Abstract
The variations among shapes, sizes, and locations of tumors are obstacles for accurate automatic segmentation. U-Net is a simplified approach for automatic segmentation. Generally, the convolutional or the dilated convolutional layers are used for brain tumor segmentation. However, existing segmentation methods of the significant dilation rates degrade the final accuracy. Moreover, tuning parameters and imbalance ratio between the different tumor classes are the issues for segmentation. The proposed model, known as Residual-Dilated Dense Atrous-Spatial Pyramid Pooling (RD2A) 3D U-Net, is found adequate to solve these issues. The RD2A is the combination of the residual connections, dilation, and dense ASPP to preserve more contextual information of small sizes of tumors at each level encoder path. The multi-scale contextual information minimizes the ambiguities among the tissues of the white matter (WM) and gray matter (GM) of the infant’s brain MRI. The BRATS 2018, BRATS 2019, and iSeg-2019 datasets are used on different evaluation metrics to validate the RD2A. In the BRATS 2018 validation dataset, the proposed model achieves the average dice scores of 90.88, 84.46, and 78.18 for the whole tumor, the tumor core, and the enhancing tumor, respectively. We also evaluated on iSeg-2019 testing set, where the proposed approach achieves the average dice scores of 79.804, 77.925, and 80.569 for the cerebrospinal fluid (CSF), the gray matter (GM), and the white matter (WM), respectively. Furthermore, the presented work also obtains the mean dice scores of 90.35, 82.34, and 71.93 for the whole tumor, the tumor core, and the enhancing tumor, respectively on the BRATS 2019 validation dataset. Experimentally, it is found that the proposed approach is ideal for exploiting the full contextual information of the 3D brain MRI datasets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Gliomas can affect the normal working of the human brain. Gliomas can be categorized into two grades: high-grade glioblastoma (HGG) and low-grade glioblastoma (LGG) [20], in which each grade has the number of classes. Based on the grading system, it is crucial to make the prediction and rate of the tumors. The MRI technique is mainly used for an in-depth analysis of the brain structure. However, the unusual variations among the shapes, size, and location of a tumor [23] in the MRIs are obstacles to developing an efficient, accurate algorithm. Feature learning has the great potential to handle such kind of problem. Classical existing methods also delivered excellent results. However, in the traditional approach, the unfeasible study of brain tumors makes impractical. In comparison to them, the process of feature learning gives an abstract representation of data. Figure 1 shows the process of feature representation with the Alexnet architecture [28]. The modified Alexnet architecture has five convolution layers to learn features. A pooling layer is employed between every pair of convolution layers to reduce the original input resolution. Furthermore, two convolutional layers of kernel size of 1 × 1 are used to reduce the features. The reduced features of the last convolution layer match the number of labels of a dataset. Finally, the output reflects the probability distribution of different classes or labels by using the softmax.
Feature representation of the modified Alexnet architecture. Each convolution layer (green) is of a kernel size of 3 × 3. A stride of size two is used with each pooling layer (yellow) to reduces the input resolution. The last two convolution layers reduce the number of channels or features before the final output
Figure 1 shows a 2D architecture in which each convolution layer, containing several 2D filters or kernels. Each filter yields the feature maps when applied to the channels of the previous layer. For example, 24 filters in the first convolution layer after convolving the input of 3 channels yield output feature maps 24 × 128 × 128. These output feature maps then input to the next convolution layer and so on. To drive the generalized formula for the output feature maps of each layer l, let Cl denoting the convolution filters or kernels and \(p_{l-1}^{z}\) denoting the 2D array corresponding to the zth input. Then the output feature map of each kernel of layer l is
where \(Weight_{l}^{kernel,z}\) denoting the weight of each kernel, \(b_{l}^{kernel}\) denoting a bias, and ⋆ representing convolution operation. f is a Leaky Rectified Linear Unit (Leaky ReLU) non-linear activation function. It has α as an extra parameter to prevent the problem of zero gradients during the training. Mathematically, it can be written as
where xj denoting the input features and \(f\left (x_{j}\right )\) representing the output features. The spatial dimensions of the output feature maps of each convolution layer reduce by a pooling layer. This spatial reduction is possible by using a stride of size 2 in the pooling layer. Mathematical expression for each pooling layer can be
where \(y_{l}^{kernel}\) denoting feature maps of pooling layer l for the kernelth input feature map \(\left (q_{l}^{kernel}\right )\), \(\max \limits \left (.\right )\) denoting the max-pool operation. Finally, the softmax activation is performed on the reduced feature maps of the last 1 × 1 convolution to generate the probability distribution of different classes. Mathematically, the softmax activation is
where the classes are denoting by the C and the last layer is representing by L.
Figure 1 reflects the deep Convolutional Neural Networks (CNNs). However layers in the deep models have complex inter-connections for better learning. A Fully Convolutional Neural Network (FCNN), especially, U-Net is very popular in biomedical image segmentation [42, 55]. The U-Net architecture gains popularity due to skip-connections [39, 56]. Skip-connections perform the concatenation operation on the maps of the different parts of a U-Net model. The potential of skip-connections can be understood from basic FCN architecture [30] (Fig. 2a) and different variations of U-Net models (Fig. 2b, and c). In Fig. 1, the feature maps of each convolution layer are reduced by the max-pooling layer. Therefore, the size of the output is smaller than that of the input. This problem can be resolved by applying the upsampling layer. For example, on upsampling the last pooling layer (in Fig. 2a) at 32, the resulting output size is equal to the input patch. Furthermore, the part with the pooling layers is known as the encoder, while the upsampling part is the decoder. However, limited contextual information is a critical issue in the deeper layers, which can be addressed by combining the predictions of the different layers (see FCN − 8s in Fig. 2a). In this way, the skip-connections combine in-depth, coarse, semantic information of the decoder part with the encoder part’s shallow, adequate location information. The FCN architecture is depicted in Fig. 2a. All the U-Net architectures, either with non-residual convolution blocks (Fig. 2b) or with residual convolution blocks (Fig. 2c), are followed the basic design of the FCN. However, the lack of non-linearities in the decoder part ruled out the use of FCN for the medical image segmentation. In the meantime, U-Net is successfully resolving the deficiency of the non-linearities by adding the convolution between the upsampling layers (see lower part of Fig. 2b and c). Furthermore, U-Net models’ strength is improved with the concatenation operation instead of the simple addition operation (see FCN − 8s in Fig. 2a). Simultaneously, depth is an essential criterion for U-Net models (for example, Fig. 2b) to improve segmentation accuracy. However, the gradients may be vanishing with such deep models. The vanishing gradients’ problem is resolved by adding the residual blocks in the U-Net (Fig. 2c). Furthermore, a deep study of U-Net architectures for medical image segmentation can be accessed by a recently published survey paper [29]. The mathematical notations of convolutions and upsampling layers are defined in Section 3.
