not have time or the skills to implement these techniques. Even they have, the networks usually output segmentation masks as images. These images in TIFF, BMP, or PNG formats cannot be uploaded back to the clinical PACS server and cannot be reused by the treatment planning systems. To resolve the dilemma, we establish a contouring system that is not only using deep neural networks to do the work but also independent from the existing radiotherapy systems. A user can download the CT image series data in DICOM format from their PACS server and input to our contouring system. In the system each organ has the corresponding trained AI models. It is replaceable in case a better organ segmentation model is released. The system automatically goes through all available contouring AI models and acquires the contours for different organs. These contouring results are written in public DICOM-RT format, which is recognizable by all the commercial treatment planning system. After uploading the contouring results back to the PACS system, the finetuning work can be finished shortly in the current treatment planning system on duty.
Except discussing the features of the system from user perspective, in the study we also proposed an organ segmentation network. It is trained by thousands of CT images and the manually-drawn contours (i.e. groundtruth) provided by Tzu Chi Hospital in Chiayi, Taiwan with IRB approval. Six models corresponding to six organs (lung, liver, stomach, esophagus, heart, and kidneys) are developed separately. The reason of training separate models for each organ in our system instead of one unified multi-organ segmentation model is because the users can retrain and update each model in case a new AI model with good performance for a specific organ is released. Although the basic architecture of the model was already released in our previous work [2], we added more organs segmentation models, performed ablation studies, completed the graphical user interface design of the system and published it on Github.
The rest of the paper is organized as follows. In Section 2 current status of multi-organ segmentation and several relevant segmentation networks were reviewed. The proposed network and its experiments are discussed in Section 3 and 4 respectively. Finally the conclusions are made in Section 5.
2 Related work
2.1 Multi-organ segmentation
Tian et al. [18] proposed an end-to-end deep neural network for multi-organ segmentation. They combined U-Net with the accuracy-complexity adjustment module (ACAM) to improve the accuracy of segmentation while reducing the model complexity and memory usage. A multiscale aggregation module (MAM) is also added to the encoder section to enrich feature representation and improve segmentation performance. This architecture used a data set from a private hospital to train segmenting the left lung, right lung, heart, and spinal cord. The accuracy in dice similarity coefficient (DSC) was 97.76%, 97.52%, 95.27% and 92.24%, respectively. Tang et al. [15] proposed an architecture that used both 2D and 3D models. First, 2D convolution was used for segmentation, and the spatial context information was extracted from the 3D model. Then a self-attention mechanism is used to control which 3D features should be used to guide 2D segmentation. They used MICCAI 2015 head and neck OAR segmentation challenge dataset. The DSC performance for 10 organs (large bowel, duodenum, spinal cord, liver, spleen, small bowel, pancreas, left and right kidneys, stomach, gallbladder) were 82.5%, 70.7%, 90.8%, 96.4%, 95.9%, 76.5%, 82.1%, 96%, 95.7%, 87.5% and 82.2% respectively.
Another Tang et al. [16] proposed a patch-based network fusion model featured on random spatial initialization and statistical fusion of the perceived overlap region (ROI). The whole network has 3 parts. First a 3D U-Net was used for preliminary segmentation. Then the surrounding area of the target was removed by random patch sampling. Finally, a majority voting mechanism was applied to perform the final segmentation. The authors used the BTCV miccai2015 challenge dataset to segment 13 organs including spleen, liver, stomach, right kidney, left kidney, esophagus, aorta, IVC, pancreas, gallbladder, portal vein and splenic vein, and adrenal glands. The corresponding DSC were 96.35%, 96.56%, 85.67%, 93.1%, 94.53%, 78.81%, 92.32%, 85.28%, 76.08%, 82.63%, 72.79%, and 73.56%, respectively.
