Abstract
Computed Tomography (CT) is the standard imaging technique for radiotherapy planning. The delineation of Organs at Risk (OAR) in thoracic CT images is a necessary step before radiotherapy, for preventing irradiation of healthy organs. However, due to low contrast, multi-organ segmentation is a challenge. In this paper, we focus on developing a novel framework for automatic delineation of OARs. Different from previous works in OAR segmentation where each organ is segmented separately, we propose two collaborative deep architectures to jointly segment all organs, including esophagus, heart, aorta and trachea. Since most of the organ borders are ill-defined, we believe spatial relationships must be taken into account to overcome the lack of contrast. The aim of combining two networks is to learn anatomical constraints with the first network, which will be used in the second network, when each OAR is segmented in turn. Specifically, we use the first deep architecture, a deep SharpMask architecture, for providing an effective combination of low-level representations with deep high-level features, and then take into account the spatial relationships between organs by the use of Conditional Random Fields (CRF). Next, the second deep architecture is employed to refine the segmentation of each organ by using the maps obtained on the first deep architecture to learn anatomical constraints for guiding and refining the segmentations. Experimental results show superior performance on 30 CT scans, comparing with other state-of-the-art methods.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
- Anatomical constraints
- CT segmentation
- Fully Convolutional Networks (FCN)
- CRF
- CRFasRNN
- Auto-context model
1 Introduction
In medical image segmentation, many clinical settings include the delineation of multiple objects or organs, e.g., the cardiac ventricles, and thoracic or abdominal organs. From a methodological point of view, the ways of performing multi-organ segmentation are diverse. For example, multi-atlas approaches in a patch based setting have been shown effective for segmenting abdominal organs [11]. Many other approaches combine several techniques, such as in [4] where thresholding, generalized hough transform and an atlas-registration based method are used. The performance of these approaches is bound to the use of separate methods that can also be computationally expensive. Usually, organs are segmented individually ignoring their spatial relationships, although this information could be valuable to the segmentation process.
In this paper, we focus on the segmentation of OAR, namely the aorta, esophagus, trachea and heart, in thoracic CT (Fig. 1), an important prerequisite for radiotherapy planning in order to prevent irradiation of healthy organs. Routinely, the delineation is largely manual with poor intra- or inter-practitioners agreement. Note that the automated segmentation of the esophagus has hardly been addressed in research works as it is exceptionally challenging: the boundaries in CT images are almost invisible (Fig. 2). Radiotherapists manually segment it based on not only the intensity information, but also the anatomical knowledge, i.e., the esophagus is located behind the trachea in the upper part, behind the heart in the lower part, and also next to the aorta in several parts. More generally, this observation can be made for the other organs as well. Our aim is to design a framework that would learn this kind of constraints automatically to improve the segmentation of all OAR and the esophagus in particular. We propose to tackle the problem of segmenting OAR in a joint manner through the application of two collaborative deep architectures, which will implicitly learn anatomical constraints in each of the organs to mitigate the difficulty caused by lack of image contrast. In particular, we perform an initial segmentation by using a first deep Sharpmask network, inspired by the refinement framework presented in [8] which allows an effective combination of low-level features and deep high-level representations. In order to enforce the spatial and intensity relationships between the organs, the initial segmentation result is further refined by Conditional Random Fields (CRF) with the CRFasRNN architecture. We propose to use a second deep architecture which is designed to be able to make use of the segmentation maps obtained by the first deep architecture of all organs, to learn the anatomical constraints for the one organ that is currently under refinement of its segmentation. We show experimentally that our framework outperforms other state-of-the-art methods. Note that our framework is also generic enough to be applied to other multi-label joint segmentation problems.
2 Method
2.1 SharpMask Feature Fusion Architecture and CRF Refinement
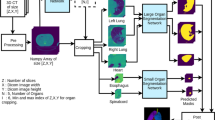
The first deep architecture performs initial segmentation, with its output as a probability map of each voxel belonging to background, esophagus, heart, aorta, or trachea. In order to alleviate the loss of image resolution due to the use of pooling operations in regular Convolutional Neural Networks (CNN); Fully Convolutional Networks (FCN) [5] and some other recent works such as the U-Net [9] and Facebooks SharpMask (SM) [8] have used skip connections, outperforming many traditional architectures. The main idea is to add connections from early to deep layers, which can be viewed as a form of multiscale feature fusion, where low-level features are combined with highly-semantic representations from deep layers.
