Abstract
Image segmentation is a popular technique that is used for extracting information from images, which has also gained a lot of interest lately due to its importance in different scientific fields such as the medical field. This paper proposes a novel image segmentation technique using Expectation-Maximization (EM) clustering algorithm and Grasshopper Optimizer Algorithm (GOA). The proposed technique and the concept of image segmentation are effectively applied on dental radiography datasets that are collected from 120 patients with an age between 6 to 60 years old. To validate the proposed technique, a comparison in terms of purity and entropy measures is conducted against K-means, X-means, EM, and Farthest First algorithms. Based on our experimental results, the proposed technique using EM and GOA achieved the best results compared to other algorithms for all three datasets in terms of entropy and purity. The best results were obtained using the second dataset, which achieved purity value of 0.7126 and entropy value of 0.3083. Further, the proposed technique also outperforms U-net and Random Forest algorithms for the selected datasets.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Dental therapy has recently obtained more attention as human beings have become well aware of their dental health. According to the oral health and dental care in the U.S.A., approximately 9,404 people died from cancer of the oral cavity and periodontal disease in the U.S.A in 2014, and the five-year survival rate of such cancers was 68% [27].
Dental radiography is commonly called X-rays, which is used by dentists to find cavities, damages, and diseases that are not visible during a clinical dental examination [19]. Consequently, they require special analysis and inspection.
Image segmentation is a popular technique that is used as the first step in medical image analysis to reinforce the diagnosis and prognoses of many diseases including dental diseases. It is also defined as the process of dividing an image into multiple segments, which is typically used to identify objects or other relevant information in digital images [55].
Image segmentation approaches can be categorized into two approaches, namely: discontinuity detection based approach and similarity detection based approach [23]. In addition, image segmentation techniques are classified into the following categories [21]:
-
Thresholding method: this method is used when the intensity distribution between the objects of a foreground and a background are very distinct. A constant value of a threshold can be used to differentiate objects apart, by replacing the intensity value of a pixel upon the value of the threshold [51].
-
Edge-based method: detection of edges in an image is a very important step that helps in understanding image features. In edge-based approach, the discovery of the partitions of an image is based on some discontinuity in the pixel values and the intensity level [24].
-
Region-based method: this approach seeks to construct segments by combining neighboring pixels according to a criterion of homogeneity [26].
-
Clustering method: this technique segments the image into clusters by assigning pixels with similar characteristics into the same cluster.
A successful segmentation method is not always applicable to work with different types of images and problem areas [44, 46]. Fundamentally, medical image processing evolves around segmenting objects in images that appear abnormal and can be an indication of a disease. However, this is a challenging task to do, and even more challenging with dental radiography segmentation due to the specific characteristics of tooth structure [16]. These challenges are, but not limited to:
-
Images often contain high level of noise due to limitations on the amount of radiation allowed during imaging.
-
The topological structure of the tooth. The teeth boundary is not a simple clear boundary with some holes appearing as inner edges that make the segmentation more difficult.
-
The difficulty of detecting a common boundary between teeth because of being in touch with adjacent teeth.
-
Tissues are very close to the teeth and even some tissues surround the teeth.
Thereupon, we propose a segmentation technique that uses the Expectation Maximization (EM) algorithm and Grasshopper Optimization Algorithm (GOA). EM is a well-known algorithm that appeared in the late 1970’s, and GOA is a recently proposed metaheuristic that showed very competitive results in a wide range of complex and challenging applications [3, 36]. Thus, the efficiency of combining both algorithms motivates us to explore their application in the dental radiography segmentation.
The rest of the paper is organized as follows. Section 2 presents the related works on image segmentation techniques. Section 3 presents the proposed methodology and some preliminary knowledge and terminology. Experimental results and analysis are provided in Section 4. The conclusions and future work are finally presented in Section 5.
2 Related works
Although there has been great efforts in the literature to develop and improve image segmentation techniques, more research is required.
Many research works have been carried out in dental radiography processing and segmentation. In [55], a segmentation approach is proposed using region, texture, and edge based techniques. The proposed technique is implemented on dental radiography images. Provided results showed that, the proposed technique outperforms traditional methods such as thresholding technique.