2.2 2D organ segmentation
When pooling is used to retain strong features in semantic segmentation tasks, it may result in the size reduction and the information loss of boundary details and target locations. Ronneberger et al. proposed the U-net [10], a symmetric network with encoder-decoder structure to solve the problem. The features of encoder layers are copied to the corresponding decoder layers to retain the missing information. Such operation is beneficial to medical image segmentation tasks. U-net achieved a high Mean Intersection over Union (mIoU), 92.03%, in the Cell Segmentation task of the 2015 ISBI challenge. Few years later, Huang et al. [3] proposed an improved U-Net called Unet3+. With full-scale skip connections, the low and high-level semantic in the multi-scale features are combined for better usage. A novel hybrid loss function was proposed to enhance extracting the boundaries of organs, and the classification-guided module (CGM) was used to avoid segmenting the images without the target organs. The performance was demonstrated using the liver data set of the ISBI LiTS 2017 Challenge and the spleen data set from a private hospital. The accuracy in DSC was 96.75% and 96.2%, respectively.
2.3 3D organ segmentation
Yu et al. [21] proposed the Volumetric ConvNet to segment the ambiguous boundaries of prostate in MR images. It is a 3D CNN model, and contains short connection paths within the residual blocks and long connection paths between the residual blocks. It greatly improved the learning efficiency from the neighboring context, and enhanced the local and global information transmission. It won the first place in MICCAI Promise12 Challenge with Dice Score 89.43% in 2017.
Novikov et al. [8] proposed a U-Net like network called Sensor3D to solve two issues shared by most concurrent 3D segmentation networks. One is that the number of input images needs to be fixed, and the other is the requirement of a whole volume of tomographic images, such as CT and MRI. Sensor3D has the Contraction block and Extraction block (C.E. blocks) for its basic segmentation capability, which are just like the down-sampling and up-sampling parts in the U-Net. The difference is that U-net takes one image as input, while Sensor3D takes three consecutive slices from a CT image series and treats each one independently in the C.E. blocks. The inherited spatial-temporal correlation between neighboring slices are learned from the Bidirectional ConvLSTMs [12], which are placed between the C.E. blocks and considered the input as a time series at time t-1, t, and t + 1. The use of two Bidirectional C-LSTM modules is to let the image processing at time t being benefited from the work at time t-1 and t + 1. By learning the local spatial information from one image and the temporal relationships from its two adjacent ones, Sensor3D resolved the whole volume requirement and the limitations on the large-memory consumption. The open data set 3Dircadb was used to evaluate its liver segmentation, and a Dice Score 95.4% was shown to outperform some other methods that also used 3Dircadb for evaluation, such as constrained level set, deformable graph cut, and cascaded FCN with random fields. Another open dataset CSI 2014 was used to demonstrate its superiority in vertebrate segmentation over some active shape model and atlas-based methods.
Wang et al. [19] proposed an organ-attention networks with reverse connections (OAN-RCs) to segment the abdominal organs. The idea behind is that the tomographic images contain abundant three-dimensional spatial information, and each of the coronal, sagittal, and axial perspective has different spatial correlation. The segmentation accuracy will be much improved if all the three kinds of correlation can be learned. When performing specific organ segmentation, in addition to the targeted organ, it also focused on the nearby background only instead of the whole image. An organ-attention map was combined with the original images under a two-stage training framework. The segmentation models using three perspectives were trained separately. Then these organ labels were fused together by expectation-maximization algorithm to derive a refined segmentation map. Due to the local structural similarity-based label fusion, OAN-RC outperformed 3D U-Net, 2D-FCN, hierarchical 3D-FCN on 13 organ or tissues segmentation tasks (aorta, colon, duodenum, gallbladder, interior vena cava (IVC), kidney (left, right), liver, pancreas, small bowel, spleen, stomach, and large veins). The corresponding mean DSC scores are 91.8%, 83%, 75.4%, 90.5%, 87%, 96.8%, 98.4%, 98%, 87.8%, 80.1%, 97.1%, 95.2%, and 80.7%.
3 Methodology
In this section, we will first introduce the flow chart of data processing in the clinical environment. Then the detail of image preprocessing and the our proposed network will be discussed.
