Abstract
MRI image segmentation and classification is one of the important tasks in medical image analysis and visualization, despite occurrence of noise makes it tough to segment the region of interest. In this paper, the MRI images are pre-processed and segmentation is carried out using modified Level set method for the tumor segmentation. Also, it is important to extract the useful features to predict the image class accurately. The proposed method operates Multi-Level wavelet decomposition features and for the wavelet coefficients modified chief descriptions like Grey Level Co-Occurrence Matrix (GLCM), Gabor and moment invariant features are extracted. The classification is carried out using the Adaptive Artificial Neural Network (AANN) methodology. In the adaptive ANN, the layer neurons are optimized using Whale Optimization Algorithm (WOA). The adaptive neural network optimizes the network structure to increase the classification accuracy and thus gives better classification results of tumors based on the segmented images. The proposed method will be executed in the working platform of MATLAB and the results are compared with the previous state of the art techniques. Finally, the proposed method results in classification accuracy of 98%.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Brain tumor is an unusual and uncontrollable growth of cells in the brain. Brain Tumors are categorized into two types namely; primary or benign brain tumors and metastatic or malignant brain tumors. A primary brain tumor begins and spreads only in the brain later. Metastatic brain tumors can originate in other parts of the body as cancer and spread to the brain. Various methods are used to diagnose the brain tumor which includes the expert opinion, human inspection, biopsy, etc. Image processing techniques can be too supportive to detect brain tumors. There are many medical imaging techniques like x-ray, magnetic resonance imaging (MRI), positron emission tomography (PET), computed tomography (CT) that are accessible for tumor detection. Owing to high resolution of MRI, it is used as most frequent modality for brain tumor growth imaging and location detection. Magnetic Resonance Imaging (MRI) is an imaging technique which non-invasively provides high contrast images of different anatomical structures. It delivers high quality images of tissues comparing to other medical imaging techniques. Evaluation and analysis of MRI images by radiologists is error-prone and time consuming [14].
Segmentation of MR images of brain is a most challenging task in medical image processing as MR images are linked with the artifacts. Segmentation in image processing is a process of segregating an image into equally exclusive regions [11]. To distinguish between cancerous and noncancerous magnetic resonance images of the brain, an automated method [2] was used to find the tumor more precisely in less processing time.
Automated segmentation methods [7, 25] based on artificial intelligence systems were suggested a method to detect tumors from MR images using fuzzy clustering technique. This algorithm uses fuzzy C-means. But the main disadvantages of this algorithm is that it requires more processing time. The MR segmentation methods have been quite effective and is in the developmental stages for pathological tissues around success verified for specific disease monitoring [1, 13, 30].
Even if there are number of general segmentation methods such as thresholding, region growing [24] and clustering [10], they are not merely relevant to the domain of brain tumor identification. This is for intensity similarities between brain tumors and certain normal tissues which can provoke misunderstanding in the segmentation algorithm.
Brain Tumor Segmentation Using Genetic Algorithm and Artificial Neural Network Fuzzy Inference System (ANFIS) [17, 25, 27] improves segmentation accuracy. Only a subset of features are chosen using Genetic algorithm and based on these features fuzzy rules and membership functions are described for segmenting brain tumor from MRI.
Various approaches used for detection of MRI Brain Tumor have been presented in subsequent sections.
2 Related works
In recent years, various techniques have been experimented for MRI Brain Tumor segmentation and classification. The most relevant works are discussed below..
V. Anitha et al. [3] have recommended a two-tier classification system with the efficient segmentation technique which labels input MRI image as normal and abnormal image. In the beginning the self-organizing map neural network processes the features obtained from the discrete wavelet transform. The resultant filter factors are subsequently qualified by the K-nearest neighbour and the testing progression was also achieved in two stages. This two-tier classification system categorizes the brain tumors in double training process which inturn improves the desirable performance.
Yse Demirhan et al. [6] have proposed a new tissue segmentation algorithm that segments brain MR images into tumor, edema, white matter (WM), gray matter (GM) and cerebrospinal fluid (CSF). The detection of the healthy tissues was functioned concurrently with the diseased tissues as investigated for the transformation triggered by the spread of tumor and edema on healthy tissues was very significant for treatment planning. They used T1, T2 and FLAIR MR images of 20 subjects suffering from glial tumor. They used an algorithm for stripping the skull before the segmentation process. The segmentation was carried out using self-organizing map (SOM) in conjunction with unsupervised learning algorithm and fine-tuned with learning vector quantization (LVQ).
Yudong Zhang et al. [29] have proposed a pathological brain detection system (PBDS) to assist neuro-radiologists to interpret magnetic resonance (MR) brain images. Initially, 12 fractional Fourier entropy (FRFE) features were obtained from each brain images. Next, they fed those features to a multi-layer perceptron (MLP) classifier. Two improvements were proposed for MLP. First improvement was the pruning technique which controls the optimal hidden neuron number. They compared three pruning techniques: dynamic pruning (DP), Bayesian detection boundaries (BDB), and Kappa coefficient (KC). The other improvement was to use the adaptive real-coded biogeography-based optimization (ARCBBO) to train the biases and weights of MLP.