An instance of gridding problems. a depicts three convolution layers with equal dilation rates (r = 2) with kernel size 3 × 3. The pixels (receptive field, denoting by blue) contribute to calculating the center pixel (representing by red). The similar dilation rates through three convolution layers reduce the local information because zeros are inserting between pixels. This effect is diminished when b different dilation rates (r = 1,2,3) are used. These rates are preventing the checkerboard patterns, which are introducing due to equal dilation in (see a) or large dilation rates in ASPP [9]. Source [48]
Traditional U-Net models either with the non-residual (see Fig. 2b) or residual (see Fig. 2c) or dense blocks [22] perform a sequence of convolution and strided convolution or max-pool operations on the original images, which can reduce the spatial dimensions of input images and increase the receptive field size in the sub-sampling process. While the upsampling operation recovers the reduced size of the input images; however, the possibility of losing the critical information of original images by the sub-sampling process can not be completely ruled out. To prevent this loss, dilated convolutions [53] can be used to learn more contextual information on the encoder path. The similarity between dilated and ordinary convolutions is that the convolution core’s size is the same. At the same time, the dilated convolution increases the receptive field size; however, the number of training parameters does not increase. Therefore, the dilated based models are not only limited to the natural images, but they are also continuously improving the segmentation accuracy in the medical domain [12, 13]. Furthermore, Devalla et al. [11] presented a U-Net model with equal or larger rates of dilation in the different parts to learn more contextual information. However, large or equal dilation rates introduces the gridding problem [54] (Fig. 3a). A similar problem of large dilation rates also exists with atrous spatial pyramid pooling (ASPP) [9]. ASPP in which multiple dilated layers have a parallel arrangement, information of these multiple-scales, also known as multi-scale information, can further boost the segmentation accuracy. Dilation rate increases the receptive field size by inserting the extra zeros between the kernel elements. This gap is continuously widening when a series of convolution layers have either similar or larger dilation rates. Here, the sparsed kernel of convolutions fails to capture any local information. In this way, the result of the gridding problem is the local information’s complete loss. This loss of local information may degrade the final accuracy. However, the gridding effect can be minimized using different dilation rates [48] (Fig. 3b). Moreover, in U-Net architecture, the mapping of information between encoder and decoder parts is different, which arises the semantic gap in the architecture. It can reduce the functionality of U-Net models. Zhou et al. [56] proposed to minimize the semantic gap by improving skip-connections. However, this improvement increases the complexity due to the multiple paths between the encoder-decoder sub-networks.
In the above discussion, U-Net architectures have followed 2D convolutions, while, several researchers extended their works into 3D convolutions [1, 20, 46] with excellent results to solve the problem of the brain MRI segmentation. In this work, we also add depth to a 3D U-Net model by employing the residual connections [15] to resolve the issue of vanishing gradients. However, residual connections used element-wise addition operations that gives a limited improvement in the segmentation accuracy. At the same time, concatenation operations are known to improve the width of the channels. Therefore, we also employed concatenations to fuse the features of different sub-networks of a 3D U-Net and in atrous-spatial pyramid pooling (ASPP) [9] blocks. However, to concatenate the features of multiple scales in ASPP, we used the recently proposed dense connections [19]. While dense connections resolve the vanishing gradients problem, they also offered features reusability property by concatenating all the layers’ feature maps. That means the input to a particular layer simultaneously has the coarse, semantic information of the deeper layers, and shallow, adequate location information of the lower layers. This information is further improved in the deeper layers. Furthermore, using an appropriate growth-rate, dense connections reduce the parameters generated by the residual networks. This reduction is essential to demonstrate maximum accuracy with minimum learned parameters. Moreover, we adopt 3D dilated convolutions and preventing the gridding problem by employing different rates of dilations. Finally, we use dense ASPP blocks on the skip-connections’ output feature maps to learn multi-scale features to improve the segmentation outcomes. This multi-scale learning from the redesigned skip-connections also minimizes the semantic gap without introducing complexity.
Inspired by the success of the residual and dense connections, dilation, and the ASPP techniques, we have proposed a variant form of 3D U-Net with the combination of the residual connections, dilation, and dense ASPP. We have offered an RD2A (Residual-Dilated Dense ASPP) 3D U-Net model. The key contributions of this study are given below:
-
A variant form of 3D U-Net. We used combined approach of residual connections and densely connected ASPP.
-
To avoid possible loss of information during training in the proposed model, we choose appropriate rates of dilation layer to gain the proper size of the receptive field on BRATS datasets. Additionally, we used dense connections among the multiple sizes of the receptive field in ASPP on the feature maps of a residual-dilated 3D U-Net model for exploiting the full contextual information of the 3D brain MRIs datasets.
-
We have worked on BRATS 2018 and BRATS 2019 datasets, where the proposed model achieved state-of-the-art performances compared to other recent methods in terms of both parameters and accuracy.
-
We have worked on iSeg-2019 datasets and achieved the best scores on the testing dataset against the best method of the iSeg-2019 validation dataset.
2 Related work
U-Net [39] introduces the concept of skip-connections. Such connections are useful to preserve the original information at each level of the encoder. At the decoder, the information concatenates with its predecessor’s level information. These connections open the door of a deep network to better understand biomedical images’ complex structure by using local and global contextual information. Marcinkiewicz et al. [33] used cascaded U-Net for brain segmentation, in which they used the first step works as a detection while the second as a multi-class classifier. Hu et al. [17] proposed a fusion method to concatenate the features of three 2D U-Net networks. Chen et al. [10] replaced the block of a residual 3D U-Net with the inception block. Two layers replace each 3D layer in a block: one for spatial information and the other for channel representation [50]. Chen et al. [10] presented their work with the parameters reduction of a 3D convolution. However, huge parameters exist due to the increased number of layers. Kermi et al. [26] proposed a 2D FCN to resolve the high memory demands of 3D brain MRI. Residual connections are used to build a profound network; therefore, the resulting model generates many parameters during training. A combination of two cost functions was used to balance the different classes. Isensee et al. [20] proposed a residual encoder-decoder architecture with a 3D UpSampling layer on the resulting maps of different blocks at decoder sub-network to extract the deeper features. Xu et al. [51] proposed a segmentation task with the combination of three 3D encoder-decoder models. The output of the first model worked like the input of the next architecture. Wang et al. [46] proposed similar approach to Xu et al. [51] without end-to-end networks. Mehta et al. [34] proposed a 3D U-Net similar to Isensee et al. [20], they used transposed convolution instead of the UpSampling layer. Roy et al. [40] implemented large dilation rates in ASPP. However, the complexity was a major problem with the combined orthogonal networks. Ensembling of different models was also proposed to improve the segmentation results. Kori et al. [27] and Kamnitsas et al. [23] implemented the idea of ensembling on different models with a majority voting scheme.