3.1 Clinical work flow of data processing
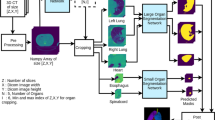
Due to the format of the current public datasets, most networks available on the internet take images in usual formats such as jpeg, png, bmp, etc., and output the masks in the same formats. To integrate the organ contouring system with the hospital information systems (i.e. PACS or RIS), generating the organ contours from an existing network is only halfway through. A complete data process is shown in Fig. 1. Assume that the CT image series in DICOM format and the manually-drawn contours in DICOM-RT format have been downloaded from the PACS server, and are ready to be used for training purpose. In the training stage, the actual image data is first extracted from the DICOM format. The Hounsfield units (HU) values in the CT images are rescaled in the preprocessing step to reduce the variability between different CT scanners. The HU value is defined based on the decay factor of X-rays passing through the human body, and closely related to the density of tissues or organs on CT images. The image is then resized in quarter before being sent to the network to ease the training loading. The weight is updated throughout the training process until the neural network converges and these model parameters are stored for being used in the prediction stage.
The test stage is similar to the training stage except that the predicted organ masks are smoothed and resized back to the original image size. Then the contouring results are extracted from the masks and converted to DICOM-RT format. During the conversion the tags ‘Media Storage SOP Instance UID’, ‘SOP Instance UID’, and ‘Implementation Class UID’ need to be distinct from the original ones so uploading them to the server will not trigger the file protection mechanism. The distinct UID tags setting allows the contours being readable and modifiable from the treatment planning system. These are the post-processing steps.
3.2 Data pre-processing in six steps
Each of the CT images for a patient downloaded from the PACS server is originally in DICOM format, and contains a number of tags that record all the important information including the UID number that identifies the file, the imaging parameters, the file storage information, etc. The pixel values of the extracted image data are usually in the range of [0, 4096]. Since it might vary with different hardware and software, the 1st step of preprocessing is to convert the pixel value to the original CT value. The pixel value-CT value conversion is shown in Eq. (1), where ‘Rescale Slope’ and ‘Rescale Intercept’ are two tags in the DICOM header and vary with the manufacturer of the CT scanner.
After the HU value is derived from Eq. (1), the 2nd step of preprocessing is to rescale the image to [0, 255] according to appropriate ‘Window Level’ and ‘Window Width’ parameters in Eqs. (2) ~ (5). The two DICOM tags represent the center value and the and the upper / lower limits of the gray scale range. Adjusting these settings can changes the brightness and contrast of the CT images. The smaller the ‘Window Level’ value, the brighter the brightness. The smaller the ‘Window Width’ value, the higher the contrast. There are several Window Level/Width settings often used clinically for diagnosing different tissues under a good image contrast, such as 40/80 for the brain, 600/1500 for the lung, and 400/1800 for the spinal bone. Here we followed the clinicians’ suggestion and used −100/400 to appropriately display the abdominal organs for most of the cases.
The 3rd step of preprocessing is applying the Contrast Limited Adaptive Histogram Equalization (CLAHE) to each image for further contrast enhancement and noise removal. As shown in Fig. 2, the image after applying the CLAHE has better contrast and more distinct organ contours, which could make it easier for the following AI model to learn the semantic features of organs. The 4th step of preprocessing is resizing the image from 512 × 512 to 256 × 256 to reduce GPU memory usage and speed up the training process. The 5th step of preprocessing is normalizing the pixel values to [0, 1] to prevent any larger feature variation from biasing the gradient descent process during model training. It will also accelerate the model convergence.
The last step of preprocessing before entering the training process is to duplicate the first and the last images of a CT image series and make them the first and the last ones in the series. In other words, the first two images are the same, and so do the last two. Since our AI model in the next section will perform organ contouring based on the current image and its previous and next ones, without such duplication the model cannot perform contouring task on the first and the last images in the CT series.
3.3 Attention-LSTM fused U-Net network
We proposed a modified Sensor3D as shown in Fig. 3. The original contraction and expansion blocks are unchanged to maintain the basic 2D-segmentation capability. The semantic information of the inputted three consecutive slices were independently extracted from the two blocks. The two Bidirectional Convolutional LSTM modules were also kept. The LSTM part explored the temporal correlation between the three slices over multiple channels. The convolutional operation made the module also sensitive to the spatial-temporal correlation over multiple channels. The bi-direction operation is equal to have both forward and backward C-LSTMs such that the prediction results of the center slice will be benefited from the ones of its neighboring slices. In other words, if the two neighboring slices do not have some organ, then the prediction result of the center slice should not have such organ as well.