Sergio Pereira et al. [15] have presented an automatic segmentation method based on Convolutional Neural Networks (CNN), exploring small kernels. The use of small kernels permits planning a deeper architecture, also having a positive effect in contradiction of over fitting agreed the rarer number of weights in the network. They have also investigated the use of intensity normalization as a pre-processing step which is not common in CNN-based segmentation methods, composed with data augmentation to be very active for brain tumor segmentation in MRI images.
Gelan Yang et al. [28] have proposed a method for automatic classification of MR Images as normal or abnormal using Wavelet-Energy, Support Vector Machine (SVM) and biogeography-based optimization (BBO). SVM was used as the classifier, and BBO was used to optimize the weights of the SVM. This automated CAD system could be used for classification of image with separate pathological condition, types and disease status. The effect of BBO-KSVM was higher compared to BP-NN, KSVM and PSO KSVM in terms of precision.
Suresh et al. [20,21,22,23] proposed the data analytics in cloud environment and distributed networks also discussed the human health care applications. Mobile data using with security proposed in [5]. Big data analytics used in vector machine [9]. Secure and optimal authentication framework in social network proposed in [4, 16].
Nidhi Gupta et al. [8] have proposed a non-invasive and adaptive method for detection of tumor from T2-weighted brain magnetic resonance (MR) images. Non-homogeneous brain MR images were improved by preprocessing and segmented across multilevel customization of Otsu’s thresholding technique. Some textural and shape features are mined from the segmented image and two prominent features are selected through entropy measure. Support vector machine (SVM) categorizes MR images using important features.
Nooshin Nabizadeh et al. [12] have proposed an integrated automated framework to detect the tumors in MR images and then the same framework was executed on T1-w and FLAIR sequences (separately). The notable accuracy of the algorithm in tumor segmentation in recital with its low computational complexity validates the efficiency of this method. Additional key benefit was its uniqueness from atlas registration, prior anatomical knowledge, or bias corrections that confine the overall application of various state-of-the-art methods. Added use of the this technique was in the use of single-spectral MRI. Although using multi-spectral MR images handle the intensity relationships between tumor and healthy tissues in various practical clinical situations.
3 Proposed MRI BRAIN tumor detection approach
The brain tumor detection and classification from MRI is decisive to reduce the rate of fatalities. Brain tumor is tough to cure as the brain has a very complex structure and the tissues are interrelated with each other in an intricate way. Even with numerous present methodologies, robust and efficient segmentation of brain tumor is even a central and challenging task. Tumor segmentation and classification is a challenging task because tumors differ in shape, appearance and location. Likewise, the brain tumor segmentation and detailed classification based on MRI images has established substantial interest over last decades. MRI requires the facility to take multiple images known as multimodality images which can deliver the detailed structure of brain to accurately categorize the brain tumor. Then, it is hard to fully segment and classify brain tumor from mono-modality scans despite its complex structure. Henceforth, to increase the accuracy of tumor segmentation and classification, multi-modality MRI images can be used in future..
Hence, in this document, we have presented the efficient way for the segmentation and classification of brain MRI’s using Cognition based Modified Level Set Segmentation method [26] and Adaptive ANN classification method. Initially few pre-processing steps are applied before the segmentation step in order to remove the noise and segmentation was carried out using cognition based modified level set algorithm. Finally to classify the segmented images into normal, benign and malignant brain MRI’s, we have extracted important image features based on Multilevel wavelet information. The original feature extraction method runs with the extraction of three level DWT features for the MRI images. Afterwards, for the wavelet coefficients collected from three levels of decomposition, important features extraction methods like GLCM [18, 19], Gabor and moment invariant feature extraction procedures are functional to extract the proper features. Finally, the classification is carried out by adaptive ANN classification method. In the adaptive ANN, the layer neurons are optimized using Whale optimization approach. The adaptive neural network optimizes the network structure in order to expand the classification implementation.
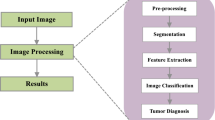
Furthermore, the Schematic Diagram of the proposed method for MRI brain tumor segmentation and classification is shown in the below Fig. 1.
Schematic Diagram of the proposed method for MRI brain tumor segmentation and classification is shown in the below Fig. 1
3.1 Outline for proposed MRI brain tumor detection approach
The stages involved in the proposed MRI Brain Tumor Detection Approach is specified as,
MRI Image Pre-Processing
Brain Tumor Segmentation by Cognition based Modified Level Set Segmentation method
Feature Extraction Using Multilevel wavelet decomposition
Brain MRI classification using Adaptive Artificial Neural Network
Discussion of proposed method is presented in detail in the subsequent sections.
3.1.1 MRI image pre-processing
Pre-Processing is the basic step of every image processing techniques. The segmentation of MRI is a tough job in medical image analysis and visualization due to occurrence of noise, with this noise it is hard to segment the region of interest. The proposed method works with three pre-processing steps: RGB to Gray conversion, Histogram equalization and Median Filtering. The conversion from RGB to Gray is carried out to reduce the computation complexity. The Histogram equalization operation is applied in order to increase the contrast of the input MRI image. The Median filtering is used to reduce the unwanted noise by image smoothing while preserving the edges. Discussion of individual preprocessing techniques are presented below.
- (i)
RGB to Grey conversion
The simplest technique to convert a RGB image to gray scale is done by taking the average of the three pixel values, given as,
- (ii)
Histogram Equalization method
The contrast of the image is enhanced with the help of Histogram equalization technique. When an image is represented by close contrast values, such as images in which both the background and foreground are bright at the same time, or else both are dark at the same time, this method is useful. This operation spreads out intensity values along with the total range of values in order to achieve higher contrast.
