Abstract
Purpose
Radiotherapy is one of most treatments for tumors. To accurately control the radiation dose distribution and lessen the radiation damage to normal tissues and organs in radiotherapy, it is essential to delineate organs at risk (OARs) precisely. However, manual delineating and some traditional methods are labor-intensive and time-consuming. There is an urgent need for fast and precise segmentation methods in radiotherapy.
Methods
This paper proposes a fully automatic segmentation method based on the 3D U-Net for multi-organ in head and neck. It introduces squeeze-and-attention blocks to gather multi-scale context information and the receptive field block to balance the performance between large-sized and small-sized organs. Furthermore, it is trained by the marginal and exclusion loss function in a partially supervised learning mode.
Results
We evaluated the model with dice similarity coefficient (DSC), 95% Hausdorff distance (95HD) and inference time. Its average DSC is 0.829, which is 4.5%, 3.2%, and 2.4% higher than AnatomyNet’s, nnU-net’s, and FocusNet’s, respectively, and its average 95HD is 2.19. Moreover, its inference time and parameters are 63% and 60% less than FocusNetv2’s.
Conclusion
For the segmentation of OARs in head and neck, our model is more accurate than AnatomyNet, faster than FocusNetv2, and better balances between segmentation accuracy and inference time. It demonstrates that our method is more applicable for clinical treatment.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
There are over 550,000 head and neck (HaN) cancer patients worldwide every year, with around 300,000 deaths [1]. Radiotherapy is one of essential treatments for them. In radiotherapy, it is necessary to accurately delineate the HaN organs to control the radiation dose distribution and lessen the damage to normal tissues and organs. Professional doctors’ manual delineation of HaN organs is inefficient, and segmentation results depend on their professional experience. Some traditional segmentation algorithms are challenging to achieve multi-organ segmentation simultaneously.
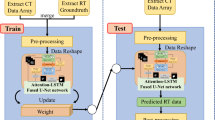
In this paper, we propose a novel model based on 3D U-Net [2] to improve the accuracy of multi-scale organs segmentation. It introduces squeeze-and-attention (SA) [3] blocks into residual blocks to gather multi-scale context information and group non-local voxels from the same organ. It also employs down-sampling only once and introduces receptive field block (RFB) [4] to balance the performance of large-sized organs and small-sized organs. Furthermore, we choose marginal loss function and exclusion loss function to train the model in partially supervised learning mode [5], which utilizes the prior knowledge among voxels to improve the generalization performance.
Related work
Methods based on 3D CNNs implement end-to-end automatic segmentation in the task of HaN OARs [6]. General 3D CNN, however, is not easy to solve imbalanced segmentation accuracy caused by excessive volume difference in OARs. Researchers improve 3D CNN to solve this problem by a variety of methods [7]. CNN with self-channel-and-spatial-attention mechanism adaptively forces the network to emphasize the meaningful features and weaken irrelevant features [8]. AnatomyNet [9] introduces 3D squeeze-and-excitation (SE) [10] blocks into 3D U-Net to enhance the feature extraction ability. It chooses dice loss function [11] and focal loss function [12] to reduce the highly imbalanced segmentation accuracy caused by the various size of OARs. The cascade of multiple models or structures is widely used in multi-target segmentation tasks, such as the cascade of two 3D U-Net models, which locates OARs from the CT image and then implements fine segmentation to obtain better results [13]. FocusNet [14] simulates the process of doctors’ delineation, combining the small-sized organs segmentation, small-sized organs location, and main segmentation network. The prior knowledge of OARs’ shape is sometimes applied in the segmentation of multi-scale organs to improve accuracy. FocusNetv2 [15] adds the adversarial shape constraint block to regularize the estimated mask, making the segmentation results consistent with the shape of small-sized organs. Imaging characteristics of multi-modality images are also utilized in segmentation tasks to improve accuracy. Liu et al. [16] exploit synthetic magnetic resonance (MR) to aid training dual pyramid network (DPN) [17]. Dai et al. [18] utilize the complementary information of cone-beam CT (CBCT) images and MR images to improve the segmentation performance.
