Abstract
Lung cancer presents the highest cause of death among patients around the world, in addition of being one of the smallest survival rates after diagnosis. In this paper, we exploit a deep learning technique jointly with the genetic algorithm to classify lung nodules in whether malignant or benign, without computing the shape and texture features. The methodology was tested on computed tomography (CT) images from the Lung Image Database Consortium and Image Database Resource Initiative (LIDC-IDRI), with the best sensitivity of 94.66%, specificity of 95.14%, accuracy of 94.78% and area under the ROC curve of 0.949.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Lung cancer is the number one cause of cancer deaths presenting an estimated value of 1.8 million new cases a year [40]. The vast majority of lung cancer cases, about 85%, are due to long-term tobacco smoking [18]. In Brazil, it is estimated an average of 28,220 cases of lung cancer in 2016, where 17,330 males and 10,890 females [21]. Early diagnosis can improve the effectiveness of treatment and increase the patients chance of survival, with a 10-year survival of almost 90% [12, 28].
A lung nodule is defined as a focal opacity whose largest diameter is between 3 mm and 3 cm in length [15]. Lung nodules are commonly found on computed tomography (CT) scan, as shown in Fig. 1, which is used in the analysis of several lesion types, including lung lesions as small as 1 mm in diameter [9]. Notwithstanding, the detection of lung nodules on CT images is still a challenging task and passive error, considering that the densities of the nodules may be similar to that of the other lung structures. Furthermore, the nodules may be characterized by low contrast and small sizes in complex anatomic regions, and they could be close or joined to blood vessels or lung border [29].
To minimize such errors, a large amount of researches has been conducted to improve computer-aided diagnosis (CADx) [6, 19, 38]. The basic concept of CADx is to provide for a second objective opinion for the assistance of medical image interpretation and diagnosis [4]. A traditional CADx commonly involves several image processing steps and then performs a classification step.
Recently, deep learning techniques have been introduced to the medical image analysis field with promising results on various applications [32, 39, 43]. Deep learning allows automatically uncovering multiple levels of features from the training images, without a complicated pipeline of image processing and classification steps [19]. In the context of CADx, deep learning-based CADx has two advantages over the traditional ones. Firstly, feature interaction and hierarchy can be exploited jointly within the intrinsic deep architecture of a neural network. Second, the three steps of feature extraction, selection and classification can be realized within the optimization of the same deep architecture, making easier the fining [4].
Thereby, this paper proposes a deep learning model of the convolutional neural network in conjunction with the genetic algorithm to classify lung nodules in whether malignant or benign on CT images. As for contributions to the computer science field, we can mention: 1) divide the nodule in subregions to incorporate local information using Otsu algorithm and particle swarm optimization, 2) combine multiple convolutional neural networks fed by the nodule and its subregions simultaneously, sharing the same fully-connected layer and 3) use the genetic algorithm in order to optimize the network parameters such as number of filters in the convolutional layers and number of neurons in the fully-connected layer.
The paper is organized as follows. Section 2 presents the related works. Section 3 presents the proposed methodology for the classification of lung nodules in whether malignant or benign. Section 4 presents and discusses the results. Finally, the Section 5 presents final remarks about this paper.
2 Related works
The earlier lung cancer is detected the higher the survival rate [28]. Accordingly, in the literature, several researches are frequently developed aiming at increasing the accuracy of lung cancer diagnosis using CADx systems [3, 4, 7]. Normally, these CADx systems use as classifiers, the artificial neural network (ANN) and support vector machine (SVM). Recently, deep learning techniques have been introduced to the CADx systems. Thereof, the researches were listed here in three groups: ANN classifiers, SVM classifiers and deep learning techniques.
In the artificial neural network-based researches, Chen et al. [3] proposed a methodology based on shape features such as size, margins, contour, among others, resulting the accuracy of 92.2%. Elizabeth et al. [13] proposed a methodology based in a combination of shape and texture features such as area, eccentricity, smoothness, among others, and radial basis function, achieving an accuracy of 94.4%, sensitivity of 92.3% and specificity of 94.9%. Dandil et al. [7] proposed a methodology based on texture features such as entropy, contrast, correlation, among others, and principal component analysis (PCA), with the accuracy of 90.6%, sensitivity of 92.3% and specificity of 89.4%. In their part, Gupta and Tiwari [14] proposed a methodology based on texture features using curvelet transform, resulting the accuracy of 90%, sensitivity of 86.6% and specificity of 93.3%. The methodology developed by Kuruvilla and Gunavathi [25] used texture features such as mean, skewness, kurtosis, among others. The authors achieved an accuracy of 93.3%, sensitivity of 91.4% and specificity of 100%.