Attention-LSTM Fused U-Net Architecture (reworked from [8])
Other than the remaining Contraction block, Expansion block, and two bidirectional C-LSTMs we introduced three changes in the modified architecture. The first one is adding the Attention Gate [9] between the Contraction and Expansion blocks. Such gate can filter out the features irrelevant to the targeted organ region when low level semantic information is passing to the high level ones through the skip connection paths between two blocks. Without the gate all of the low level information in the image extracted in the contraction block could be forwarded to the expansion block. Oktay et al. compared a plain U-Net with the Attention U-Net, and demonstrated the improved accuracy by segmenting the pancreas, spleen, and kidney [9].
After the second bidirectional C-LSTM module, a segmentation mask can be derived by passing the output to a 1 × 1 convolution layer with a sigmoid activation. This is the original operation of Sensor3D. Our second change is to let such segmentation result being multiplied by the slice at time t again. The operation is considered as deriving an attention map. The reason to introduce this change is that we found in some cases the original segmentation result of Sensor3D may over segment some small organs (small size and occupy less slices), even the CLAHE has been applied in the preprocessing to enhance the boundaries of organs. Such attention map derivation could remove irrelevant tissues and preserve the area of the target organ. To process the attention map, we used U-Net again to perform the final image segmentation. The U-Net here plays a role similar to a clinician that receives the initial segmentation result from an automatic organ contouring tool, and then fine-tunes it according to his clinical experience and anatomical knowledge.
The third change is using a weighted summation of loss functions that assigns equal weight 0.5 to the existing generalized Dice loss (GDL) and Binary cross entropy (BCE). GDL can handle the data imbalance issue [13] and avoid overfitting toward the background when the number of pixels in the foreground is far less than the ones in the background. Such property makes it suitable for small organs segmentation. However, there are also large organs such as the lung and liver that could occupy more than 50% of the body region in the transverse plane, and their feature learning may not be efficient under the influence of GDL. Therefore, to make a segmentation model work for both large and small organs, we combined both GDL and BCE loss to reach the balance. The BCE and GDL [13] functions are shown in Eqs. (6) and (7)
where l is the number of class, n is the number of pixels, w is the weight of corresponding class, r and p represent groundtruth and prediction respectively. The class weight wl is determined according to the number of groundtruth pixels in that class, and defined as \( {w}_l=1/{\left({\sum}_{n=1}^N{r}_{ln}\right)}^2 \). In GDL the less pixels the groundtruth has, the larger the class weight is. In the following ablation study we will compare the models’ performance when the loss function is either the GDL only or the combined version, which is denoted as GDL&BCE and defined as 0.5 GDL + 0.5 BCE.
4 Experimental results and evaluation
In this section, we will introduce the data sets used in our experiments, the amount of data used for each organ, as well as the evaluation methods. Finally, the performance of different models as a ablation study for each organ is tabulated.
4.1 Experimental dataset
The dataset used in our experiments is CT images series retrospectively collected by the Radiation Oncology Department of Tzu Chi Hospital in Dalin with IRB approval (B10804010-1). The CT scanner used for the lung, liver, stomach, esophagus, and heart dataset is manufactured by SIEMENS, and the one for the kidney dataset is manufactured by GE MEDICAL SYSTEMS. The original image size is 512 × 512. The contours of six organs were manually drawn by the physicians and have been applied to actual treatment planning. The number of training and testing datasets varies with different organs and are shown in Table 1.
4.2 Evaluation index
The Dice similarity coefficient (DSC) is the most commonly used index in the field of medical image segmentation, and is used here for evaluation. It calculates the ratio of overlap between two areas. According to [14], the formula is shown in Eq. (8) and the terms in the numerator and denominator are the elements in the Confusion Matrix where TP = True Positive, FP = False Positive, TN = True Negative, FN = False Negative.