Data
The dataset scale is vital for image segmentation based on supervised learning; therefore, we use 3 public datasets to train our model. The public domain database for computational anatomy (PDDCA) dataset [19] contains 25 training samples, additional 8 training samples added after the MICCAI 2015 Head and Neck Auto Segmentation Challenge (MICCAI 2015), 10 offsite test samples, and 5 onsite test samples. It contains the whole-volume CT images of HaN and binary labels, including the brainstem, mandible, chiasm, optic nerve left (Optic. L), optic nerve right (Optic. R), parotid gland left (Paro. L), parotid gland right (Paro. R), submandibular gland left (Subm. L), and submandibular gland right (Subm. R). According to the data processing method provided by AnatomyNet, we expanded the training dataset by the public available dataset of head and neck cetuximab [20] (46 samples) and institutions in Québec, Canada [21] (177 samples). Finally, the training dataset includes 261 samples, and the test dataset includes 10 offsite test samples.
The PDDCA test dataset contains 9 annotated labels, but the expanded training dataset does not include all. To maintain dataset consistency, we cropped the original CT images to the same size, retaining the basic organs information, and resampled them to 3 mm \(\times \) 1.2 mm \(\times \) 1.2 mm.
Method
Squeeze-and-attention block
The principle of pixel-by-pixel prediction is usually applied in semantic segmentation tasks based on CNN. We introduce the pixel grouping mechanism implemented by SA block to improve the segmentation performance. Figure 1 illustrates its structure. The average pooling layer gathers non-local spatial attention of feature maps in SA block by increasing the receptive field, encoding global features, and grouping non-local voxels from the same organ. Its average pooling layer, of which kernel size and stride are both 2, scales the size of its feature map to 1/8 of the original size. Next, two successive convolution blocks with kernel size of 3 and stride of 1 extract its feature map information. Then, the up-sampling layer recovers the size of feature map. Finally, the residual connection fuses the local and non-local information.
Receptive field block
The down-sampling operation increases the receptive field of the model but loses some details. Therefore, we employ down-sampling only once and introduce RFB [4], which increases the receptive field and balances segmentation accuracy between large-sized and small-sized organs. Figure 2 illustrates the structure of RFB based on the inception [22] block, which is improved by the atrous convolution layers [23] to extract multi-scale features. Three branches calculate the input feature map to extract multi-scale features, and each branch comprises convolutions with kernel sizes of \(1\times 1\times 1\), \(3\times 3\times 3\), or \(5\times 5\times 5\), followed by an atrous convolution with the rate of 1, 3, or 5 to increase the receptive field. In addition, the shortcut connection fuses the branches’ concatenation features and input features.
Network architecture
Based on the 3D U-Net, we propose an HaN segmentation network as illustrated in Fig. 3; X and \(X^{\prime }\) denote input image and segmentation result, respectively. Its encoder and decoder have three SA blocks that classify and group voxels from the same organ. The down-sampling increases the receptive field but loses some details of the feature map and reduces the accuracy of small-sized organs. Therefore, we only employ down-sampling and up-sampling once in the model. In addition, we introduce an RFB to balance segmentation accuracy between large-sized and small-sized organs and to learn multi-scale features, which increases the receptive field and improves the segmentation accuracy of small-sized organs.
Loss function
The MICCAI 2015 dataset contains 9 annotated labels for each sample, but other datasets contain fewer labels. To deal with various labels of training datasets, we introduce a vector denoted by M(c) (\(M \in R^{1\times 10}\), \(c=0\) denotes the index of background information for CT images, \(c \in [1,9]\) denotes the index of 9 OARs in HaN) to mark missed annotated data, which indicates the presence of a class in training sample with 1 or 0 otherwise. The output of the last convolution layer is the prediction probability of voxels, denoted by P, of which the dimension is \(N_c\times H\times W\times D\), where \(N_c\) represents its channels, corresponding to the type of OARs, and \(H\times W\times D\) represents the size of CT image.