Histogram equalization defines a mapping of gray levels ‘x’ into gray levels ‘b’ such that the distribution of gray level ‘a’ is uniform. This mapping stretches contrast (expands the range of gray levels) for gray levels near the histogram maxima. Since contrast is expanded for most of the image pixels, the transformation improves the detectability of many image features.
The probability density function of a pixel with intensity level ‘Pi’ can be given by:
Where, 0 ≤ Pi ≤ 1, i = 0, 1, …, 255; fi is the number of pixels with intensity level ‘pi’ and f denotes the total number of pixels.
Then, the histogram can be derived by plotting pdfP(Pi) against pi. Now a new intensity level gi is calculated as
This results in expanding the contrast locally, and changing the intensity of each pixel according to its local neighborhood. In general, the image has high contrast when the image has a large difference in intensity between the highest and lowest intensity levels. Hence, by equalizing the Histogram of image, the areas of lower local contrast could gain a higher contrast without affecting the global contrast. After, enhancing the image contrast, the images are subjected for Median filtering process.
- (iii)
Median Filtering
Median filtering works by replacing the center pixel with the median of gray levels in a window which slides pixel by pixel over the entire image. Let us consider a filter with (y × y|y = 3, 5, ..), i.e. square windows of odd size as it is the most widely used form of median filter. For a given (P × Q) image, I(l, m) with (l, m) ∈ {1, 2, . ., P} × {1, 2, . ., Q}, a 2-Dimensional (y × y) median filter can be defined as:
Where, \( t,v\in \left(-\left(\frac{y-1}{2}\right),\dots, \left(\frac{y-1}{2}\right)\right) \) and y(i, j) is the pixel value at point (i, j).
An important feature of median filtering technique is that it is known for smoothing the images while preserving the edges. Therefore, the median filter is frequently used as a denoising filter. After pre-processing by Histogram Equalization and Median filtering approaches, the images were subjected to segmentation process.
3.1.2 Brain tumor segmentation by Cognition based modified level set segmentation method
After pre-processing the brain MRI’s, the proposed model uses cognition based modified Level Set segmentation method for the segmentation of tumor. The major significance of the segmentation is to isolate the region of interest from its background. The level set methods have been broadly used for spotting image limitations. The elementary knowledge of the level set method is that the initial contour is denoted by the zero-level set of a higher dimensional function called as level set function. The motion of the contour is expressed based on the development of the level set function.
The steps involved in the proposed modified Level Set segmentation algorithm are as follows:
- 1)
In the beginning, the image entropy is designed to locate how much data is delivered by the image and then entropy is calculated by:
$$ E=-\sum \limits_j{P}_j\left({\log}_2{P}_j\right) $$(5)Where Pj is the probability of the difference between the adjacent pixels.
- 2)
Next, the PMD (persona Malik diffusion) filter is used in place of Gaussian filtering for smoothing the images and is denoted as:
$$ {\displaystyle \begin{array}{c}\frac{\partial L}{\partial x}=\mathit{\operatorname{div}}\left(d\left(m,n,t\right)\nabla L\right)\\ {}=\nabla d.\nabla L+d\left(m,n,x\right)\nabla L\end{array}} $$(6)Where ∆ denotes laplacian, ∇ denotes gradient, div (……) denotes divergence operator and d (m, n, x) is the diffusion coefficient which controls the rate of diffusion to preserve edges in the image.
- 3)
After smoothing, the image gradient magnitude is calculated by:
$$ {G}_{norm}=\frac{\left|\nabla A\left(m,n\right)\right|-\min \left(\left|\nabla A\left(m,n\right)\right|\right)}{\max \left|\nabla A\left(m,n\right)\right|-\min \left|\nabla A\left(m,n\right)\right|} $$(7) - 4)
Once again, a modified speed function is calculated based on the exponent of a negative product of a parameter monitoring the motion of contour and the image gradient magnitude. It is denoted as:
where s is a parameter for controlling the motion of contour.
- 5.
When computing the speed function, the histogram of each pixel locations in every image is processed. For all image, the peak intensity pixel is agreed as the initial contour.
- 6.
In the end, the new contour is located from the contour evolution.
- 7.
The conclusion of new contour is repetitive till it converges or check-out the maximum iteration is acquired.
In the output we get two images; one covers the tumor part and the other covers the remaining portion. But the segmentation of tumor part is ended in unverified way but founded on the pixel intensity range (i.e., the segmented portion will be found for tumor other than non-tumor MRI’s also). Hereafter, for predicting the normal, benign and malignant class of brain MRI’s, the proposed method uses an adaptive ANN classification technique. For the AANN to predict the MRI image classes, certain relevant features were to be extracted.
3.1.3 Feature extraction using multilevel wavelet decomposition
Multilevel wavelet decomposition gives the information about frequency components of the image and improves the information about the image for further processing. Mainly for fully smooth images the subbands of the first level of decomposition doesn’t encompass edges at all. Likewise, for noisy image, the first level subbands contain edges but it is quite tough to identify them regardless of noise, though the succeeding levels will comprise more and more valuable data and less noise. Mostly, a number of levels of decomposition allows evaluating and practice image features of different scales which can be significant in certain applications. Thus, three level DWT is useful in the implementation of feature extraction stage.