A study of dental image segmentation and feature extraction is presented in [46]. K-means algorithm is used to segment images and texture statistics techniques by gray-level co-occurrence matrix for feature extraction. Experimental results showed that it is a promising technique for segmentation, however, more enhancement is recommended for better accuracy.
Amer and Aqel [5] proposed a preprocessing method for diagnosing problems with the wisdom teeth. Further, their work presented a method to segment wisdom teeth from panoramic images. The proposed work showed good potential for accurately classifying wisdom teeth that can be used later in the diagnosis of related impactions.
Lira et al. [30] proposed a segmentation approach for dental X-ray images using a Bayesian classifier that classifies pixels into two classes: inside the teeth and outside the teeth. The X-ray images were provided by the Odontological Department of Federal University of Rio de Janeiro, Brazil.
Patanachai et al. [43] used wavelet transformation for dental radiography segmentation. The segmentation of dental radiography results in a teeth feature marking each pixel. Such as, edges detection from the segmented image. The authors used a dataset provided by the faculty of dentistry hospital, Chulalongkorn University, Bangkok, Thailand.
Authors in [52] used active contour algorithm to extract the contour of a teeth in dental images. The algorithm depends on the intensity of the tooth region and does not require the existence of a sharp boundary between teeth. The dental images used were granted from the Criminal Justice Information Services (CJIS) division’s digitized radiographic images database of the Federal Bureau of Investigation, Washington.
In [25], the authors used a texture-based fuzzy inference system to effectively apply segmentation to dental radiography.The proposed approach achieved 83% accuracy on the CJIS division’s digitized radiographic images database.
Dental characteristics help with the identification of individuals. Authors in [40] used a fusion of matching algorithms including segmentation using adaptive thresholding. The proposed algorithm achieved an accuracy value of 82.5%.
A multi-step method based on mean shift algorithm for automatic tooth segmentation from Computerized Tomography (CT) images is presented in [38]. The proposed methods first classify bony tissues from non-bony tissues. Secondly, the general region of the teeth structure is separated from the other bony structures. After that, individual tooth region is detected and segmentation using mean shift algorithm is applied on the individual tooth. The proposed mean shift algorithm achieved an accuracy of 97%on a 3D dataset of dental Cone Beam Computed Tomography (CBCT) images.
Silva and Oliveira present a deep study on automatic teeth segmentation in x-ray images [54]. The study reviews different methods used to tackle this problem and most importantly it answers the following question:“Which category of segmentation method is most used in the reviewed works?”. The provided study states that the revised literature has been increasingly focused on research to segment dental x-ray images based on thresholding.
Among geometric models, level set method was used in [16] for root segmentation. In [13], a thresholding segmentation based on maximum fuzzy entropy method was proposed. The threshold values were automatically generated by the fuzzy entropy theory.
There has been a considerable amount of research aimed at developing image segmentation using deep learning models such as Convolutional Neural Network (CNN), Recurrent Neural Network (RNN) and Fully Convolutional Networks (FCN). However, there is no doubt that the use of deep learning poses several challenges such as: the need for more challenging and different types of datasets, speed of the models and memory efficient models [35].
A summary of the dental radiography analysis is provided in Table 1, which shows the techniques used for each work, the purpose behind applying the analysis, and the concluding results for each study. Based on Table 1, it is observed that different techniques are used for dental radiography segmentation, which indicates that there are no dominant technique for this purpose and that research are yet not mature to solve the problem. It is also observed that, Bayesian classifier and wavelet transformation achieved better results among the others.
However, clustering is used by many researchers for image segmentation as an unsupervised learning method. Bora and Gupta [6] proposed a color based segmentation by predetermining colors importance in the image. K-means clustering algorithm was then applied using a cosine distance measure.
In [12], image segmentation is performed using k-means algorithm, and subtractive clustering algorithm to generate the initial centroid points. A review of applying clustering techniques for image segmentation can be found in [39]. The review includes fuzzy c-mean algorithm, hierarchical clustering, K-means algorithm, and self-organizing map.