The marginal probability fuses the probability of unlabeled organs and background to drive the model to learn information of unlabeled organs, and it is formulated as Eq. (1).
where \( P_M\) and c denote the marginal probability and the type of organs, respectively; \(M(c)=0\) represents the organ c is not annotated. The marginal probability and the annotated organs’ probability consist of a new vector; it is denoted by Q and formulated as Eq. (2).
The binary mask data of annotated organs in the training dataset are denoted by Y, on which one-hot encoding is performed and formulated as Eq. (3).
The dice loss function and the focal loss function are plugged into the marginal loss to solve imbalanced segmentation accuracy caused by huge volume differences, formulated in Eqs. (4)–(9).
where \(\mathrm{TP}_m(t)\), \(\mathrm{FP}_m(t)\), and \(\mathrm{FN}_m(t)\) denote true positive, false positive, and false negative of marginal probability for the organ t, respectively. \(Q_{n}(t)\) denotes the marginal probability of voxel n for organ t, and \(Z_{n}(t)\) denotes the one-hot encode of voxel n for organ t. T and N denote the total number of annotated organs and voxels for one sample, respectively, and C denotes the total number of OARs, which is 9 in our task. \(L_{\mathrm{mDice}}\), \(L_{\mathrm{mFocal}}\), and \(L_m\) denote dice loss function, focal loss function, and marginal loss function, respectively, where \(\lambda \) trades off the dice loss function and focal loss function; \(\alpha \) and \(\beta \) trade off weights for false negative and false positive. For the best performance, \(\lambda \) is set to 0.2; \(\alpha \) and \(\beta \) are set to 0.5.
The exclusion vector exploits the principle that each voxel can only belong to one organ and mutual exclusion of voxels between OARs. Its calculation is negated to the one-hot vector of the binary mask and formulated in Eq. (10).
The exclusion vector is denoted by E(c), of which the dimension is \(N_c \times H \times W \times D\). In this paper, the dice loss function is plugged into the exclusion loss, denoted by \(L_{\mathrm{eDice}}\), where \(P_{1n}(c)\) and \(P_{0n}(c)\) indicate the probability that voxel n is predicted to be organ c or not be organ c, respectively; \(E_{0n}(c)\) and \(E_{1n}(c)\) indicate that the value of exclusion vector represented by the voxel n of organ c is 0 or 1, respectively. \(\alpha \) and \(\beta \) are set to 0.5 for the best performance. The exclusion loss is formulated as Eq. (11).
Verified by many experiments, the model gets the best performance when the weight of exclusion loss is 2. Finally, the loss function denoted by \(L_{\mathrm{loss}}\) is formulated in Eq. (12).
Comparisons of the model performance with the original challenge dataset. (a, b) represent the DSC and 95HD of models, respectively; AC, TA, ET, and OUR represent results of Antong Chen et al. [25], Thomas Albrecht et al. [26], Tappeiner et al. [13], and our model, respectively. ET does not provide results of the submandibular gland left and right. In addition, we cropped the ET’s error bars of the brainstem and parotid gland left for visualization purposes, and their standard deviations are 14.3 and 33.3, respectively
Results
Implementation details and evaluation metrics
Experiments were run on the platform with NVIDIA RTX 2080Ti GPU and INTER I7-10700 CPU, and the model was implemented by PyTorch. The apex mixed precision released by the NVIDIA platform accelerated the training process and saved hardware resources. The RMSprop algorithm optimized the gradient of the loss function, of which the learning rate was 0.001, the number of epochs was 200, and the batchsize was 1 caused by the vector M in the loss function. Dice similarity coefficient (DSC), 95% Hausdorff distance (95HD) [19], and inference time were used to evaluate the performance.