The DWT divides the image segments into four non-covering sub-groups. Each sub-group containing a portion of the original data about the image. The conversion profits the approximation subband (LL), the horizontal detail subband (LH), the vertical detail subband (HL) and the diagonal detail subband (HH). Therefore, the LL indicates the low-frequency components composed with the horizontal and vertical directions, the LH implies the low-frequency components in the horizontal direction and high-frequency components in the vertical direction, the HL means the high-frequency components in the horizontal direction and low-frequency components in the vertical direction and the HH high-frequency components in both directions. Through next levels of decomposition, the LL sub-band will be seperated.
In the proposed work the three levels of DWT features are extracted. In the first level of decomposition, the image component is seperated to high and low frequency sub-bands (i.e. LL1, LH1, HL1and HH1). Then the LL1 is decomposed to an extra set of high and low frequency sub-bands (i.e. LL2, LH2, HL2and HH2). Again the LL2 sub-band is decomposed to alternative set of high and low frequency sub-bands (i.e. LL3, LH3, HL3and HH3).
At this point, the wavelet coefficient values achieved for the LH, HL and HH sub-bands through each level of decomposition, the GLCM, Gabor and Moment invariant features extracted.
- I.
Gray-level co-occurrence matrix Feature
The GLCM (Gray-level co-occurrence matrix) is a method of statistical image analysis which is used to estimate image properties related to second-order statistics. GLCM reflects the relation among two neighboring pixels in one offset as the second order texture where the first pixel is called reference and the second pixel as the neighbor pixel. GLCM with two position pixels of joint probability density to describe not only reveals the distribution characteristics of the brightness also replicates with similar brightness or near to the brightness of the distribution characteristics amongst pixel positions is a type of actual data tools of second order image texture statistics. GLCM is well-defined as: the element point, (m, n) in GLCM signifies the frequency of two pixels in an accurate window with one pixel grayscale value is m, other pixel grayscale value is n and the adjacent distance to ‘d’ in the ‘θ’ direction. Generally, ‘d’ takes 1 or 2, and θ takes the four directions value of 0 °, 45 °, 90 ° and 135°. In this work the offset ‘d’ is preferred as 2. Hence, two sets of GLCM matrix values will be originated. For all GLCM matrix, 22 possible characteristic features can be collected. So, 44 features are extracted.
All element values of GLCM are determined as follows:
In the equation (9), q(m, n, d, θ) is the frequency of the double element point, one of which the pixel grayscale value is m, other pixel grayscale value of n and the adjacent distance to ‘d’ in the ‘θ’ direction.
Characteristic parameter of GLCM
GLCM deceptively reflects the image gray information such as level rank, directions step and change of range, to more directly designate the texture status based on GLCM. The subsequent four parameters are used for quantitative description of texture condition based on GLCM.
Entropy
Image entropy is a main index of calculating the high degree of the image information and the size of the entropy exemplifies the average image information. It holds non-uniformity or complexity of the textures in the image. If the image does not have any texture, the GLCM nearly convert to zero matrix. To be exact, more multifaceted the texture is greater the entropy grows.
Second Moment
The second moment is variance and the central statistics characterization parameter of the partial image texture. It centers on the partial characteristics of the image.
Contrast
Contrast gradient is also called the principal diagonal moment of inertia and it shows the change in total quantity of partial Gray gradation in the image. Contrast exposes the clarity of the image and the depth of the furrow wrinkles texture. It can ideally detect images contrast and extract edge information of the objects. Contrast is higher, the result is stronger.
Correlation
Correlation is the gray-scale measure of the linear relationship and it explains the relationship among the elements of Columns or rows in GLCM.
Where, λmλn, μm, μn are calculated from the following equations.
- II.
Gabor Feature Extraction
Gabor filters have been used widely in image processing, texture analysis for their excellent properties: optimal joint spatial\spatial-frequency localization and capability to pretend the receptive fields of simple cells in the visual cortex. Gabor filter is a linear filter used for edge detection termed later as Dennis Gabor. They are instigated to render distortion tolerance space for near with optimal resolution in spatial and frequency domain. The 2D Gabor filter can be denoted as a complex sinusoidal signal controlled by a Gaussian kernel function as given in eq. 10.
βy and βz are the standard deviations of the Gaussian envelope along y and z dimensions, c is the central frequency of the sinusoidal plane wave and orientation θ. The rotation of the y-z plane by an angle θ will answer in a Gabor filter at the orientation θ. The angle θis defined by
q € n where q implies the number of orientation. Let c(y,z) be the intensity at the coordinate (y, z) in a gray scale face image, its density with a Gabor filter that extract feature accuracy is defined as
The Gabor feature is found at θ equivalent to 300. At 300orientation, the mean and variance are measured for the Gabor feature matrix. Therefore, the Gabor feature extraction method conclusions with two features (i.e., mean, variance) for every image.
- III.
Moment Invariant Features:
The moment invariant features were presented by Hu, based on the normalized central moments. Two-dimensional (u, v)th order moment is described as follows
(Where,) u, v = 0, 1, 2, …
Also, the two-dimensional (u, v)th order central moment from which the invariant features are reached is defined as follows
Where, \( \left(\overline{i},\overline{j}\right) \) is the image centroid pixel point.