As mentioned earlier, the profound variations of U-Net architectures can learn significant features about unhealthy brain structures. Hence, the most straightforward strategy with deep U-Net architectures is residual learning. The residual networks have several direct connections between layers to prevent the problem of vanishing gradients. Therefore, nearly all the above-discussed methods used residual connections. However, the generation of huge parameters is a severe problem with residual networks. Furthermore, designing skip-connections with the traditional approach hinders U-Net architectures’ potential from learning sophisticated information. Moreover, the system’s complexity increases with the combination of various architectures, such as cascaded U-Nets, ensembling, etc. In our work, we also used the residual network for the encoder sub-part. Here, dilated convolutional layers are employed. In this way, our redesigned encoder sub-network can learn more contextual information from input brain MRIs than the traditional U-Net architectures. Furthermore, we used dense ASPP blocks to design the skip-connections to allow our network to learn more fine-grained multi-scale features. The dense connections reduced the parameters and offered more scaling [52] to each dilated convolution by improving the channels’ width. This scaling factor in multi-scale features minimizes the semantic gap. In this way, our proposed architecture without adding any complexity can solve all the previously proposed methods’ problems. Furthermore, a comparison between the proposed dense ASPP blocks and the architecture of Zhou et al. [56] is depicted in Fig. 4.
Outline of skip-connections variations, a illustrates the proposed dense ASPP blocks on the skip-connections. Conv in an oval shape denoting the standard convolution layers. Simultaneously, the solid red and blue lines represent the pooling and the upsampling operations while their dashed forms denote the repetitive pooling and the upsampling processes in b. In the meantime, the concatenation operations are indicating in gray. Furthermore, small circular shapes (blue) representing the different dilation rates of the convolutions, and the black oval shapes denoting the 1 × 1 × 1 convolutions. The concatenated operations of multiple dilated convolutions deduced more fine-grained multi-scale features from high and low-level input resolutions (indicating by the encoder). These multi-scale features further improved in the decoder part. b illustrates the UNet++ in detail. The numbers represent the concatenation operations between multiple encoder-decoder sub-networks, which will be continuously increasing by adding depth to the network. As a consequence, redesigned skip-connections in UNet++ increases complexity. Moreover, the solid orange line in a and b represent the output
3 RD2A (residual-dilated dense ASPP 3D U-Net)
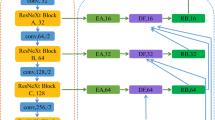
Figure 5 shows our proposed architecture. The combined approach of the residual connections, dilation, and dense ASPP consists of a Residual-Dilated and Dense ASPP blocks. Residual-Dilated blocks are in the first part of the RD2A 3D U-Net model and the application of dense ASPP on the feature maps of the Residual-Dilated blocks. The residual-dilated block shares a common idea of a dilated convolutional layer. Figure 6 shows the design of the residual-dilation block. To learn more contextual information, we have used dilated convolutional with the rates 1 and 2 in each residual-dilation block. Unlike Wang et al. [46], we implement the different dilation rates within each residual-dilation block because the same rates introduce the gridding problem [48]. In the proposed architecture, we have used 4 residual-dilation blocks. After each block, strided convolutional is used to reduce the input resolution. The dense ASPP block is applied to the feature maps of the first three residual-dilation blocks before the sub-sampling layers. Therefore, the reduced size of feature maps is processed via the multiple parallels dilated layers on different rates with dense connections to exploit the multi-scale contextual information, and non-dilated layer implemented to deduce global contextual information. Figure 7 exhibits the dense ASPP block of our proposed architecture. Here, we used four different dilated layers with rates of 1, 2, 3, and 5. The residual-dilation and dense ASPP blocks exist at the encoder part of our proposed architecture. In the decoder path, the input resolution at the corresponding level’s predecessor of encoder path recovers by using a 3D UpSampling layer of size 2 × 2 × 2. In our proposed approach, each level of the encoder path preserves more contextual information with the concatenation of generated features at each level of the decoder path.
Proposed architecture. RD2A 3D U-Net model is divided into a lower part (encoder sub-network) and a higher part (decoder sub-network). The lower part has the residual-dilated blocks (red). For each residual-dilated block (Fig. 6), two different rates of dilated convolutional layers are used. The dense ASPP blocks (purple) are employed after each upsampling layer (violet) on the maps of the lower part in the higher part before each concatenation operation (symbol C in an oval shape). For each Dense ASPP block (Fig. 7), dense connections are used among the three different parallel dilated convolutional layers
Outline of a dense ASPP block, a illustrates a dense ASPP block in detail, b demonstrate the idea of the concatenation operations (denoting by a symbol c in oval shape) in the parallel dilated layers. The input feature maps to a layer is the combination of output feature maps of all previous layers. For example, the input to a last dilated convolution (blue) includes the output feature maps, x− 1, x0, x1 and x2 because of the dense connections
We divide the proposed architecture into two parts: residual-dilation blocks at encoder RE and the upsampling process at decoder RU. We use 3D convolutional filters or kernels to transform the 3D raw brain MRIs into our architecture features. For the first convolutional layer, when a kernel of size 3 × 3 × 3 (denoting by \({Weight_{1}^{1, 4}}\)) is applied to the input patch of size 128 × 128 × 128 (representing by [IF]− 1), then feature map of size 1 × 128 × 128 × 128 is extracted. 4 are the number of modalities or channels which are used to extract the input patches. Since 24 filters (denoting by \({\sum }_{z=1}^{4} Weight_{1}^{24, 4}\)) are in use. Hence, 24 × 128 × 128 × 128 (denoting by [IF]0) will be the final size of the features for the first convolutional layer. Mathematically, the procedure of features extraction for the first weighted layer can be written as
where \(b_{1}^{kernel}\) is bias term. ⋆ representing convolution operation. f is a Leaky Rectified Linear Unit (Leaky ReLU) non-linear activation function which was defined in (2).