In this work, we use an SM architecture that has been shown superior to the regular FCNs for thoracic CT segmentation [12]. The CRF refinement is done subsequently with the CRFasRNN architecture, which formulates the mean field approximation using backpropagable operations [15], allowing the operation to be part of the network (instead of a separated postprocessing step) and even to learn some of its parameters. Thus, a new training is performed for fine-tuning the learned weights from the first step, and also for learning some parameters of the CRF [12]. In the second deep architecture as described below, the segmentation initial results of the surrounding organs by the first deep architecture will be used to refine the segmentation of each target organ separately.
2.2 Learning Anatomical Constraints
The second deep architecture, using SharpMask, is trained to distinguish between background and each target organ under separate refinement. This architecture has two sets of inputs, i.e., (1) the original CT image and (2) the initial segmentation results of the neighbouring organs around the target organ under refinement of segmentation. The main difference of this second deep architecture, compared to the first deep architecture with multiple output channels representing different organs and background, is that it only has two output channels in the last layer, i.e., a probability map representing each voxel belonging to background or a target organ under refinement of segmentation. The basic assumption is that the second deep architecture will learn the anatomical constraints around the target organ under refinement of segmentation and thus help to produce better segmentation for the target organ. In Fig. 3 we show the full framework with both the first deep architecture (top) and the second deep architecture.
Note that our framework (using two deep architectures) shares some similarity with a refinement step, called AutoContext Model (ACM) [13], which has been successfully applied to brain image segmentation [14], by using traditional classifiers such as Random Forests. The main idea of ACM is to iteratively refine the posterior probability maps over the labels, given not only the input features, but also the previous probabilities of a large number of context locations which provide information about neighboring organs, forcing the deep network to learn the spatial constrains for each target organ [14]. In practice, this translates to train several classifiers iteratively, where each classifier is trained not only with the original image data, but also with the probability maps obtained from the previous classifier, which gives additional context information to the new classifier. Comparing our proposed framework with the ACM, we use a deep architecture. Overall, our method has three advantages: (1) it can avoid the design of hand-crafted features, (2) our network can automatically learn the selection of context features, and (3) our method uses less redundant information. Note that, in the classical ACM, the selection of these features must be hard-coded; that is, the algorithm designer has to select a specific number of sparse locations (i.e., using sparse points from rays at different angles from a center point [13]), which makes the context information limited within a certain range by the algorithm designer. On the other hand, in our method, the context information can be automatically learned by the deep network, and limited only by the receptive field of the network (which can even be the whole range of the image in deep networks). Regarding the redundancy, ACM uses the probability maps of all organs as input, which is often very similar to the ground-truth label maps. In this way, the ACM is not able to further refine the results. In our method, we use only the complementary information, the label probability maps of the neighboring organs around the target organ under refinement of segmentation.
3 Experiments
In our implementation, the full slices of the CT scan are the inputs to our proposed framework. Both the first and second architectures use large filters (i.e., 7 \(\times \) 7, or 7 \(\times \) 7 \(\times \) 7), as large filters have been shown beneficial for CT segmentation [2]. Both 2D and 3D settings are implemented in our study. We have found that, different from MRI segmentation [7], small patches are not able to produce good results for CT segmentation. Thus, we use patches of size \(160\,\times \,160\,\times \,48\) as the training samples. Specifically, we first build a 3D mesh model for each organ in all the training CT images, and then define each vertex as the mean of a certain Gaussian distribution with diagonal covariance, from which we can sample points as the centers of the respective patches. In this way, the training samples will contain important boundary information and also background information. In particular, the elements in the diagonal are chosen to be 5, in such a way, the kernel size used would include them when centered in the boundary. In addition, it is also important to sample inside the organs and thus, we also sample in an uniform grid.
3.1 Dataset and Pre-processing
The dataset used in this paper contains 30 CT scans, each with lung cancer or Hodgkin lymphoma and 6-fold cross validation is performed. Manual segmentations of the four OAR are available for each CT scan, along with the body contour (which can used to remove background voxels during the training). The scans have \(512\,\times \,512\,\times \,(150 \sim 284)\) voxels with a resolution of 0.98 \(\times \) 0.98 \(\times \) 2.5 mm\(^3\). For each CT scan, its intensities are normalized to have zero mean and unit variance, and it is also augmented to generate more CT samples (for improving the robustness of training) through a set of random affine transformations and random deformation fields (generated with B-spline interpolation [6]). In particular, an angle between −5 to 5\(^{\circ }\) and a scale factor between 0.9 and 1.1 were randomly selected for each CT scan to produce the random affine transformation. These values were selected empirically trying to produce realistic CT scans similar to those of the available dataset.