The image centroids are calculated as follows,
It is visited that the eq. (23) and (24) equivalent to each other as the center of moment, Iuv is equal to the image centroids.
The scale invariance is achieved by normalization and the normalized central moments can be defined as follows,
(Where,) u + v = 2, 3, 4…
The moment invariant features are as,
The seven moment invariant features are extracted for the coefficients gained from the wavelet decomposition. So, 477 features (i.e. (9 wavelet coefficient matrices*(44 GLCM+2Gabor + 7Moment invariant))) are acquired for every image. The features extracted are fed as input to the AANN.
3.1.4 Brain MRI classification using artificial neural network
Artificial neural system (ANN) is an artificial intelligence technique that resembles the operation of human brain which can reflect the basic linear or non-linear connections between input and target information. Locating knowledge from an outside source, ANN supplies data to its inner processing units and transmits them by approaches for the communication of the transfer functions and the association parameters among adjacent layers.
Artificial neural network (ANN) normally consist of many layers, i.e. a collection of three layers’ network includes of huge number of neurons that performs the task of processing input information into required output. A Multi Layer Perceptron (MLP) feed-forward network is generally utilized which contains three layers to be precise of input and output layer with one hidden layer. These layers attach with specific number of neurons. The neurons in all layer are linked to various synaptic weights. The input parameters are agreed to pass through layers and number of neurons in the layer associates with input parameters.
The data from input layer (i.e. the extracted brain image features) is passed to hidden layer. The number of neurons in input layer will be equal to number of features extracted from the input image. The output neurons are predetermined. Now, the output must synchronize the objective (1 = malignant, 2 = benign and 3 = Normal). The quantity of hidden layers’ neurons is randomly varied till the error reaches to least value. Henceforth, to progress the training performance, we have included the Whale optimization algorithm for controlling the optimal number of hidden nodes for the artificial neural network. Likewise, the Flowchart for proposed Adaptive ANN is given in below 2(a) and 2(b).
In the above figure, {x1, x2, …xp} indicates the input features assigned to each node of the input layer and {y1, y2, …yq} denotes the hidden nodes.
The benefit of proposed technique is that it is not roughly determining number of hidden nodes but based on accuracy of outcomes. In the proposed method, number of hidden nodes are not predefined but determined in the training time. The training algorithm is utilized as a part of the proposed brain tumor diagnosis framework also based on the Back propagation learning algorithm where the weights of the neurons are modified based on the network output error.
- a)
Whale optimization approach
Recently a new optimization algorithm called whale optimization algorithm (Mirjalili 2016) has been familiarized to metaheuristic algorithm by Mirjalili and lewis. The whales are reflected to be as highly intelligent animals with motion. The WOA is inspired by the distinctive hunting behavior of humpback whales. Typically, the humpback whales prefer to hunt krills or small fishes which are near to the surface of sea. Humpback whales use a unique hunting method called bubble net feeding method. In this method they swim around the prey and create distinctive bubbles along a circle or 9-shaped path.
The mathematical model of WOA is explained in the following phases
- 1.
Encircling prey.
- 2.
Bubble net hunting method.
- 3.
Search the prey.
The steps involved in the proposed Whale optimization algorithm for creating the optimal network structure are given as follows,
- Step 1:
Initialization
The algorithm is formed by randomly producing the solution (i.e. the network structure) that interconnects to the result. At this point the network structure covering the number of hidden layers and its consistent nodes are signified by the random value in the search space is given as:
Where, N signifies the Initial population of the fish at ‘P’ dimension (i.e, the random network structure with random number of hidden layers along with its neurons). In the above equation, n1 denotes the number of layers required and (n2, n3, …nx) represents the number of neurons equivalent to each hidden layer. The dimension of the solution differs consistently with the selected number of hidden layers. For example, if the number of hidden layer is three then the dimension of solution will be four. And too initialize the coefficient vectors of whale such as, \( \overrightarrow{d},\overrightarrow{D}, and\overrightarrow{E} \).
- Step 2:
Fitness Calculation
Assess the fitness of the input solution on the base of the eq. (34). To get the best network structure, the fitness value of the solution is computed. It is given as,
In above equation, the classification accuracy represents how correct the network predicted classes are (i.e. the minimum value of the network error). Later, the initial solution with maximum accuracy is selected.
- Step 3:
Update the position of current solution
Encircling prey
The position of prey is accepted by humpback whale and then it encircles the prey. For the best search operator, the other search operators will subsequently try to update their positions when the best search agent is categorized. The updated method is assessed by the below equations:
Where ‘i’ shows a current iteration, \( \overrightarrow{D} \) and \( \overrightarrow{E} \) specifies a Coefficient vector, \( {\overrightarrow{N}}^{best} \) directs a position vector for best solution, \( \overrightarrow{N} \) represents a current position Vector and || denotes an absolute value.
The vectors \( \overrightarrow{D} \) and \( \overrightarrow{E} \) are considered as follows:
Where, \( \overrightarrow{d} \) is linearly reduced from 2 to 0 through the course of iterations (in both exploration and exploitation phases), \( \overrightarrow{j}\in \left(0,1\right) \).
Bubble-net attacking method (exploitation phase)
To model the bubble-net behavior of humpback whales mathematically two methodologies are upgraded as follows:
- 1.
Shrinking encircling mechanism
- 2.