[IF]0 will be input to our first RE to generate the feature maps \({[R_{E}]}_{F_{0}}\) in (6). Since RE denoting a residual-dilated block of two convolutional layers. Hence, for each layer of RE, we use a similar idea of feature extraction, which was explained in (5).
where \({Weight_{2}^{kernel,z}}\) and \(Weight_{3}^{kernel,z}\) are the filters of the second and third convolutional layers, respectively. [IF]0 and [IF]1 are the input features of sizes 24 × 128 × 128 × 128.
By taking inspiration from (6), the generalized formula of the feature maps for the remaining RsE of the encoder sub-part can be written as
where D = 1,2,3 denoting the current RE of the encoder. For each RE, Cl representing number of filters of a layer l where l= 1,2, \(b_{l}^{kernel}\) representing the bias term of layer l, and \({[R_{E}]}_{F_{D}}\) denoting feature maps. In the meantime, \([R_{E}]_{{F}_{\left (D-1\right )}}\) representing the input maps of previous RsE.
After the feature extraction from the encoder part, we perform the up-sampling operation RU in the decoder part. Here, the final input resolution of encoder path upsamples to its predecessor’s input size. To explain the current RU, consider a tensor of the shape 192 × 16 × 16 × 16 representing the previous convolutional layer’s feature maps. These feature maps are resized on applying an UpSampling layer of factor 2 × 2 × 2. After this step, the size of feature maps at current RU is twice than that of its corresponding RE, i.e., 192 × 32 × 32 × 32 at RU and 96 × 32 × 32 × 32 at RE. To exactly matches the sizes of features at both ends, a 3D convolution with filter size 3 × 3 × 3 is applied on RU. After then, a dense ASPP block of three parallel dilated layers is applied on RE. Finally, two tensors (one of RU and other of RE) of the shapes 96 × 32 × 32 × 32 are concatenated. A 3D convolutional layer is then applied to the combined tensor of the shape 96 × 32 × 32 × 32 for the remaining RsU. The general formula for each level of RU can be summarized in (??)
where \({{[R]}_{U_{D}}}\) denoting current RU and \({[R_{E}]}_{F_{D}}\) representing current RE, at D= 1,2,3. Equation (8a) denoting a upsampling process of size 2 × 2 × 2 on RE. \({{[R_{E}]}_{F}}_{\left (D-1\right )}\) representing the previous RE. Equation (8b) representing a dense ASPP block of three parallel layers where layer l= 1,2,3. Equation (8c) denoting a single convolution layer after the concatenation layer. Finally, the last RU is reduced to match the brain MRIs’ labels, followed by the softmax activation. The shape of the tensor after the softmax function is 3 × 128 × 128 × 128. After resampling, this shape is changed to the size of the original MRI for the submission purpose.
4 Experiments
We have used three benchmark datasets to validate our proposed architecture: BRATS 2018 and 2019 datasets [3,4,5,6, 35], and the six-month infant brain MRI iSeg-2019 dataset [44]. BRATS dataset is for brain tumor segmentation and the iSeg dataset for infant brain tissue segmentation. The difference for each subject among the BRATS and the iSeg datasets is the number of modalities; BRATS datasets have four different modalities, while the iSeg-2019 dataset has two different modalities. The description of BRATS datasets is discussed in the Section 4.1.1. The information of the iSeg-2019 dataset is given in the Section 4.2.1. The essential metrics used by the organizers in the MICCAI BRATS and iSeg competitions are explained in Section 4.4. Furthermore, we have worked on quantitative and qualitative analysis of BRATS and iSeg-2019 datasets.
4.1 Brain tumor segmentation challenge
4.1.1 Data description
BRATS 2018 and 2019 training sets contain 285 and 335 patients, respectively. According to the gliomas classification, both high-grade and low-grade patients are available in the BRATS training sets. We used 210 high-grade and 75 low-grade patients from the BRATS 2018 training dataset. In the meantime, 259 patients of high-grade and 76 patients of low-grade are selected from the BRATS 2019 training set. Each patient has four types of MRI: native (T1), post-contrast T1-weighted (T1ce), T2-weighted (T2) and Fluid Attenuated Inversion Recovery (FLAIR). The organizers performed different pre-processing steps on the entire data, such as skull-stripping, re-calculation to the equal 1mm3 resolution, and all the scans of each case were co-registered to magnify the unhealthy tissues. The code of all pre-processing steps is now publicly availableFootnote 1. The manual segmentation of the entire training dataset was performed by the experts and provided by the organizers. 240 × 240 × 155 is the dimension of each MRI modality. For each subject, the annotated labels has the values of 1 for the necrosis and non-enhancing tumor (NCR/NET), 2 for peritumoral edema (ED), 4 for enhancing tumor (ET), and 0 for background. The segmentation accuracy is measured by several metrics, where the predicted labels are evaluated by merging three regions, namely whole tumor (Whole Tumor or Whole: label 1, 2 and 4), tumor core (Tumor Core or Core: label 1 and 4), and enhancing tumor (Enhancing Tumor or Enhancing: label 1). We have evaluated our proposed model on validation datasets, 66 patients in the BRATS 2018, and 125 patients in BRATS 2019. Each patient in the validation datasets has no truth label.
4.2 Infant brain MRI segmentation challenge
4.2.1 Data description
In the MICCAI 2019 infant brain MRI competition, each team has access to three different datasets. 10 subjects are available in the iSeg-2019 training set. The validation dataset contains 13 subjects of one location, while 16 cases of three sites are available in the testing set. Each subject consists of two different MRI modalities: T1 and T2. The ground truth values are available with the training dataset. The dimension of each modality is 144 × 192 × 256. For each subject, the annotated labels has the values of 1 for the cerebrospinal fluid (CSF), 2 for gray matter (GM), 3 for white matter (WM), and 0 for background. At around 6 months of age, the intensity ranges of voxels in GM and WM in structural MRI images are largely overlapping (especially around the cortical regions), leading to the ambiguities creating the most challenge for tissue segmentation. The subjects of the validation and testing sets have no truth labels.