In the support vector machine-based researches, Nascimento et al. [31] proposed a methodology based on texture features using shannon and simpson indexes and linear discriminant analysis (LDA), with the accuracy of 92.78%, sensitivity of 85.6% and specificity of 97.8%. Krewer et al. [23] proposed a methodology based in a combination of shape and texture features such as, longest diameter, length, Law features, among other, using correlation-based feature subset selection, resulting the accuracy of 87.8%, sensitivity of 85.7% and specificity of 89.4%. Orozco et al. [33] proposed a methodology based on texture features such as, angular second moment, contrast, correlation, among others, using best first and k-means, obtaining an accuracy of 84%, sensitivity of 83.3% and specificity of 84.8%. In their part, Huang et al. [20] proposed a methodology using texture features based on fractional Brownian motion model, with the accuracy of 83.1%. The methodology proposed by Parveen and Kavitha [35] used texture features such as, contrast, energy, homogeneity, among others, with the sensitivity of 91.38% and specificity of 89.5%.
In the deep learning-based researches, Hua et al. [19] proposed a methodology based on two techniques, deep belief network (DBN) and convolutional neural network (CNN), comparing to explicit feature computing such as SIFT and fractal analysis. The best results were achieved using DBN, with sensitivity of 73.4% and specificity of 82.2%. Kumar et al. [24] proposed a methodology based on autoencoder (AE), resulting the accuracy of 75%. In their part, Cheng et al. [4] proposed a deep learning-based CADx using stacked denoising autoencoder (SDAE), comparing to the lung CADx system, denoted as CURVE [38], with the best accuracy of 95.6%, sensitivity of 92.4% and specificity of 98.9% using SDAE. The methodology developed by Sun et al. [39] used three techniques, CNN, DBN and SDAE, achieving the best accuracy of 81.1% using DBN.
The aforementioned researchers present good results. However, some researches use small samples, low complexity databases, individual nodule analysis and classifiers with parameters and architecture chosen manually. Thus, we explore the effectiveness of deep learning combined with genetic algorithm, optimizing the network parameters, in order to improve the performance of the model, applying in large samples, using local information of subregions of nodule.
3 Material and methods
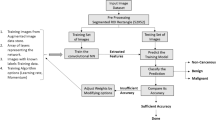
The proposed methodology is divided into four steps as described in Fig. 2. In summary, the first step details the materials used as images of CT scans in the LIDC-IDRI database and the nodules segmentation. In the second step, nodules preprocessing is performed. Finally, the diagnostic is completed using the evolutionary convolutional neural network following of the results evaluation.
3.1 Materials
The image database used in this research is the LIDC-IDRI [1], which is available online as a result of an association between the Lung Image Database Consortium and the Image Database Resource Initiative, including 1018 CT scans along with annotations from up-to four radiologists. Nonetheless, two factors have had negative impact over 185 CT scans so that they were inappropriate to be used for this methodology. Firstly, only scans including nodules with a diameter larger than 3 mm were annotated with diagnostic information. The second factor included the divergence of information found in the annotations file of a scan versus the information present in the DICOM header of the same scan [6, 8]. For that reason, the annotation is invalidated. The proposed methodology was applied to 833 CT scans.
Every single nodule larger than 3 mm annotated by the radiologists is taken into account and extracted, which leads to up-to 4 annotations per nodule in some cases. The diagnostic information for each nodule presents ratings between 1 and 5. In this research, rating 1 and 2 were considered as benign nodule, rating 3 as indeterminate nodule and rating 4 and 5 as malignant nodule. In the cases with more than one annotation per nodule, the diagnosis was calculated by computing the histogram to benign, malignant and indeterminate classes. The final diagnosis is attributed to the class with the highest absolute frequency. If there is not only a class with the highest absolute frequency, the nodule is not considered. A total of 3243 nodules (1413 malignant and 1830 benign) were obtained.