Spiral updating position
Shrinking encircling mechanism
The value of \( \overrightarrow{d} \) in the eq. (37) is reduced to realize this behavior. Note that \( \overrightarrow{d} \) is used to reduce the variation range of, \( \overrightarrow{D} \). In other words, where \( \overrightarrow{d} \) is shortened from 2 to 0. The new position of a search agent can be defined wherever by setting the random value, \( \overrightarrow{D} \) from [−1,1].
Spiral updating position
A spiral equation among the position of whale and prey is formed to imitate the helix-shaped movement of humpback whales is as follows:
Where \( {S}_{dist}=\left|{\overrightarrow{N}}_q^{best}(i)-\overrightarrow{N}(i)\right| \) and designates the distance of the qth whale (which is the best solution attained so far) to the prey, m is the random value from, [−1,1], y signifies the shape of the logarithmic spiral and it is a constant value. During optimization the position of whales is updated by supposing a probability of 50% by taking either the spiral model or shrinking encircling mechanism to model this simultaneous behavior. The mathematical model is given by eq. (40).
Where, J ∈ [0, 1]. In addition to bubble-net method, The humpback whales search for prey arbitrarily to procedure bubble net.
Search for prey (exploration phase)
To search for prey in exploration phase, the equivalent search approach used in the exploitation phase based on the variation of the \( \overrightarrow{D} \) vector can be used. Actually, along with the position of each other humpback whales search indiscriminately. So, to force search agent to move so far from a reference whale we use \( \overrightarrow{D} \) with the random values greater than 1 or less than −1. The position of the search agent is restructured in exploitation phase. To make a global search, this mechanism and \( \left|\overrightarrow{D}\right|>1 \) emphasize exploration agree the WOA algorithm. The mathematical model is given below:
Where, \( {\overrightarrow{N}}_{rand}(i) \) is a current population random position vector. Search agents renew their positions at each iteration with reference to either the best solution obtained or randomly preferred search agent. So as to deliver exploration and exploitation the parameter ‘d’ is decreased from 2 to 0,consistently. For updating, a random search agent is selected when \( \left|\overrightarrow{D}\right|>1 \), whereas the best solution is preferred when \( \left|\overrightarrow{D}\right|<1 \) for the position of the search agents. Contingent on the value of ‘J’, WOA is capable to switch between either a circular or spiral movement.
Throughout every solution updating, the fitness evaluation can be completed to locate the best solution among them. Again, at each time of making new network structure, the newer weights are allotted based on the back propagation error processed from the back propagation training algorithm.
- Step 4:
Termination criteria
The WOA algorithm is ended whilst best network structure is acquired for the approval of a termination criterion.
Using the optimal network originated, the input data are qualified. When the training procedure is fixed, the proposed Adaptive ANN can be simplified to predict the classes of the input brain MRI based on its typical image features.
4 Result and discussion
In this section, the results of proposed method for brain tumor classification is discussed which includes preprocessing using histogram equalization and median filtering techniques, segmentation using cognition based modified level set method, results of various feature extraction methods and finally classification using adaptive ANN is presented. The proposed algorithm is implemented in the MATLAB platform and configuration of the system includes 4gb RAM and Intel i3 processor clocked at 2.10 giga Hz is choosen for experimentation.
The pre-processing steps are carried out to eliminate noise before segmentation, then separation of ROI is done by segmentation, finally to predict the segmented images to normal, benign and malignant class of brain MRI’s, we have extracted relevant image feature established on Multi-Level wavelet decomposition in order to diagnosis the brain tumor efficiently.
Here, the network structure obtained for default ANN is illustrated below (Figs. 3),
The above neural network structure in Figure 3 determines the Default ANN method, the default arrangement for ANN network consist of 477 neurons in the input layer which is equal to the number of features extracted from each input image and one neuron in the output layer. In the proposed classification approach the layer neurons and hidden layers are maximized by the application of Whale optimization Algorithm.
From the maximization of layer neurons, the above neural network structure in Figure 4 determines the WOA based ANN derived. In the proposed method the whale optimization algorithm increases the number of hidden layer to two layers. The first hidden layer consist of 24 neurons and second hidden embodies 22 neurons in it.
The accuracy of the ANN classifier can be increased by applying various optimzation algorithms available in the literature. In order to assess the performance of proposed WOA- ANN classifier, the Grey Wolf Optimizer based ANN is selected. The above figure 5 shows network structure obtained from GWO- ANN network, which consist of three hidden layers. The first hidden layer consist of 18 neurons, the second hidden layer consists of 15 neurons and third hidden layer consists of 23 neurons in it.
For analysis, the brain tumor MRI images were collected from the standard database MICCAI-BRATS 2015. The BRATS database contains four sequences of MRI images for every patient which includes FLAIR, T1, T1 with Contrast and T2 weighted images. The BRATS 2015 database consist of two types of datasets viz., Training and Testing. The Training dataset consist of 220 High Grade Glioma (HGG) images and 54 Low Grade Glioma (LGG) images. The Testing dataset consist of 110 images which includes both HGG and LGG. Figure 6 shows some of the images from BRATS 2015.
The input images are considered are of Malignant, Benign and Normal classes in the Brain MRI database. Moreover, the GUI designed for the proposed brain tumor classification method is as shown in below figure,
In the above figure ‘LGG_12.jpg’ image is given as the input. The input image gets pre-processed and classified. Then it will obtain the relevant ‘Benign’ output class established on the extracted features. In the GUI, moreover, the evaluations are found and also tabulated (Fig. 7).