4.3 Implementation details
In this work, bias field correction and normalization steps are performed on each training dataset. During training, we performed the five-fold validation on each dataset. 4 × 128 × 128 × 128 is the input size for BRATS datasets while 2 × 32 × 32 × 32 is the size for the iSeg-2019 dataset. The batch size for the BRATS training datasets is 1, while the batch size is 4 for the iSeg-2019 dataset. We used Keras to build the proposed architecture. We have used Adam optimizer with 4 × 10− 5 learning rate and a weight decay of 1 × 10− 5. We train our architecture for 60000 iterations with the BRATS training datasets. In contrast, 112800 iterations with the iSeg-2019 training dataset. Data augmentation have been undertaken on the fly for each patch, including flipping horizontally and rotating a random 90∘ to avoid the over-fitting problem during training. We used the Leaky ReLU non-linearity during training. We implemented instance normalization [20] because of a small batch size. The loss function is an important hyper-parameter during the training process. It helps balance the classes; in the BRATS and the iSeg training datasets, healthy tissues are bigger than unhealthy tissues. Different loss functions were previously proposed [14, 21, 24, 41]. We found that cross-entropy loss is not ideal with such kind of highly unbalanced datasets. Multi-label dice loss function has shown the remarkable results in highly imbalanced datasets [20, 36, 46]. We have used the loss function [36], while the number of samples per batch is one. (9) shows the mathematical representation of loss function.
where predj,d and truthj,d are the prediction obtained by softmax activation and ground truth at voxel j for class d, respectively. D is the total number of classes.
4.4 Evaluation metrics
For evaluating the BRATS and the iSeg datasets, we use the various metrics: the Dice Similarity Coefficient (DSC), the sensitivity, the specificity, the Hausdorff95 distance or modified Hausdorff distance (H95), and the average surface distance (ASD). Mathematically, each metric can be written as:
where TP, FP, TN, and FN are the number of true positive, false positive, true negative, and false negatives voxels, respectively. For both of H95 and ASD, G and S are truth and segmented sets of voxels, respectively. For H95, d(y,S) is point-to-set distance defined by: d(y,S)=\(\displaystyle \min \limits _{x \in s} \|{y-x}\|\), with ∥.∥ denoting euclidean distance. We use the similar notation of H95 for ASD and \(\left |.\right |\) denoting the cardinality of a set.
In the BRATS and the iSeg challenges, the ranking of the teams depends on the dice scores. We also report the best method based on the highest average dice scores in our work. DSC, sensitivity, and specificity evaluate the voxel-wise overlap between the truth and the segmented MRIs. H95 and ASD are the spatial distance-based metrics. The earlier is used for the BRATS datasets while the last one is used for the iSeg datasets. Furthermore, another name of metric H95 is modified hausdorff distance, commonly used in iSeg competitions.
5 Quantitative analysis
5.1 Brain tumor segmentation challenge
To check the capability of proposed architecture, we build four different architectures based on our proposed model. For the first architecture, Residual 3D U-Net, we replaced the residual-dilated blocks with residual blocks. Also, the dilated convolutional layers are replaced with non-dilated convolutions within each block. Moreover, the dense ASPP blocks are removed from the proposed architecture. For the second architecture, Residual-Dilated, we keep the residual-dilated blocks and remove the dense ASPP blocks from our proposed architecture. For the third architecture, Residual-Dense-Dilated, we implement the dense connections within each residual-dilated block and remove the proposed architecture’s dense ASPP blocks. For the fourth architecture, Residual-Dilated-ASPP, we keep the residual-dilated blocks and remove the dense connections from the ASPP blocks. Table 1 depicts the details of the different models, including the proposed architecture. All the architectures are trained and validated with the BRATS 2018 datasets. To train each architecture, we used the BRATS datasets’ training schemes, which were explained in the implementation details Section 4.3. The best fold of each architecture is used on the full training and the validation sets for the predicted MRIs. These predicted MRIs are then submitted to the organizersFootnote 2 for final scores. Each architecture’s scores are based on several metrics: the Dice Similarity Coefficient, the sensitivity, the specificity, and the Hausdroff95 distances (H95). These metrics were explained in the evaluation metrics Section 4.4.
Table 2 depicts the results of all the models that include our proposed architecture. The number of parameters with the Residual-Dense-Dilated model is lowest compared to the algorithms: Residual 3D U-Net, and Residual-Dilated. We do not reach the Residual-Dense-Dilated model in terms of parameters with our proposed model and Residual-Dilated-ASPP architecture due to the presence of three parallel dilated convolutional layers in ASPP. Our proposed model’s number of parameters is reduced compared to the Residual-Dilated-ASPP architecture by introducing the dense connections with a growth rate of 12 in ASPP. For the Residual-Dilated model, the dice scores of the three types of cancer tumors increase (on validation dataset) compared to the Residual 3D U-Net model. The increased dice scores reaffirm the potential of the dilation blocks with residual connections. For Residual-Dilated-ASPP and Residual-Dense-Dilated models, the whole tumor’s scores for the sensitivity and the H95 distances increase, and the specificity decreases; thereby, the occurrence of the false positives increases.
The Residual-Dilated model compared to the models: Residual-Dense-Dilated and Residual-Dilated-ASPP, the sensitivity scores and the H95 distances of the whole tumor decrease, and the specificity increases; thereby, the occurrence of false negatives increases. Our proposed model can balance the events of false positives and false negatives based on the combined approach of the residual network, dilation, and dense ASPP. For the Residual-Dilated, Residual-Dense-Dilated, and Residual-Dilated-ASPP models, the tumor core’s dice scores and enhancing tumor increases. Also, the sensitivity and the H95 distances decrease for the tumor core and enhance tumor but increase the specificity. Thus, the aggregation process improves the dice scores of the tumor core and the enhancing tumor. In comparing our proposed architecture, the Residual 3D U-Net model obtained the Dice similarity coefficient’s low scores, a deciding metric for the best methods in BRATS competitions. The combined approach of the residual connections, dilation, and ASPP gives excellent results, as shown in Table 2 in terms of parameters.