4.3 Ablation study
Comparing to the original sensor3D architecture, we introduced three kinds of changes: attention gate, generating new attention map and followed by U-net segmentation, and sum of two loss functions (generalized Dice loss and Binary Cross Entropy loss). The performances in DSC scores over the six organs were tabulated in Table 2. The highest DSC scores in the ablation study were marked in bold face. The state-of-art performance with ‘*’ symbol derived from other datasets were also documented in Table 2 as a reference, although they are not comparable with our DSC scores due to different training conditions and datasets.
Table 2 shows that our proposed combinations of changes reached the highest DSC score only when segmenting the Lung, Liver, and Kidney. The original Sensor3D network still has the highest DSC scores in the other three organs. This suggested us that applying separate models for different organs is a more flexible way to maintain the highest segmentation accuracies for our automatic contouring system. With the same dataset and parameter settings, a missing value happened when the BCE loss was applied to Esophagus. This is because the model cannot be trained normally. The training loss kept declining but the testing loss remained the same, which indicated that the current network might be overfitting. The other 4 models from ‘Sensor3D’ to ‘Sensor3D + U-Net + Attention’ have values higher than 80% also showed that Esophagus occupies the least area and therefore is much more suitable for using the GDL function only.
Except the overall DSC scores, we also selected some example images and displayed both the predicted contours of the six organs generated by different models and the gold standard delineated by the clinicians in Fig. 4. The green line represented the ground truth, and the red line represented the results predicted by AI. Most of the time the two contours were largely overlapped with each other.
Although our modifications did not show overall significant improvement over the original Sensor3D in Table 2, its leading gap is only to the first decimal place for three organs. Such tiny difference less than 1% could change their directions by using different testing datasets. In addition, when an organ has complex shapes and large variability over different subjects, such as the liver and stomach shown in Fig. 5 (case 3) and Fig. 6 (case 1), our modification demonstrated better contouring accuracy over Sensor3D. This can be found only by examining the actual contouring results. Choosing the best model by simply comparing the DSC scores may miss some models that can bring practical clinical values and benefits.
4.4 Graphical user Interface for the clinicians
In order to integrate the automatic organ contouring system with the hospital information systems (i.e. PACS or RIS), we need to first extract the CT image data from the DICOM format, perform pre-processing, initiate model training, and finally write the prediction results back to DICOM-RT format. To reduce the effort typing python commands for the clinicians, these steps are integrated into a graphical user interface (GUI) shown in Fig. 7. This is convenient for the clinicians to perform the automatic contouring tasks and save the results back to their treatment planning system. The GUI and the automatic organ contouring system have been released on Github (https://github.com/chenpin627/Organ-Segmentation-UI). The steps to use the GUI are listed as follows:
-
Step 1:
Click the “Load Data” button and select the CT image folder of 1 patient pre-downloaded from the PACS server.
-
Step 2:
Click the “Load DicomRT” button and select the patient’s DicomRT file.
-
Step 3:
Check the organ to be predicted (can check multiple organs at once).
-
Step 4:
Click “Prediction Start” button to start prediction, and finally a DicomRT file will be generated.
-
Step 5:
Upload the DicomRT file back to the server, then the contours can be read or refined in the treatment planning system.
5 Conclusions
In this work a modified Sensor3D is proposed to perform organ contouring on CT images. We compare the results of several versions of modified Sensor3D with the original model. The DSC scores of different models for each organ may not be significantly different, but when we tested with difficult cases such as liver and stomach that have more complicated shapes, our proposed model generated better contour than Sensor3D. In addition to comparing the accuracy between different models, a GUI for our automatic contouring system was also established to allow the AI-generated contours compatible and modifiable under the current hospital information systems. The tool is open sourced, available on the Github, and is extremely helpful for the clinicians. To our best knowledge the work might be the first non-commercial model-integrated system compatible with the commercial treatment planning systems and ready to be used by the clinicians.
Data availability
No.