Preprocessed results
The input MRI image is subjected to preprocessing in order to remove the noise which aids in proper segmentation of tumor in the next stages, the results of preprocessing is shown in (Fig. 8):
-
Segmentation results
The input image after preprocessing is then subjected to segmentation for the exact identification of the tumor region. The segmented region of the image is given below (Fig. 9):
From the above figure, (a) is the segmented tumor region of the brain, (b) is the segmented tumor region by the proposed modified level set method and (c) is the tumor segmented by ground truth and proposed method.
Feature extraction results
The segmented regions of the brain tumor images are then subjected to feature extraction. The three level wavelet decomposed intermediate images are given below (Fig. 10):
From the above figure ‘a’, ‘b’, ‘c’ and ‘d’, denotes LL1, LH1, HL1 and HH1 respectively for the first level decomposed images. The second level decomposed images LL2, LH2, HL2 and HH2 are denoted as ‘e’, ‘f’, ‘g’ and ‘h’ respectively. Then the third level decomposed images LL3, LH3, HL3 and HH3 are represented as ‘i’, ‘j’, ‘k’ and ‘l’.
Again, for the wavelet decomposed image, GLCM, Gabor and Moment invariant features were extracted, which is tabulated in the below Tables 1, 2 and 3.
4.1 Evaluation metrics
The evaluation metrics considered are Sensitivity, Specificity, Accuracy and False Acceptance Rate. The standards applied are True Positive (TP), True Negative (TN), False Positive (FP) and False Negative (FN) which are determined in below sections.
Sensitivity
The ratio of a number of true positives to the sum of true positive and false negative is named as sensitivity.
Specificity
Specificity is the ratio of a number of true negative to the sum of true negative and false positive.
Accuracy
Accuracy is computed by the results of sensitivity and specificity. It is determined as follows,
Dice
The Dice similarity coefficient is a metric to measure the similarity of two samples. It can be written as,
4.2 Performance analysis
The performance evaluation of the Sensitivity, Specificity and Accuracy values for modified level set + WOA-ANN, Grey Wolf Optimized (GWO)-ANN, ANN, Level set-ANN and Region Growing- ANN methods are calculated and are tabulated in the Table 4.
It is clear that the proposed method is more accurate than any other being techniques by testing the tables. The proposed method WOA-ANN for sensitivity, specificity, and accuracy is 0.96 and 0.98 and 0.98% respectively from Table 4. Likewise, the sensitivity, specificity and accuracy values for existing methods like GWO-ANN, ANN, Level set-ANN and Region Growing- ANN are also tabulated (Table 5).
The dice coefficient and area under curve values for the existing and the proposed methods from the above table clearly shows that the proposed method is better in classification as it shows 94% accurate results as compared with other methods.
The comparison plots for Sensitivity, Specificity, Accuracy, Dice and Area under Curve values are demonstrated as below (Figs. 11, 12, 13 and 14):
The sensitivity, specificity and accuracy results for WOA-ANN, GWO-ANN, ANN, Levelset- ANN and Region Growing- ANN methods are established in comparison result. Moreover, the sensitivity and specificity values are more reliable for the proposed WOA-ANN technique when compared with other techniques.
5 Conclusion
An efficient approach for automatic segmentation and classification the brain MRI images by applying Modified Level Set approach and Adaptive ANN is presented in this paper. In the hidden layer, the proposed Adaptive ANN applies Whale optimization algorithm for determining the optimal layer neuron sets. Moreover, for predicting the image classes accurately, it applies the Multi level wavelet based features. The performance of the proposed AANN is compared with GWO-ANN and ANN methods in terms of sensitivity, specificity and accuracy measures. The result outcome demonstrates that around 98% accuracy is achieved for the proposed method while it is 92% for GWO-ANN and 68% for default ANN; which establishes that the proposed classification approach performs well when compared to the other methods. In future, the proposed method can be enhanced to classify the tumors into edema, non-enhancing solid core, necrotic/cystic core, enhancing core classes.
Change history
03 July 2018
In the original publication, equation 46 was incorrect. The original article has been corrected.