5.1.1 Comparison with the best methods
Table 3 shows the comparison of our proposed work with the state-of-art methods of the BRATS 2018 validation dataset [2, 7, 8, 10, 14, 17, 18, 25, 27, 32, 33, 37, 38, 43, 45, 47, 49]. Here, we compared the results based on the dice scores. The proposed architecture secures best average scores against all the other algorithms, even obtained the higher scores than the ensembling of several architectures [2, 25, 27]. Furthermore, our work can save the time which is spent in performing the complex post-processing Conditional Random Field (CRF) Chandra [ResNet + CRF, V-Net + CRF] et al. [8] and test-time augmentations (TTA) Wang [3D UNer + TTA, Multi-class WNet + TTA] et al. [47], common strategies for removing the false-positive voxels. Based on the higher dice scores, our proposed model is more generalized on the unseen validation dataset.
We choose three different algorithms from Table 3 to justify our proposed approach of the residual connections, dilation, and dense ASPP. These three algorithms are: Chen et al. [10], Sun [DFKZ Net] et al. [43], and Chandra et al. [8]. Chen et al. [10] performed the division operation on a 3D weighted layer within each block of a residual network; the resulting layers were increased the number of parameters. Our proposed architecture scores exhibit the necessity of a 3D Convolutional layer to process the 3D brain MRIs. Sun [DFKZ Net] et al. [43] used a residual-based 3D U-Net model of Isensee et al. [20] with BRATS 2018 datasets; non-dilated convolution layers were used in the first part of the architecture. To reaffirm the potential of enhanced sizes with the residual part, we removed the dense ASPP blocks from our proposed architecture; the resulting architecture became a Residual-Dilated model (see row number 2 of Table 2). Its scores reaffirm our contribution of implementing the different dilation rates to preserve more information about the tumor’s small sizes. Chandra et al. [8] enhances the sizes of the receptive field to extract the complete information of an image [31] just before the softmax activation. For this, an atrous spatial pyramid pooling (ASPP) was used. The number of parameters was increased as three residual U-Nets were used. Moreover, Chandra et al. [8] used the big rates of dilation, thereby introducing the gridding problem; the vital local information was lost. Our proposed architecture scores with only 4.53 M of parameters exhibit the combined residual-dense connections’ necessity.
In summary, our proposed architecture achieved excellent results in all types of tumors. Furthermore, all the predicted MRIs of the BRATS 2019 datasets were submitted to the organizer’s webpageFootnote 3 for the online evaluation. The evaluation scores are shown in Table 4.
5.2 Infant brain MRI segmentation challenge
To ensure the proposed architecture’s capability, we also validate our architecture on the iSeg-2019 datasets. The training dataset (10 subjects) is divided into five-folds. In each fold, 8 subjects are selected for the training and remaining for the validation. We have chosen the best fold to evaluate the iSeg-2019 validation (13 subjects) and the testing datasets (16 subjects). Table 5 shows the results of all methods for iSeg-2019 validation and testing datasets. The scores without brackets are related to the validation dataset while the remaining to the iSeg-2019 testing dataset. The average score of each metric, especially DSC, is best with our proposed model than the MASI (baseline), long, and UBC001 methods. In the meantime, the CSF average scores for the metrics DSC and the ASD are lower with our presented work compared to the lyh and the tiantian methods. Furthermore, the scores of the Brain_Tech method are higher than our proposed work. In short, the Brain_Tech method is best with the iSeg-2019 validation dataset. At the same time, we secure the best scores on the testing dataset than the validation dataset’s top method. The validation dataset subjects belong to only one site, while the 16 subjects of the testing dataset are collected from three different locations. Our proposed model can perform better generalization on the unseen dataset of two or more sites based on the higher testing scores. Furthermore, the best testing dice scores are successfully distinguishing the contrast between the gray matter (GM) and white matter (WM) tissues.
6 Qualitative analysis
6.1 Brain tumor segmentation
The segmentation results of our proposed architecture are shown in Fig. 8. We choose two different patients from the BRATS 2018 training dataset. For these two patients, we only visualize the T1ce modality with ground-truth and prediction. Figure 8a and b represent the ground-truth and prediction with the T1ce modality of one patient, respectively. Figure 8c and d of another patient represents the T1ce modality with the ground-truth and prediction, respectively. Moreover, Fig. 8a and c exhibit the variations among the shape, size, and location of the tumors in different patients. As depicted by the predicted T1ce modality in Fig. 8d, our proposed algorithm has the potential to segment the big size of the whole tumor and the small size of the tumor core.
Segmentation results using our proposed architecture. a and b represent the ground-truth and prediction on T1ce modality, respectively. c and d denote the ground-truth and prediction on the T1ce modality of another patient, respectively. Each color represents a different tumor: red for Tumor core, green for the Whole tumor, and yellow for Enhancing tumor
Our proposed algorithm failed for some patients of the BRATS 2018 training dataset. A case of wrong segmentation is depicted in Fig. 9. We only visualize the T1ce modality with ground-truth and the prediction. A long orange arrow in Fig. 9b exhibits the instance of the wrong segmentation, in which our proposed algorithm wrongly predicted the background label as a tumor core. We will investigate to solve the wrong prediction in the future work through the combined loss functions.
An instance of wrong segmentation results. a and b represent the ground-truth and prediction on T1ce modality of a patient, respectively. b represents an instance of the wrong segmentation; our proposed algorithm is wrongly predicted the background label as a tumor core that is shown by a long orange arrow
6.2 Infant brain MRI segmentation
Figure 10 shows the segmentation results of our proposed architecture. We choose a subject from the iSeg-2019 training dataset. We demonstrated ground-truth and prediction of the selected subject on a T1 modality. Figure 10a and b represent the ground-truth and prediction, respectively. Predicted visualization exhibits the potential of the proposed algorithm for infant brain MRI segmentation.
7 Discussion and conclusion
We have proposed a model with the combination of the residual connections, dilation, and dense ASPP. Different atrous rates are chosen in the residual-dilation blocks to avoid the gridding problem. In the meantime, dense connections are employed to reduce the parameters. Dense ASPP blocks exploit the multi-scale information to avoid the ambiguities among brain MRIs’ labels and tissues. The multi-label cost function is used to prevent the imbalanced data problem. Augmentation techniques such as flipping and rotations are used to avoid the over-fitting problem during training. Finally, the combined approach achieved outstanding results with different brain MRI datasets.