Adaptive k-means algorithm is proposed in [58] to segment images based on adaptive k values. In this approach, the image is first normalized, then it is converted to LAB color space, and finally it is segmented based on adaptive values of k that are determined using maximum connected domain algorithm [59] .
Carson et al. [7] used the Expectation-Maximization (EM) algorithm to segment images. The proposed algorithm was experimented on 10,000 images. To segment each image, the joint distribution of color, texture, and position features are modeled. Liu et al. [31] integrated EM clustering algorithm with the total variation regularization scheme to segment a variety of vector-valued images. Experiments showed that the combination of the EM algorithm and the total variation regularization scheme gives a good segmentation of images.
A method called Mask R-CNN was introduced by Facebook AI Research (FAIR) [18] that is used for predicting segmentation masks on each region of interest in an image. The method was tested on COCO dataset [29], which provided better generalization of various tasks with average precision value of 37.1.
In a more recent study on segmentation techniques, Panoptic Segmentation (PS) was introduced [22]. PS focuses on two tasks; assigning each pixel in an image to a semantic label and segmentation of each object instance. Every object instance detected is set an id. Hence, only one semantic label and one instance id are allowed to be assigned to each pixel. This approach achieves a more rich and complete segmentation process according to panoptic quality measure.
Geus et al. [9] proposed a Fast Panoptic Segmentation Network (FPSNet). The authors use a convolutional neural network module that does not require any prior predictions, which are computationally costly. FPSNet showed superior results in terms of speed compared to existing state-of-the-art panoptic segmentation networks. In addition to being fast, FPSNet also achieved a competitive panoptic quality score value of 55.1 on the Cityscapes dataset [8].
There is no recognized nor unified standard for image segmentation. Therefore, image segmentation has become an optimization problem. Optimization problems have been tackled by meta-heuristic algorithms as they have demonstrated effectiveness and efficiency in finding optimal solutions [45]. Mohsen et al. [37] presented a new image segmentation technique based on Particle Swarm Optimization (PSO). The proposed PSO based segmentation approach solves the thresholding problem using the search optimization function of PSO. The proposed method was evaluated on different datasets and has shown efficient results. Another implementation of PSO based segmentation mammography images is presented in [47].
Segmentation techniques require the definition of an optimization framework that addresses the critical issues including choosing segmentation algorithms’ parameters, choosing the representation for the image, and the parameter configuration of some previous image processing step [34]. For example, optimization techniques involve the use of Genetic Algorithm (GA). The main use of GA in image segmentation is the modification of parameters in existing segmentation algorithms and pixel-level segmentation [14].
GA has been widely used in image processing and segmentation. Amelio and Pizzuti [4] proposed a GA for color image segmentation based on color and texture. Also, Sahu and Bhurchandi [49] used GA-based algorithm to segment color images based on their histograms. Moreover, GA is used to maximize the entropy to efficiently segment the image into objects and background [1].
Nature inspired algorithms have been used with multi-thresholding medical image segmentation [28, 41, 50]. Multi-thresholding based methods segment an image into two or more regions depending on a threshold value that may increase significantly, which becomes computationally inefficient [11]. Thus, requiring algorithms that maintain robustness with high dimensional problems.
In our work, we will combine a recent optimization algorithm called Grasshopper Optimization Algorithm (GOA) and the well known EM clustering algorithm for dental radiography segmentation, which to the best of our knowledge is not yet experimented in this domain.
3 Proposed methodology
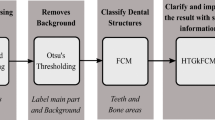
This section describes the methodology used for constructing the proposed EM-GOA technique, which is given in Fig. 1.
Based on Fig. 1, the obtained radiography images were pre-processed and converted into matrices. After pre-processing the instances in the dataset, teeth are detected using Level-1 EM. Level-1 EM includes applying EM algorithm on the radial image to detect each tooth separately. Then, segmentation is applied on each detected tooth. The segmentation operation of teeth also uses EM algorithm and is optimized via the GOA optimizer. GOA optimizes the parameters by which the EM operates to further enhance the clustering results. Also, other clustering techniques are applied for the purpose of comparison. Lastly, the performance of the algorithms is evaluated.