Experimental results and analysis
We compared the model’s DSC with previous state-of-the-art methods, as illustrated in Table 1. With the same training dataset, the DSC of our model is 4.5% higher than AnatomyNet’s [9], which also uses one down-sampling layer to avoid loss of information of small organs. The RFB expands the receptive field and addresses the conflict between receptive field and small organs. Moreover, the marginal loss function handles data without labels, and the exclusion loss function improves performance according to prior knowledge among voxels. Our model is also superior to the best results in MICCAI 2015 [19] (It just gives average DSC for symmetrical organs). Compared with nnU-net [24], it shows better performance of imbalanced organs, and nnU-net gets the poor performance of small-sized organs. Compared with the state-of-the-art models, it is close to the performance of FocusNetv2 [15] for large-sized organs and is slightly worse for small-sized organs, but FocusNetv2 is a larger model with more parameters and trained by more private data. In addition, FocusNet and FocusNev2 are not end-to-end models, trained separately by three sub-networks followed by a combined network to implement segmentation of OARs.
DSC is sensitive to internal details of organs, and 95HD is sensitive to boundaries. Experimental results in Table 2 demonstrate that our method is not better than FocusNetv2 [15] in the metric of 95HD and much better than others. Our model has better performance on boundaries of organs because \(L_\mathrm{eDice}\) employs mutual exclusive information among voxels of different organs. In addition, we performed Kruskal–Wallis test on the offsite test dataset. Their p-value and test statistic of DSC are 0.9998 and 2.0630, respectively, and their 95HD is 0.5293 and 12.9636, respectively.
To evaluate the performance of the model more credible, we trained our model with the original training samples (0522c001 to 0522c0328 of PDDCA) and tested on 15 samples, including offsite samples and 5 onsite samples. With the same dataset, we also compared our model with method of Tappeiner et al. [13] and some participants who provided full experimental results of MICCAI 2015 [25, 26], and Figure 4 illustrates their DSC score and 95HD.
We compared the number of parameters and the average inference time on the same hardware platform. Experimental results in Table 3 show that the parameters of our model are 60% less than FocusNetv2’s, and the inference time is 63% less, which means our model requires fewer hardware resources and less time. Compared to AnatomyNet, our model has higher accuracy with the same order of magnitude of inference time and the model’s parameters.
Visualization
Figure 5 illustrates the visualization of segmentation results, of which the legend shows the correspondence between colors and organs. In the cross-sectional view, predicted contours match ground truth quite well for large-sized organs, such as the mandible, but there is a slight difference in size and shape for small-sized organs. In the 3D view, there are very tiny differences in volume size and shape between the predicted mask and ground truth.
Conclusion
In conclusion, our model delineates OARs in HaN to better balance inference time and accuracy. SA blocks are introduced into the model, which aggregates multi-scale context information and encourages voxel grouping of the same organ. Our model only employs down-sampling once and introduces a receptive field block to balance the segmentation accuracy between large-sized and small-sized organs. In addition, its loss function combines the marginal loss and the mutual exclusion loss, which trains the model by partially supervised learning and exploits the prior information among voxels. Compared with natural images, there are more relatively fixed shapes and stable spatial structures in HaN CT images. The prior knowledge of OARs, such as shape, symmetry, and similarity, should be considered in the following research.