We cannot train our network on big patch sizes due to memory limitations, especially with the BRATS datasets. Chen et al. [9] implemented ASPP on a big patch size with improved results. In the future, we will try our proposed approach on multiple medical imaging problems, especially for the kidney tumor segmentation using big patch sizes. Kidney tumor segmentation is a very challenging problem due to lack of information from only one modality in the MICCAI KiTS 2019 dataset [16]. In our work, we experimentally proved that the parameters could be efficiently reduced with improved results. Our proposed architecture has the potential to solve the problem of other medical imaging tasks. Furthermore, we will propose an architecture with weighted majority schemes and the study on the different normalization layers with varying batch sizes.
Availability of data and material
BRATS 2018 datasets
The description of the datasets and procedures to download them can be accessedFootnote 4.
BRATS 2019 datasets
The description of the datasets and procedures to download them can be accessedFootnote 5.
iSeg-2019 datasets
The description of the datasets and procedures to download them can be accessedFootnote 6.
References
Ahmad P, Qamar S, Hashemi SR, Shen L (2020) Hybrid labels for brain tumor segmentation. In: Crimi A, Bakas S (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 158–166
Albiol A, Albiol A, Albiol F (2019) Extending 2D deep learning architectures to 3D image segmentation problems. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 73–82
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C (2017) Segmentation labels and radiomic features for the pre-operative scans of the TCGA-GBM collection. The Cancer Imaging Archive (2017)
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann J, Farahani K, Davatzikos C (2017) Segmentation labels and radiomic features for the pre-operative scans of the TCGA-LGG collection. The Cancer Imaging Archive 286
Bakas S, Akbari H, Sotiras A, Bilello M, Rozycki M, Kirby J, Freymann JB, Farahani K, Davatzikos C (2017) Advancing The Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci Data 4:170117. https://doi.org/10.1038/sdata.2017.117
Bakas S, Reyes M, Jakab A, Bauer S, Rempfler M, Crimi A, Shinohara RT, Berger C, Ha SM, Rozycki M, Prastawa M, Alberts E, Lipková J, Freymann JB, Kirby J, Bilello M, Fathallah-Shaykh HM, Wiest R, Kirschke J, Wiestler B, Colen RR, Kotrotsou A, LaMontagne P, Marcus DS, Milchenko M, Nazeri A, Weber M, Mahajan A, Baid U, Kwon D, Agarwal M, Alam M, Albiol A, Albiol A, Varghese A, Tuan TA, Arbel T, Avery ABP, Banerjee S, Batchelder T, Batmanghelich K, Battistella E, Bendszus M, Benson E, Bernal J, Biros G, Cabezas M, Chandra S, Chang YJ et al (2018) Identifying the Best Machine Learning Algorithms for Brain Tumor Segmentation, Progression Assessment, and Overall Survival Prediction in the {BRATS} Challenge. CoRR arXiv:1811.02629
Carver E, Liu C, Zong W, Dai Z, Snyder JM, Lee J, Wen N (2019) Automatic brain tumor segmentation and overall survival prediction using machine learning algorithms. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 406–418
Chandra S, Vakalopoulou M, Fidon L, Battistella E, Estienne T, Sun R, Robert C, Deutsch E, Paragios N (2019) Context Aware 3D CNNs for Brain Tumor Segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 299–310
Chen LC, Papandreou G, Schroff F, Adam H (2017) Rethinking Atrous Convolution for Semantic Image Segmentation. CoRR arXiv:1706.05587
Chen W, Liu B, Peng S, Sun J, Qiao X (2019) S3d-UNet: Separable 3D U-Net for Brain Tumor Segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 358–368
Devalla SK, Renukanand PK, Sreedhar BK, Perera SA, Mari JM, Chin KS, Tun TA, Strouthidis NG, Aung T, Thiery AH, Girard MJA (2018) {DRUNET:}{A} Dilated-Residual U-Net Deep Learning Network to Digitally Stain Optic Nerve Head Tissues in Optical Coherence Tomography Images. CoRR arXiv:1803.00232
Dolz J, Ayed IB, Desrosiers C (2018) Dense Multi-path U-Net for Ischemic Stroke Lesion Segmentation in Multiple Image Modalities. CoRR arXiv:1810.07003
Dolz J, Xu X, Rony J, Yuan J, Liu Y, Granger E, Desrosiers C, Zhang X, Ayed IB, Lu H (2018) Multi-region segmentation of bladder cancer structures in {MRI} with progressive dilated convolutional networks. CoRR arXiv:1805.10720
Feng X, Tustison N, Meyer C (2019) Brain tumor segmentation using an ensemble of 3D U-Nets and overall survival prediction using radiomic features. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 279–288
He K, Zhang X, Ren S, Sun J (2015) Deep Residual Learning for Image Recognition. CoRR arXiv:1512.03385
Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, Rosenberg J, Blake P, Rengel Z, Oestreich M et al (2019) The kits19 challenge data: 300 kidney tumor cases with clinical context, ct semantic segmentations, and surgical outcomes. arXiv:1904.00445
Hu Y, Liu X, Wen X, Niu C, Xia Y (2019) Brain Tumor Segmentation on Multimodal MR Imaging Using Multi-level Upsampling in Decoder. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 168–177
Hua R, Huo Q, Gao Y, Sun Y, Shi F (2019) Multimodal brain tumor segmentation using cascaded V-Nets. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 49–60
Huang G, Liu Z, Weinberger KQ (2016) Densely Connected Convolutional Networks. CoRR arXiv:1608.06993
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2018) Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the {BRATS} 2017 Challenge. CoRR arXiv:1802.10508
Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH (2019) No New-Net. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, pp 234–244
Jégou S, Drozdzal M, Vázquez D, Romero A, Bengio Y (2016) The One Hundred Layers Tiramisu: Fully Convolutional DenseNets for Semantic Segmentation. CoRR arXiv:1611.09326
Kamnitsas K, Bai W, Ferrante E, McDonagh SG, Sinclair M, Pawlowski N, Rajchl M, Lee MCH, Kainz B, Rueckert D, Glocker B (2017) Ensembles of Multiple Models and Architectures for Robust Brain Tumour Segmentation. CoRR arXiv:1711.01468
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78
Kao PY, Ngo T, Zhang A, Chen JW, Manjunath BS (2019) Brain tumor segmentation and tractographic feature extraction from structural MR images for overall survival prediction. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 128–141