Algorithm 1 translates the methodology of pre-processing the radiography images, applying Level-1 EM, and applying Level-2 EM-GOA. The algorithm accepts four parameters which are the radiography image, the number of clusters (k), the population size, and the number of iterations. The algorithm first pre-process the image into a pixel matrix. The pixel matrix is then converted to a feature-pixel matrix. This is further explained in Section 3.1. EM algorithm segments the feature-matrix into predicted clusters where each cluster reflects a tooth, which is further explained in Section 3.2. The algorithm then iterates over each segmented tooth and the GOA algorithm is applied by simulating repulsion and attraction forces between the grasshoppers to find the best grasshopper at each iteration. The last iteration of GOA returns the best grasshopper having the best fitness value along with the predicted assignment of points for the grasshopper. The role of GOA is further described in Section 3.3. The rest of this section describes detailed knowledge and terminology used in the proposed algorithm and corresponding methodology.

3.1 Dataset Description and Pre-processing
The datasets were created for a grand challenge for automated detection of anatomical landmarks and analysis for diagnosis in cephalometric X-ray Images [56]. Moreover, it was used for a second challenge for computer-automated detection of caries in Bitewing radiography. The datasets consist of dental anatomy data repository of bitewing radiographs.
There are 120 bitewing images collected from 120 patients aged between 6 to 60 years. The cephalograms were acquired in TIFF format and the image resolution was 1935 x 2400 pixels. For evaluation, 19 land-marks were manually marked in each image and reviewed by two experienced medical doctors. Figure 2 provides three datasets of bitewing images. Dataset 1, dataset 2, and dataset 3 consist of 88614, 280251, and 108368 pixels, respectively [56].
Datasets are pre-processed to obtain the features of the data set which are used to detect teeth using Level-1 EM. The pre-processing includes converting the bitewing image of the dataset into a matrix where each pixel in the image is represented by a matrix element having the value as the color of the pixel. The matrix is then converted to a feature-pixel matrix where features are represented by columns and pixels are represented by rows. The features include the x-coordinate, y-coordinate, and gray-scale color.
3.2 Expectation-maximization algorithm
The idea of EM is to assign data points partially to different clusters instead of assigning them to only one cluster. To do this partial assignment, EM models each cluster using a probabilistic distribution. So, initially, a data point is associated with a cluster with certain probability and then it is reassigned to the cluster with the highest probability in the final assignment [15]. In practice, the algorithm is used to find the mixture of Gaussians that can model the dataset. Given a model of a mixture Guassians, we have the following parameters:
-
X: a set of observed values.
-
Z: a set of estimates for unobserved values (missing values).
-
𝜃: a vector of unknown parameters for the model.
-
L(𝜃,X,Z): a likelihood function.
The core idea of EM is to use the set of available observed data (X) to estimate the missing values (Z) and upon this, update the values of the parameters.
EM works as follow. First, initialize the unknown parameters 𝜃 to random values. Next, use the values of observed data to generate an estimate for the missing data, this is called the expectation step . Then, use the complete data generated in the previous step to update the values of the parameters. This is known as the maximization step. Finally, iterate until convergence. Mathematically, the algorithms works as follows [10]:
-
1.
Initialization. Set the parameters 𝜃 to some random values:
$$ \theta=\{\mu_{1},\mu_{2},...,\mu_{t}\} $$(1) -
2.
Expectation step.Define Q(𝜃|𝜃(t)) as the expected value of the log likelihood function of 𝜃 as:
$$ Q(\theta|\theta^{t})=E_{Z|X,\theta^{(t)}} [Log L(\theta;X,Z)] $$(2) -
3.
Maximization step. Find the parameters that maximize Q:
$$ \theta^{(t+1)}=argmax~Q(\theta|\theta^{(t)}) $$(3) -
4.
Evaluate the log likelihood function and check for convergence. If not converged, go to step 2.
EM is a very popular and powerful algorithm used in many applications including estimating motion models for tracking, hidden Markovian models, and in image segmentation.