3.2 Training
For organ segmentation in CT images, the data samples are often highly imbalanced, i.e., with more background voxels than the target organ voxels. This needs to be considered when computing the loss function in the training. We utilize a weighted cross-entropy loss function, where each weight is calculated as the complement of the probability of each class. In this way, more importance will be given to small organs, and also each class will contribute to the loss function in a more equally way. We have found that this loss function leads to better performance than using a regular (equally-weighted) loss function. However, the results are still not reaching our expected level. For further improvement, we use our above-obtained weights as initialization for the network, and then fine-tune them by using the regular cross-entropy loss. This new integrated strategy always outperforms the weighted or the regular cross-entropy loss function. In our optimization, stochastic gradient descent is used as optimizer, with an initial learning rate of 0.1 that is divided by 10 every 20 epochs, and the network weights are initialized with the Xavier algorithm [3].
3.3 Results
In Fig. 4, we illustrate the improvement on the esophagus segmentation by using our proposed framework with learned anatomical constraints. The last column shows the results using the output of the first network as anatomical constraints. We can see how the anatomical constraints can help produce a more accurate result on the segmentation of the esophagus, even when having air inside (black voxels inside the esophagus). Interestingly, the results obtained by using the output of the first network or the ground-truth manual labels as anatomical constraints are very similar, almost with negligible differences. Similar conclusions can also be drawn for segmentations of other organs. In Fig. 5, we show the segmentation results for the aorta, trachea, and heart, with and without anatomical constraints. In the cases of segmenting the aorta and trachea, the use of anatomical constraints improves the segmentation accuracy. For the trachea, our network is able to generalize to segment the whole part on the right lung (i.e., left side of the image) even when it was segmented partially in the manual ground-truth. On the other hand, for the heart, there are some false positives when using anatomical constraints, as can be seen in the third column. However, accurate contours are obtained, which are even better than those obtained without anatomical constraints, as can be seen in the fourth column.
Segmentation results for the esophagus. (a) Input data to the second network, with the anatomical constraints overlapped; results using (b) only the first network without anatomical constraints, (c) manual labels on the neighboring organs as anatomical constraints, (d) the output of the first network as anatomical constraints.
Segmentation without (1st row) and with (2nd row) anatomical constraints. Green contours denote manual ground-truths, and red contours denote our automatic segmentation results. Right panel shows the 3D rendering for our segmented four organs, i.e., aorta (blue), heart (beige), trachea (brown), and esophagus (green). (Color figure online)
In Table 1, we report the Dice ratio (DR) obtained using each of the comparison methods, including a state-of-the-art 3D multi-atlas patch-based method (called OPAL (Optimized PatchMatch for Near Real Time and Accurate Label Fusion [10])) and different deep architecture variants using 2D or 3D as well as different combinations of strategies. Specifically, SM2D and SM3D refer to the use of the Network 1 in Fig. 3 using 2D or 3D respectively. We also tested their refinement with ACM and CRF, and finally, the proposed framework is denoted as SM2D + Constraints. As OPAL mainly compares patches for guiding the segmentation, OPAL should be effective in segmenting the clearly observable organs, such as the trachea (an identifiable black area), which is true as indicated by the table. But, for the organs with either low contrast or large intensity variation across slices and subjects, which is the case for the esophagus, the respective performance is seriously affected, as the table shows. The highest performance for each organ is obtained by the SM2D-based architectures, while all 3D-based architectures do not improve the segmentation performance. This is possibly due to large slice thickness in the CT scans, as noticed also in [1], where the authors preferred to handle the third dimension by the recurrent neural networks, instead of 3D convolutions. Another observation is that the ACM model is not able to outperform the CRF refinement. We believe that this is mainly due to the fact that the CRF used is fully connected and not based on the neighboring regions. The latter has been used as comparison in the ACM [13], for claiming that the advantage is coming from the context range information that the framework can reach. On the other hand, our proposed framework is able to improve the performance for all the organs, except the heart whose quantitative results are very similar to those obtained by the first network, and which can be well-segmented by it, by leveraging the large heart size and also the good image contrast around it. However, the quality of the obtained contours with the proposed framework is better as shown in Fig. 5. Although room for improvement is still left for the esophagus (with mean DR value of 0.69), the experimental results show that our proposed framework does bring an improvement, compared to the other methods.