Kermi A, Mahmoudi I, Khadir MT (2019) Deep convolutional neural networks using U-Net for automatic brain tumor segmentation in multimodal MRI volumes. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 37–48
Kori A, Soni M, Pranjal B, Khened M, Alex V, Krishnamurthi G (2019) Ensemble of fully convolutional neural network for brain tumor segmentation from magnetic resonance images. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 485–496
Krizhevsky A, Sutskever I, Hinton GE (2012) Imagenet classification with deep convolutional neural networks. In: Advances in neural information processing systems, pp 1097–1105
Liu L, Cheng J, Quan Q, Wu FX, Wang YP, Wang J (2020) A survey on U-shaped networks in medical image segmentations. Neurocomputing 409:244–258. https://doi.org/10.1016/j.neucom.2020.05.070. http://www.sciencedirect.com/science/article/pii/S0925231220309218
Long J, Shelhamer E, Darrell T (2014) Fully Convolutional Networks for Semantic Segmentation. CoRR arXiv:1411.4038
Luo W, Li Y, Urtasun R, Zemel RS (2017) Understanding the Effective Receptive Field in Deep Convolutional Neural Networks. CoRR arXiv:1701.04128
Ma J, Yang X (2019) Automatic Brain Tumor Segmentation by Exploring the Multi-modality Complementary Information and Cascaded 3D Lightweight CNNs. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 25–36
Marcinkiewicz M, Nalepa J, Lorenzo PR, Dudzik W, Mrukwa G (2019) Segmenting Brain Tumors from MRI Using Cascaded Multi-modal U-Nets. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 13–24
Mehta R, Arbel T (2019) 3D U-Net for brain tumour segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 254–266
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber M, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Criminisi A (2015) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024. https://doi.org/10.1109/TMI.2014.2377694
Milletari F, Navab N, Ahmadi SA (2016) V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation. CoRR arXiv:1606.04797
Nuechterlein N, Mehta S (2019) 3d-ESPNet with Pyramidal Refinement for Volumetric Brain Tumor Image Segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 245–253
Rezaei M, Yang H, Meinel C (2019) voxel-GAN: Adversarial Framework for Learning Imbalanced Brain Tumor Segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 321–333
Ronneberger O, Fischer P, Brox T (2015) U-Net: Convolutional Networks for Biomedical Image Segmentation. CoRR arXiv:1505.04597
Roy Choudhury A, Vanguri R, Jambawalikar SR, Kumar P (2019) Segmentation of Brain Tumors Using DeepLabv3+. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 154–167
Samanta A, Saha A, Satapathy SC, Fernandes SL, Zhang YD (2020) Automated detection of diabetic retinopathy using convolutional neural networks on a small dataset. Pattern Recogn Lett 135:293–298. https://doi.org/10.1016/j.patrec.2020.04.026. http://www.sciencedirect.com/science/article/pii/S0167865520301483
Sarker MMK, Rashwan HA, Akram F, Banu SF, Saleh A, Singh VK, Chowdhury FUH, Abdulwahab S, Romani S, Radeva P, Puig D (2018) SLSDeep: Skin Lesion Segmentation Based on Dilated Residual and Pyramid Pooling Networks. CoRR arXiv:1805.10241
Sun L, Zhang S, Luo L (2019) Tumor segmentation and survival prediction in glioma with deep learning. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 83–93
Sun Y, Gao K, Wu Z, Lei Z, Wei Y, Ma J, Yang X, Feng X, Zhao L, Phan TL, Shin J, Zhong T, Zhang Y, Yu L, Li C, Basnet R, Ahmad MO, Swamy MNS, Ma W, Dou Q, Bui TD, Noguera CB, Landman B, Gotlib IH, Humphreys KL, Shultz S, Li L, Niu S, Lin W, Jewells V, Li G, Shen D, Wang L (2020) Multi-Site Infant Brain Segmentation Algorithms: The iSeg-2019 Challenge
Tuan TA, Tuan TA, Bao PT (2019) Brain Tumor Segmentation Using Bit-plane and UNET. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 466–475
Wang G, Li W, Ourselin S, Vercauteren T (2017) Automatic Brain Tumor Segmentation using Cascaded Anisotropic Convolutional Neural Networks. CoRR arXiv:1709.00382
Wang G, Li W, Ourselin S, Vercauteren T (2019) Automatic brain tumor segmentation using convolutional neural networks with test-time augmentation. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), vol 11384 LNCS. Springer, pp 61–72. https://doi.org/10.1007/978-3-030-11726-9_6
Wang P, Chen P, Yuan Y, Liu D, Huang Z, Hou X, Cottrell GW (2017) Understanding Convolution for Semantic Segmentation. CoRR arXiv:1702.08502
Weninger L, Rippel O, Koppers S, Merhof D (2019) Segmentation of Brain Tumors and Patient Survival Prediction: Methods for the braTS 2018 Challenge. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 3–12
Xie S, Sun C, Huang J, Tu Z, Murphy K (2017) Rethinking Spatiotemporal Feature Learning For Video Understanding. CoRR arXiv:1712.04851
Xu Y, Gong M, Fu H, Tao D, Zhang K, Batmanghelich K (2019) Multi-scale Masked 3-D U-Net for Brain Tumor Segmentation. In: Crimi A, Bakas S, Kuijf H, Keyvan F, Reyes M, van Walsum T (eds) Brainlesion: glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. Springer International Publishing, Cham, pp 222–233
Yang M, Yu K, Zhang C, Li Z, Yang K (2018) DenseASPP for Semantic Segmentation in Street Scenes. 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp 3684–3692
Yu F, Koltun V (2016) Multi-scale Context Aggregation by Dilated Convolutions. coRR arXiv:1511.0
Yu F, Koltun V, Funkhouser TA (2017) Dilated Residual Networks. CoRR arXiv:1705.09914
Zhang J, Jin Y, Xu J, Xu X, Zhang Y (2018) MDU-Net: Multi-scale Densely Connected U-Net for biomedical image segmentation. CoRR arXiv:1812.00352
Zhou Z, Siddiquee MMR, Tajbakhsh N, Liang J (2018) UNet++: {A} Nested U-Net Architecture for Medical Image Segmentation. CoRR arXiv:1807.10165
Acknowledgements
This work is supported by the National Natural Science Foundation of China under Grant No.61672250 and the Hubei Provincial Development and Reform Commission Project in China.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
The authors declare that there is no conflict of interest/competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ahmad, P., Jin, H., Qamar, S. et al. RD2A: densely connected residual networks using ASPP for brain tumor segmentation. Multimed Tools Appl 80, 27069–27094 (2021). https://doi.org/10.1007/s11042-021-10915-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-021-10915-y