3.3 Grasshopper Optimization Algorithm (GOA)
Although in the last couple of years a huge number of nature inspired algorithms have been proposed, credible theories in literature such as the No Free Lunch (NFL) theorem answers the question “why we need more algorithms despite the many algorithms proposed so far?” [57]. The answer to this question based on the NFL theorem logically has proven that there is no optimization technique for solving all optimization problems. In optimization problems, the main goal is to search a space to find an optimal or near optimal solution.
The GOA optimizer is a population-based meta-heuristic inspired from the grasshopper insects in nature. Grasshopper form a large swarm that migrate over large distances. The main characteristics of grasshopper is that they tend to move slowly and in small steps in the larval phase. In adulthood, they move in a long range and abrupt movement. Repulsion forces allow grasshoppers to explore the search space, whereas attraction forces encourage them to exploit promising regions. This movement of grasshopper agents over a maximum iteration is modeled by (4) [32].
where UB and LB are the lower and upper bounds of the search space dimensions, d = 1,2,...,D. p is an individual in the population. Parameter c is the coefficient that reduces the comfort zone for p in the search space and is decreased according to (5).
where L is the maximum number of iterations. The attraction forces between individuals in the swarm is modeled using function s in (6).
with l = 1.5 and f = 0.5 as specified in the original paper of GOA.
4 Experimental results and analysis
This section presents the experiments conducted to evaluate the proposed dental radiography image segmentation technique. The following sections present the performance metrics, the quantitative and the qualitative results obtained from the conducted experiments, and a comparison with other Dental Radiography techniques.
4.1 Performance measures
In this section, we present the performance metrics used to evaluate our proposed approach. These metrics are well-known measures used to evaluate clustering approaches.
-
1.
Entropy
Entropy is a quality measure used to measure the extent to which cluster labels match the ground truth. The lower entropy value means better clustering. When the objects in the cluster are more diverse, the entropy value grows. Entropy is found by (7) [2].
$$ Entropy=\sum\limits_{j=1}^{k}\frac{(|C_{j}|)}{n}E(C_{j}) $$(7)where Cj contains all data instances assigned to cluster j, n is the number of instances in the dataset, k is the number of clusters, and E(Cj) is the individual entropy of a cluster. Individual cluster entropy is calculated by (8)
$$ E(C_{j})=- \frac{1}{log q} \sum\limits_{i=1}^{q} \frac{|C_{j} \cap L_{i}|}{C_{j}} log(\frac{|C_{j} \cap L_{i}|}{C_{j}}) $$(8)where Li denotes the ground truth assignments of data instances in cluster i, and q represents the number of actual clusters in the dataset.
-
2.
Purity The purity metric measures to what extent a cluster contains a single class. Basically, it reflects the quality of clustering. The purity is computed using (9) for each individual cluster [2].
$$ Purity = \frac{1}{n} \sum\limits_{j=1}^{k} max_{i}(|L_{i} \cap C_{j}|) $$(9)
4.2 Quantitative analysis
The characteristics of the three aforementioned datasets, and the results obtained from running the proposed algorithm, are displayed in Table 2. The number of points and the number of teeth can be recognized for each dataset. The number of points represents the number of pixels for each teeth image. The results are evaluated using the purity and entropy clustering evaluation measures. Higher purity and lower entropy values are considered of a better segmentation quality. The proposed technique is evaluated against common clustering algorithms including K-means, X-means, EM, and FF.
Table 2 shows that the proposed technique outperforms all the other algorithms for all the three datasets having the highest values for purity and the lowest values for entropy. On the other hand, X-means is the worst algorithm for all the datasets having the lowest values for purity and the highest values for entropy. In addition, k-means has the nearest values for both purity and entropy to the proposed algorithm for all the datasets.
The complexity analysis of the proposed technique can be analyzed as follows:
-
Level-1 EM for segmenting each tooth can be considered as O(nmi) [42] where i is the number of iterations, n is the number of points in the image, and m is the unknown elements of the discrete source distribution with f1,,ft,fm, where ft is the expected number of photons emitted from source bin t per unit time.