References
Jemal A, Bray F, Center MM (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90. https://doi.org/10.3322/caac.20107
Çiçek Ö, Abdulkadir A, Lienkamp S.S., Brox T, Ronneberger O (2016) 3d u-net: Learning dense volumetric segmentation from sparse annotation. In: Ourselin, S., Joskowicz, L., Sabuncu, M.R., Unal, G., Wells, W. (eds.) Int. Conf. on Medical Image Computing and Computer-assisted Intervention, pp. 424–432. Springer, Cham . https://doi.org/10.1007/978-3-319-46723-8_49. https://springerlink.bibliotecabuap.elogim.com/chapter/10.1007%2F978-3-319-46723-8_49
Zhong Z, Lin Z.Q., Bidart R, Hu X, Daya I.B., Li Z, Zheng W.S., Li J, Wong A (2020) Squeeze-and-attention networks for semantic segmentation. In: 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), pp. 13062–13071 . https://doi.org/10.1109/CVPR42600.2020.01308
Liu S, Huang D, Wang Y (2018) Receptive field block net for accurate and fast object detection. In: Ferrari, V., Hebert, M., Sminchisescu, C., Weiss, Y. (eds.) Computer Vision –ECCV 2018, pp. 404–419. Springer, Cham . https://doi.org/10.1007/978-3-030-01252-6_24
Shi G, Xiao L, Chen Y, Zhou S.K. (2021) Marginal loss and exclusion loss for partially supervised multi-organ segmentation. Medical Image Analysis 70, 101979 . https://doi.org/10.1016/j.media.2021.101979
Ren X, Lei X, Dong N, Shao Y, Zhang H, Shen D, Qian W (2018) Interleaved 3d-cnns for joint segmentation of small-volume structures in head and neck ct images. Medical Physics 45(5), 2063–2075 . https://doi.org/10.1002/mp.12837
Vrtovec T, Močnik D, Strojan P, Pernuš F, Ibragimov B (2020) Auto-segmentation of organs at risk for head and neck radiotherapy planning: From atlas-based to deep learning methods. Medical Physics 47(9), 929–950 . https://doi.org/10.1002/mp.14320
Gou S, Tong N, Qi S, Yang S, Chin R, Sheng K (2020) Self-channel-and-spatial-attention neural network for automated multi-organ segmentation on head and neck CT images. Physics in Medicine & Biology 65(24), 245034 . https://doi.org/10.1088/1361-6560/ab79c3
Zhu W, Huang Y, Liang Z, Chen X, Yong L, Zhen Q, Nan D, Wei F, Xie X (2018) Anatomynet: Deep learning for fast and fully automated whole-volume segmentation of head and neck anatomy. Medical Physics 46(2), 576–589 . https://doi.org/10.1088/1361-6560/abd953
Hu J, Shen L, Albanie S, Sun G, Wu E (2020) Squeeze-and-excitation networks. IEEE Transactions on Pattern Analysis and Machine Intelligence 42(8), 2011–2023 . https://doi.org/10.1109/TPAMI.2019.2913372
Shelhamer E, Long J, Darrell T (2017) Fully convolutional networks for semantic segmentation. IEEE Transactions on Pattern Analysis and Machine Intelligence 39(4), 640–651 . https://doi.org/10.1109/TPAMI.2016.2572683
Lin T-Y, Goyal P, Girshick R, He K, Dollár P (2020) Focal loss for dense object detection. IEEE Transactions on Pattern Analysis and Machine Intelligence 42(2):318–327. https://doi.org/10.1109/TPAMI.2018.2858826
Tappeiner E, Pröll S, Hönig M, Raudaschl P.F., Zaffino P, Spadea M.F., Sharp G.C., Schubert R, Fritscher K (2019) Multi-organ segmentation of the head and neck area: an efficient hierarchical neural networks approach. International Journal of Computer Assisted Radiology and Surgery 14, 745–759 . https://doi.org/10.1007/s11548-019-01922-4
Gao Y, Huang R, Chen M, Wang Z, Deng J, Chen Y, Yang Y, Zhang J, Tao C, Li H (2019) FocusNet: Imbalanced Large and Small Organ Segmentation with an End-to-End Deep Neural Network for Head and Neck CT Images. https://arxiv.org/abs/1907.12056v1
Gao Y, Huang R, Yang Y, Zhang J, Shao K, Tao C, Chen Y, Metaxas D.N., Li H, Chen M (2021) Focusnetv2: Imbalanced large and small organ segmentation with adversarial shape constraint for head and neck ct images. Medical Image Analysis 67, 101831. https://doi.org/10.1016/j.media.2020.101831