Code availability
References
Bilic P, Christ PF et al (2019) The Liver Tumor Segmentation Benchmark (LiTS). arXiv:1901.04056
Chen PH et al (2020) Attention-LSTM fused U-Net architecture for organ segmentation in CT images. International Symposium on Computer, Consumer and Control (IS3C)
Huang H et al (2020) UNet 3+: A Full-Scale Connected UNet for Medical Image Segmentation. ICASSP 2020–2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), Barcelona, Spain, pp 1055–1059
Liu Y, Lei Y, Fu Y, Wang T, Tang X, Jiang X, Curran WJ, Liu T, Patel P, Yang X (2020) CT-based multi-organ segmentation using a 3D self-attention U-Net network for pancreatic radiotherapy. Med Phys 47:4316–4324. https://doi.org/10.1002/mp.14386
Macchia ML et al (2012) Systematic evaluation of three different commercial software solutions for automatic segmentation for adaptive therapy in head-and-neck, prostate and pleural cancer. Radiat Oncol 7:160
Maitre AL et al (2012) Impact of the accuracy of automatic tumour functional volume delineation on radiotherapy treatment planning. Phys Med Biol 57(17):5381–5397
Mattiucci GC, Boldrini L, Chiloiro G, D’Agostino GR, Chiesa S, de Rose F, Azario L, Pasini D, Gambacorta MA, Balducci M, Valentini V (2013) Automatic delineation for replanning in nasopharynx radiotherapy: what is the agreement among experts to be considered as benchmark? Acta Oncol 52(7):1417–1422
Novikov AA et al (2019) Deep sequential segmentation of organs in volumetric medical scans. IEEE Trans Med Imaging 38(5):1207–1215
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K et al ( 2018) Attention u-net: Learning where to look for the pancreas. arXiv preprint arXiv:180403999
Ronneberger O et al (2015) U-net: convolutional networks for biomedical image segmentation. In: International Conference on Medical image computing and computer-assisted intervention. Springer, Cham
Shen C, Milletari F et al (2019) Improving V-Nets for multi-class abdominal organ segmentation. Proc. SPIE 10949, Medical Imaging 2019: Image Processing, 109490B
Shi X et al (2015) Convolutional LSTM Network: a machine learning approach for precipitation nowcasting. In: Proceedings of the 28th International Conference on Neural Information Processing Systems—volume 1 (NIPS’15). MIT Press, Cambridge
Sudre CH et al (2017) Generalised dice overlap as a deep learning loss function for highly unbalanced segmentations. In: Deep learning in medical image analysis and multimodal learning for clinical decision support. Springer, Cham, pp 240–248
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging 15(1):29
Tang H et al 2021 Spatial context-aware self-attention model for multi-organ segmentation. arXiv:2012.09279, accepted by WACV
Tang Y et al (2021) High-resolution 3D abdominal segmentation with random patch network fusion. Med Image Anal 69:101894
Thomson D et al (2014) Evaluation of an automatic segmentation algorithm for definition of head and neck organs at risk. Radiat Oncol 9:173
Tian Z et al (2020) A multiscale-based adjustable convolutional neural network for multiple organ segmentation. Wirel Commun Mob Comput 2020:Article ID 9595687 13 pages
Wang Y, Zhou Y, Shen W, Park S, Fishman EK, Yuille AL (2019) Abdominal multi-organ segmentation with organ attention networks and statistical fusion. Med Image Anal 55:88–102
Yang J, Beadle BM, Garden AS, Schwartz DL, Aristophanous M (2015) A multimodality segmentation framework for automatic target delineation in head and neck radiotherapy. Med Phys 42(9):5310–5320
Yu L et al (2017) Volumetric ConvNets with mixed residual connections for automated prostate segmentation from 3D MR images. Thirty-first AAAI conference on artificial intelligence
Acknowledgments
The work is supported by the grants from Ministry of Science and Technology, Taiwan (No.: MOST 108-2221-E-194 -042, MOST 110-2221-E-194-011).
Funding
Supported by the grant from Ministry of Science and Technology, Taiwan (No.: MOST 108–2221-E-194 -042).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/competing interests
No conflict of interest regarding to the manuscript.
Ethics approval
IRB number B10804010-1 approved by Tzu Chi Hospital in Chiayi, Taiwan.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chen, PH., Huang, CH., Chiu, WT. et al. A multiple organ segmentation system for CT image series using Attention-LSTM fused U-Net. Multimed Tools Appl 81, 11881–11895 (2022). https://doi.org/10.1007/s11042-021-11889-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-11889-7