-
Level-2 EM-GOA considers applying EM algorithm for each grasshopper at each iteration for a single tooth. This requires O(cgknmi) for each segmented tooth, which are generated from Level-1 EM, where k is the number of clusters, g is the number of generations/iterations, c is the number of grasshoppers at each iteration, nmi reflects the EM complexity.
In comparison with the other algorithms, EM requires O(nmi) [42] as indicated previously, FF requires O(nk) [53], k-means requires O(nkdi) [17, 33], and x-means requires O(dinlogk) [20] where d is the dimension value. The additional complexity of the EM-GOA is due to the additional computation of the GOA which are added to the EM algorithm to further optimize the results, which are recognized in Table 2 as discussed previously.
4.3 Qualitative analysis
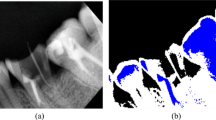
This section presents a visual representation of the segmented images as shown in Figs. 3, 4, and 5. The proposed EM-GOA detects most of the teeth boundaries, enamels, dentins, crowns, and restoration.
Most of the other algorithms could not detect the teeth boundaries and considered multiple teeth as a single tooth. X-means got the worst results among the others and it could not detect the teeth boundaries nor segment the tooth correctly. The results of EM is very similar to that for the K-means. FF on the other hand has some problems in detecting the teeth boundaries for the three datasets.
4.4 Comparisons of the proposed EM-GOA technique with other dental radiography techniques
In this section, EM-GOA is compared with two other techniques found in literature that have used the same benchmark dataset experimented in this work.
The first technique was proposed by Ronneberger et al. [48]. The authors used a u-shaped convolutional neural network (u-net) to segment dental x-ray images. The second technique was proposed by Lee et al. [56]. In this work, the authors used a random forest for the automation of dental image segmentation. Table 3 presents the quantitative evaluation of the three techniques (EM-GOA, u-net, and random forest). Three evaluation measures are used to compare the techniques in the table. These measures are: precision, sensitivity and F-score.
As shown in Table 3, the average precision values of EM-GOA, u-net and random forest are 0.533, 0.437, and 0.211, respectively. EM-GOA high precision indicates positive segmentation relative to the ground truth image. The average sensitivity of EM-GOA, u-net, and random forest are 0.799, 0.554, and 0.523, respectively. A sensitivity of 0.799 for EM-GOA means that the proposed EM-GOA algorithm detected most of the teeth boundaries, enamels, dentins, crowns, and restoration positively. In terms of F-score values,the best value was achieved by EM-GOA with an average of 0.639 indicating a more robust performance against the other two techniques. Based on the results presented in Table 3, our proposed algorithm outperforms u-net and random forest.
5 Conclusions and future work
In medical research, image segmentation plays a vital role in the process of extracting and classifying features in an image. However, medical image segmentation is a challenging task due to poor image contrast, noise and artifacts. Therefore, advanced techniques are required such that they incorporate different dimensions of knowledge to image segmentation.
This paper implemented a segmentation technique using EM algorithm and GOA. The proposed technique was tested on dental radiography datasets and evaluated in terms of purity and entropy, which consists of a two-level EM process. During the first level, objects were detected whereas during the second level the objects of interest were segmented. GOA was used to optimize the search process of EM algorithm by searching for the best density value of clustering.
Based on our experimental results, the proposed EM-GOA technique was compared to some well-known clustering algorithms including K-means, EM, Farthest First, and X-means. Our proposed technique is very competitive to these well-known algorithms and was able to achieve better results in terms of purity with values of 0.6833, 0.7126, and 0.6375 for the three datasets, respectively. It was also able to achieve better results in terms of entropy with values of 0.3582, 0.3083, and 0.3810 for the same datasets, respectively. Furthermore, the proposed technique was compared with a u-shaped CNN and random forest techniques. EM-GOA outperformed the two techniques in terms of precision, sensitivity and F-score with values of 0.533, 0.799 and 0.639, respectively. These results show that the proposed EM-GOA is precise and robust in segmenting dental radiography.