Liu, Y., Lei, Y., Fu, Y., Wang, T., Zhou, J., Jiang, X., McDonald, M., Beitler, J.J., Curran, W.J., Liu, T., Yang, X.: Head and neck multi–organ auto-segmentation on ct images aided by synthetic mri. Medical Physics 47(9), 4294–4302 (2020). https://doi.org/10.1002/mp.14378
Xu, X., Chen, J., Zhang, H., Han, G.: Dual pyramid network for salient object detection. Neurocomputing 375, 113–123 (2020). https://doi.org/10.1016/j.neucom.2019.09.077
Dai, X., Lei, Y., Wang, T., Dhabaan, A.H., McDonald, M., Beitler, J.J., Curran, W.J., Zhou, J., Liu, T., Yang, X.: Head-and-neck organs-at-risk auto-delineation using dual pyramid networks for CBCT-guided adaptive radiotherapy. Physics in Medicine & Biology 66(4), 045021 (2021). https://doi.org/10.1088/1361-6560/abd953
Raudaschl, P., Zaffino, P., Sharp, G., Spadea, M., Chen, A., Dawant, B.M., Albrecht, T., Gass, T., Langguth, C., Lüthi, M., Jung, F., Knapp, O., Wesarg, S., Mannion-Haworth, R., Bowes, M., Ashman, A., Guillard, G., Brett, A., Vincent, G., Orbes-Arteaga, M., Cárdenas-Pen̄a, D., Castellanos-Dominguez, G., Aghdasi, N., Li, Y., Berens, A., Hannaford, B., Schubert, R., Fritscher, K.D.: Evaluation of segmentation methods on head and neck ct: Auto-segmentation challenge 2015. Medical Physics 44(5), 2020–2036 (2017). https://doi.org/10.1002/mp.12197
Clark, Vendt, Smith, Freymann, Kirby, Koppel, Moore, Phillips, Maffitt, and, P. (2013) The cancer imaging archive (tcia): Maintaining and operating a public information repository. Digit Imaging 26, 1045–1057 . https://doi.org/10.1007/s10278-013-9622-7
Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts H, Khaouam N, Nguyen-Tan PF, Wang CS, Sultanem K (2017) Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Scientific Reports 7(1):10117. https://doi.org/10.1038/s41598-017-10371-5
Szegedy C, Vanhoucke V, Ioffe S, Shlens J, Wojna Z (2016) Rethinking the inception architecture for computer vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), 2818–2826 . https://doi.org/10.1109/CVPR.2016.308
Chen L.-C., Papandreou G, Kokkinos I, Murphy K, Yuille A.L. (2018) Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Transactions on Pattern Analysis and Machine Intelligence 40(4), 834–848 . https://doi.org/10.1109/TPAMI.2017.2699184
Isensee F, Jaeger P.F., Kohl S.A.A., Petersen J, Maier-Hein K.H. (2021) nnu-net: a self-configuring method for deep learning-based biomedical image segmentation. Nature Methods 18, 203–211 . https://doi.org/10.1038/s41592-020-01008-z
Chen A, Dawant B (2016) A multi-atlas approach for the automatic segmentation of multiple structures in head and neck ct images . https://doi.org/10.54294/hk5bjs
Albrecht T, Gass T, Langguth C, Lüthi M (2015) Multi atlas segmentation with active shape model refinement for multi-organ segmentation in head and neck cancer radiotherapy planning . https://doi.org/10.54294/kmcunc
Funding
We would like to thank the financial supports from National Natural Science Foundation of China (62175156, 61976140, 61675134, 81827807), Shanghai Committee of Science and Technology (1944190 5800), and Wenzhou Medical University Key Laboratory Open Project (K181002).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
For the retrospective studies formal consent is not required.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hongdong, M., Guogang, C., Shu, Z. et al. Multi-scale organs image segmentation method improved by squeeze-and-attention based on partially supervised learning. Int J CARS 17, 1135–1142 (2022). https://doi.org/10.1007/s11548-022-02632-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02632-0