4 Conclusions
We have proposed a novel framework for joint segmentation of OAR in CT images. It provides a way to learn the relationship between organs which can give anatomical contextual constraints in the segmentation refinement procedure to improve the performance. Our proposed framework includes two collaborative architectures, both based on the SharpMask network, which allows for effective combination of low-level features and deep highly-semantic representations. The main idea is to implicitly learn the spatial anatomical constraints in the second deep architecture, by using the initial segmentations of all organs (but a target organ under refinement of segmentation) from the first deep architecture. Our experiments have shown that the initial segmentations of the surrounding organs can effectively guide the refinement of segmentation of the target organ. An interesting observation is that our network is able to automatically learn spatial constraints, without specific manual guidance.
References
Chen, J., Yang, L., Zhang, Y., Alber, M., Chen, D.Z.: Combining fully convolutional and recurrent neural networks for 3d biomedical image segmentation. In: NIPS, pp. 3036–3044 (2016)
Dou, Q., Chen, H., Jin, Y., Yu, L., Qin, J., Heng, P.: 3d deeply supervised network for automatic liver segmentation from CT volumes. CoRR abs/1607.00582 (2016)
Glorot, X., Bengio, Y.: Understanding the difficulty of training deep feedforward neural networks. In: AISTATS (2010)
Han, M., Ma, J., Li, Y., Li, M., Song, Y., Li, Q.: Segmentation of organs at risk in CT volumes of head, thorax, abdomen, and pelvis. In: Proceedings of SPIE, vol. 9413 (2015). Id: 94133J-6
Long, J., Shelhamer, E., Darrell, T.: Fully convolutional networks for semantic segmentation. In: CVPR (2015)
Milletari, F., Navab, N., Ahmadi, S.: V-net: fully convolutional neural networks for volumetric medical image segmentation. CoRR abs/1606.04797 (2016)
Nie, D., Wang, L., Gao, Y., Shen, D.: Fully convolutional networks for multi-modality isointense infant brain image segmentation. In: ISBI, pp. 1342–1345 (2016)
Pinheiro, P.H.O., Lin, T., Collobert, R., Dollár, P.: Learning to refine object segments. CoRR abs/1603.08695 (2016)
Ronneberger, O., Fischer, P., Brox, T.: U-Net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) MICCAI 2015. LNCS, vol. 9351, pp. 234–241. Springer, Cham (2015). doi:10.1007/978-3-319-24574-4_28
Ta, V.-T., Giraud, R., Collins, D.L., Coupé, P.: Optimized PatchMatch for near real time and accurate label fusion. In: Golland, P., Hata, N., Barillot, C., Hornegger, J., Howe, R. (eds.) MICCAI 2014. LNCS, vol. 8675, pp. 105–112. Springer, Cham (2014). doi:10.1007/978-3-319-10443-0_14
Tong, T., et al.: Discriminative dictionary learning for abdominal multi-organ segmentation. Med. Image Anal. 23, 92–104 (2015)
Trullo, R., Petitjean, C., Ruan, S., Dubray, B., Nie, D., Shen, D.: Segmentation of organs at risk in thoracic CT images using a sharpmask architecture and conditional random fields. In: ISBI (2017)
Tu, Z., Bai, X.: Auto-context and its application to high-level vision tasks and 3d brain image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 32(10), 1744–1757 (2010)
Wang, L., et al.: Links: learning-based multi-source integration framework for segmentation of infant brain images. NeuroImage 108, 160–172 (2015)
Zheng, S., et al.: Conditional random fields as recurrent neural networks. In: ICCV (2015)
Acknowledgment
This work is co-financed by the European Union with the European regional development fund (ERDF, HN0002137) and by the Normandie Regional Council via the M2NUM project.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this paper
Cite this paper
Trullo, R., Petitjean, C., Nie, D., Shen, D., Ruan, S. (2017). Joint Segmentation of Multiple Thoracic Organs in CT Images with Two Collaborative Deep Architectures. In: Cardoso, M., et al. Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support . DLMIA ML-CDS 2017 2017. Lecture Notes in Computer Science(), vol 10553. Springer, Cham. https://doi.org/10.1007/978-3-319-67558-9_3
Download citation
DOI: https://doi.org/10.1007/978-3-319-67558-9_3
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-67557-2
Online ISBN: 978-3-319-67558-9
eBook Packages: Computer ScienceComputer Science (R0)