For future work, EM-GOA is recommended to be experimented with other domains such as document categorization, cancerous data, and financial risk analysis. In addition, other dental radiography datasets could be gathered from dental clinics, which are not labeled and might have noise. EM-GOA could be further experimented with such datasets considering further pre-processing of data using manual labeling of points and denoising techniques.
References
Abdel-Khalek S, Ishak AB, Omer OA, Obada AS (2017) . Optik-Int J Light Electron Opt 131:414
Aljarah I, Ludwig SA (2013) In: 2013 IEEE Congress on Evolutionary Computation (CEC). IEEE, pp 2642–2649
Aljarah I, Ala’M AZ, Faris H, Hassonah MA, Mirjalili S, Saadeh H (2018) Cognitive Computation 10(3):478
Amelio A, Pizzuti C (2013) In: European Conference on the Applications of Evolutionary Computation. Springer, pp 314–323
Amer YY, Aqel MJ (2015) . Procedia Comput Sci 65:718
Bora DJ, Gupta AK (2015) arXiv:1506.01710
Carson C, Belongie S, Greenspan H, Malik J (2002) . IEEE Trans Pattern Anal Mach Intell 24(8):1026
Cordts M, Omran M, Ramos S, Rehfeld T, Enzweiler M, Benenson R, Franke U, Roth S, Schiele B (2016) Proceedings of the IEEE conference on computer vision and pattern recognition (CVPR)
De Geus D, Meletis P, Dubbelman G (2020) IEEE Robotics and Automation Letters
Dempster AP, Laird NM, Rubin DB (1977) Journal of the Royal Statistical Society, Series B (methodological) 39:1–38
Dhal KG, Das A, Ray S, Gálvez J, Das S (2019) Archives of Computational Methods in Engineering 1:1–34
Dhanachandra N, Manglem K, Chanu YJ (2015) . Procedia Comput Sci 54(2015):764
Dong-ri S, Fu-yuan G (2010) In: 2010 International Conference on Bioinformatics and Biomedical Technology (ICBBT). IEEE, pp 74–78
Farmer ME, Shugars D (2006) In: 2006. CEC 2006. IEEE Congress on Evolutionary Computation. IEEE, pp 1300–1307
Forsyth DA, Ponce J (2002) Computer vision: a modern approach. Prentice Hall Professional Technical Reference
Gao H, Chae O (2010) . Pattern Recogn 43(7):2406
Hartigan JA, Wong MA (1979) . J R Stat Soc Series C (Appl Stat) 28(1):100
He K, Gkioxari G, Dollár P, Girshick R (2017) In: Proceedings of the IEEE international conference on computer vision, pp 2961–2969
Institute AS (2018) X-rays. https://www.ada.org/en/member-center/oral-health-topics/x-rays/. [Online; accessed 27-April-2018]
Ishioka T et al (2005) In: Proceedings of International Conference on Computational Intelligence, vol 2, pp 91–95
Kaur D, Kaur Y (2014) . Int J Comput Sci Mob Comput 3(5):809
Kirillov A, He K, Girshick R, Rother C, Dollár P (2019) In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 9404–9413
Kumar R, Arthanari M, Sivakumar M (2011) . Int J Multimed Image Process 1:72
Kumar R, Arthanariee A (2013) . World Acad Sci Eng Technol Int J Math Comput Nat Phys Eng 7(3):320
Lai Y, Lin P (2008) In: International Conference on Advanced Concepts for Intelligent Vision Systems. Springer, pp 936–947
Lalaoui L, Mohamadi T (2013) International journal of advanced computer science and applications. (IJACSA) 4(6):211–219
Laporte J (2017) Oral health and dental care in the u.s. - statistics and facts. https://www.statista.com/topics/3944/oral-health-and-dental-care-in-the-us/. [Online; accessed 27-April-2018]
Li Y, Bai X, Jiao L, Xue Y (2017) . Appl Soft Comput 56:345