References
Ahmadvand A, Daliri MR, Zahiri SM (2017) Segmentation of brain MR images using a proper combination of DCS based method with MRF. Multimed Tools Appl, Springer
Amin J, Sharif M, Yasmin M, Fernandes SL (2017) A distinctive approach to brain tumor detection and classification using MRI. Pattern Recogn Lett
Anitha V, Murugavalli S (2016) Brain tumour classification using two-tier classifier with adaptive segmentation technique. IET Comput Vision 10(Issue: 1):2
Arularasan AN, Suresh A, Seerangan K (2018) Identification and classification of best spreader in the domain of interest over the social networks. Cluster Comput. https://doi.org/10.1007/s10586-018-2616-y
Chinnasamy A, Sivakumar B, Selvakumari P, Suresh A (2018) Minimum connected dominating set based RSU allocation for smartCloud vehicles in VANET. Cluster Comput. https://doi.org/10.1007/s10586-018-1760-8
Demirhan A, Toru M, Guler I (2015) Segmentation of tumor and edema along with healthy tissues of brain using wavelets and neural networks. IEEE J Biomed Health Info 19:4
Fletcher-Heath LM, Hall LO, Goldgof DB, Reed Murtagh F (2001) Automatic segmentation of non-enhancing brain tumors in magnetic resonance images. Artif Intell Med 21(1):43–63
Gupta N, Khanna P (2017) A non-invasive and adaptive CAD system to detect brain tumor from T2-weighted MRIs using customized Otsu’s thresholding with prominent features and supervised learning. Signal Process Image Commun 59:18–26
Kannan N, Sivasubramanian S, Kaliappan VS, Suresh A (2018) Predictive big data analytic on demonetization data using support vector machine. Cluster Comput. https://doi.org/10.1007/s10586-018-2384-8
Liew AWC, Yan H (2003) An adaptive spatial fuzzy clustering algorithm for 3-D MR image segmentation. IEEE Trans Med Imag 22:1063–1075
Meshram VA (2016) Automatic segmentation of brain MRI images and tumor detection using morphological techniques. Int Conf Elect, Electron, Commun, Comput Optim Techn: 6–11
Nabizadeh N, Kubat M (2015) Brain tumors detection and segmentation in MR images: Gabor wavelet vs. statistical features. Comput Elect Eng 45:286–301
Nayak DR, Dash R, Majhi B (2016) Pathological brain detection using curvelet features and least squares SVM. Multimed Tools Appl, Springer
Patil PG, Karande KJ (2016) Brain tumor detection techniques: a survey. Int Res J Eng Technol (IRJET) 03(Issue):10
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5)
Selvarani P, Suresh A, Malarvizhi N (2018) Secure and optimal authentication framework for cloud management using HGAPSO algorithm. Cluster Comput. https://doi.org/10.1007/s10586-018-2609-x
Sharma M, Mukharjee S (2013) Brain tumor segmentation using genetic algorithm and artificial neural network fuzzy inference system (ANFIS). Adv Comput Info Technol: 329–339
Suresh A, Shunmuganathan KL (2012) Feature fusion technique for colour texture classification system based on gray level co-occurrence matrix. J Comput Sci, ISSN 1553–3468 @ Sci Publ. Vol. 8, No.12, December 2012, pp. 2106–2111
Suresh A, Shunmuganathan KL (2012) Image texture classification using gray level co-occurrence matrix based statistical features. Eur J Sci Res, ISSN 1450-216X Vol. 75 No.4 (2012), April 2012. pp. 591–597
Suresh A, Varatharajan R (2017) Competent resource provisioning and distribution techniques for cloud computing environment. Cluster Comput. https://doi.org/10.1007/s10586-017-1293-6
Suresh A, Varatharajan R (2018) Recognition of pivotal instances from uneven set boundary during classification. Multimed Tools Appl. https://doi.org/10.1007/s11042-018-5905-9
Suresh A, Reyana A, Varatharajan R (2018) CEMulti-core architecture for optimization of energy over heterogeneous environment with high performance smart sensor devices. Wireless Pers Commun. https://doi.org/10.1007/s11277-018-5504-0
Suresh A, Kumar R, Varatharajan R (2018) Health care data analysis using evolutionary algorithm. J Supercomput. https://doi.org/10.1007/s11227-018-2302-0
Tang H, Wu E, Ma Q, Gallagher D, Perera G, Zhuang T (2000) MRI brain image segmentation by multi-resolution edge detection and region selection. Comput Med Imag Graph 24:349–357
Vasuda P, Satheesh S (2010) Improved fuzzy C-means algorithm for MR brain image segmentation. Int J Comput Sci Eng 02(issue 05):1713–1715
Virupakshappa, Amarapur B (2018) Cognition based MRI brain tumor segmentation technique using modified level set method. Cogn Technol Work, Springer
Yang G, Zhang Y, Yang J, Ji G, Dong Z, Wang S, Feng C, Wang Q (2015) Automated classification of brain images using wavelet-energy and biogeography-based optimization. Multimed Tools Appl, Springer
Yang G, Zhang Y, Yang J, Ji G, Dong Z, Wang S, Feng C, Wang Q (2016) Automated classification of brain images usingwavelet-energy and biogeography-based optimization. Multimed Tools Appl 75(23):15601–15617
Zhang Y, Sun Y, Phillips P, Liu G, Zhou X, Wang S (2016) A multilayer perceptron based smart pathological BrainDetection system by fractional Fourier entropy. J Med Syst: 1–11
Zhu W, Huang W, Lin Z, Yang Y, Huang S, Zhou J (2015) Data and feature mixed ensemble based extreme learning machine for medical object detection and segmentation. Multimed Tools Appl, Springer
Acknowledgments
The authors would like to thank Dr. Nagendra Patil, Head of the Department of Radio Diagnosis K B N Institute of Medical Sciences Kalaburagi, for the validation of the obtained results with respect to the ground truth samples of BRATS-2015 database. Also for their valuable suggestions during the design and implementation of the proposed work.
Author information
Authors and Affiliations
Corresponding author
Additional information
The original version of this article was revised: Equation 46 was incorrect.
Rights and permissions
About this article
Cite this article
Virupakshappa, Amarapur, B. Computer-aided diagnosis applied to MRI images of brain tumor using cognition based modified level set and optimized ANN classifier. Multimed Tools Appl 79, 3571–3599 (2020). https://doi.org/10.1007/s11042-018-6176-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-018-6176-1