Lin TY, Maire M, Belongie S, Hays J, Perona P, Ramanan D, Dollár P, Zitnick CL (2014) In: European conference on computer vision. Springer, pp 740–755
Lira PHM, Giraldi GA, Neves LAP, Feijoo RA (2014) . IEEE Lat Am Trans 12(4):694
Liu J, Ku YB, Leung S (2012) . J Vis Commun Image Represent 23(8):1234
Łukasik S, Kowalski PA, Charytanowicz M, Kulczycki P (2017) In: 2017 Federated Conference on Computer Science and Information Systems (FedCSIS). IEEE, pp 71–74
Manning CD, Raghavan P, Schütze H (2008) Introduction to information retrieval. Cambridge University Press
Mesejo P, Ibánez O, Cordón O, Cagnoni S (2016) . Appl Soft Comput 44:1
Minaee S, Boykov Y, Porikli F, Plaza A, Kehtarnavaz N, Terzopoulos D (2020) arXiv:2001.05566
Mirjalili SZ, Mirjalili S, Saremi S, Faris H, Aljarah I (2018) Applied Intelligence 48(4):805
Mohsen FM, Hadhoud MM, Amin K (2011) IJACSA) International journal of advanced computer science and applications special issue on image processing and analysis
Mortaheb P, Rezaeian M, Soltanian-Zadeh H (2013) In: 2013 8th Iranian Conference on Machine Vision and Image Processing (MVIP). IEEE, pp 121–126
Naik D, Shah P (2014) . Int J Comput Sci Inform Technol 5(3):3289
Nomir O, Abdel-mottaleb M (2008) . IEEE Trans Inf Forensic Secur 3(2):223
Panda R, Agrawal S, Samantaray L, Abraham A (2017) . Appl Soft Comput 50:94
Parra L, Barrett HH (1998) . IEEE Trans Med Imaging 17(2):228
Patanachai N, Covavisaruch N, Sinthanayothin C (2010) In: 2010 8th International Conference on ICT and Knowledge Engineering. IEEE, pp 103–106
Qaddoura R, Faris H, Aljarah I (2020) . Int J Mach Learn Cybern 11 (3):675
Qaddoura R, Faris H, Aljarah I, Castillo PA (2020) In: International conference on the applications of evolutionary computation (Part of EvoStar). Springer, pp 20–36
Rad AE, Mohd Rahim MS, Rehman A, Altameem A, Saba T (2013) . IETE Tech Rev 30(3):210
Raju NG, Rao PN (2013) . Int J Eng Res Appl 3(6):1572
Ronneberger O, Fischer P, Brox T (2015) In: International Conference on Medical image computing and computer-assisted intervention. Springer, pp 234–241
Sahu M, Bhurchandi K (2016) International Journal of Computer Applications 140(5)
Sandhya G, Babu Kande G, Savithri TS (2017) BioMed research international 2017
Senthilkumaran N, Vaithegi S (2016) . Comput Sci Eng Int J 6(1):1
Shah S, Abaza A, Ross A, Ammar H (2006) In: 2006 Biometrics Symposium: Special Session on Research at the Biometric Consortium Conference. IEEE, pp 1–6
Sharma N, Bajpai A, Litoriya MR (2012) . Facilities 4(7):78
Silva G, Oliveira L, Pithon M (2018) . Expert Syst Appl 107:15
Subramanyam RB, Prasad KP, Anuradha B (2014) . Int J Eng Res Appl 4(7):173
Wang CW, Huang CT, Lee JH, Li CH, Chang SW, Siao MJ, Lai TM, Ibragimov B, Vrtovec T, Ronneberger O et al (2016) . Med Image Anal 31:63
Yang XS (2010) In: Nature inspired cooperative strategies for optimization (NICSO 2010). Springer, pp 65–74
Zheng X, Lei Q, Yao R, Gong Y, Yin Q (2018) . EURASIP J Image Video Process 2018(1):68
Zuo W (2006) . Comp Appl Softw 23(1):97
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Qaddoura, R., Manaseer, W.A., Abushariah, M.A.M. et al. Dental radiography segmentation using expectation-maximization clustering and grasshopper optimizer. Multimed Tools Appl 79, 22027–22045 (2020). https://doi.org/10.1007/s11042-020-09014-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-020-09014-1