3.2 Preprocessing
Prior to the nodule classification step, three preprocessing have been performed. The first step consists of the application of the Otsu algorithm [34] based on particle swarm optimization (PSO) [11]. The goal of this step is to divide each nodule into two subregions with maximum inter-class variance. The nodule and its subregions were analyzed simultaneously for the classification in whether malignant or benign. This process is illustrated in Fig. 3, generating, therefore, three databases.
The PSO algorithm is an evolutionary technique based on swarm intelligence [11]. Each particle in the swarm is a potential solution for a targeted problem. In this research, the first particle is initialized with the two centroids of the subregions generated by the Otsu algorithm. These centroids represent the particle positions (xi). The remaining particles are initialized with two numbers, ranging between the minimum and maximum Hounsfield unit (HU) in the nodule. At each iteration, the particles traverses toward its new best position by altering another parameter called velocity (vi). The updating equations are as follows.
where, w is the inertia weight which controls the impact of previous history of velocities on the current velocity, C 1 is the cognitive parameter which pulls each particle towards local best position, C 2 is the social parameter which pulls the particle towards global best position, Pbest is the best position of particle, Gbest is the best position in the whole swarm and r 1 , r 2 are two independent random numbers between 0 and 1.
The fitness of each particle is evaluated minimizing the intra-class variance between the subregions of the nodule. The subregions are generated clustering all the voxels in the nodule to the particle positions which the Euclidean distance is minimal. The process is repeated until the number of iterations is reached. Once terminated, the particle positions represented by Gbest are the optimal centroids to the final subregions. The PSO algorithm parameters are assigned based on Table 1.
The second preprocessing consists of using each two-dimensional CT slice as individual sample. Since CT scans are three-dimensional images presenting a high slice thickness variation (0.6-5 mm), three-dimensional features may sometimes be inaccurate due to low resolution on the z axis [4, 19]. Moreover, it increases the amount of samples in the three databases. Resulting in 21,631 slices of the nodule for each database (13,321 malignant and 8310 benign).
The third preprocessing is the resizing of all nodules and subregions’ slices (regions of interest - ROIs) in 28 × 28 dimension. This process is due to fully-connected layer on top of the CNN architecture, which needs fixed input neurons. Slices with smaller dimensions, Fig. 4a, were extracted and put into background images of 28 × 28. Slices with larger dimensions, Fig. 4b, were rescaled by setting the higher axis in the dimension 28 and reducing the less axis proportionally, finally, they were extracted and put into background images of 28 × 28.
3.3 Classification
Convolutional neural networks (CNN) are a biologically-inspired trainable architecture that can learn multi-level hierarchies of features [27, 30]. The network extracts implicitly features of visual patterns presented as input and classifies patterns from the extracted features. Typical CNN usually consist of convolutional layers, pooling layers, neuron layers and fully-connected layers [22]. Fig. 5 shows the architecture of a basic CNN.
Convolutional layers, as shown in Fig. 6, have trainable filters that are applied across the entire input, generating feature maps [26]. Each filter detects a particular feature that occurs in any part of the input [27]. Once a feature has been detected, its exact location becomes irrelevant.
Pooling layers are non-linear down-sampling layers that yield maximum or average values in each sub-region of input image or feature maps. Pooling layers increase the robustness of translation and reduce the number of network parameters [22]. Fig. 7 shows the max-pooling layer.
Neuron layers apply non-linear activations on input neurons. Common activations are softmax function, sigmoid function, rectified linear unit, etc. Fully-connected layers are responsible for classifying patterns presented to CNN, generally a multilayer perceptron (MLP).
Supervised training is performed using a form of stochastic gradient descent (SGD) to minimize the discrepancy between the desired output and the current output of the network, based on some loss function [27]. All the weights of all the filters in all the layers are updated simultaneously with the backpropagation algorithm [16].
Such neural networks trained with backpropagation admit a large variety of specific architectures applicable to a wide range of applications [2, 37, 42]. However, not all network architectures can be expected to learn a given task successfully. Therefore, we propose an evolutionary approach to this problem. Genetic algorithm (GA) [17] was used to optimize the CNN parameters such as number of filters in the convolutional layers and number of neurons in the MLP. The proposed network is described below.
3.3.1 Evolutionary convolutional neural network
In order to analyze the malignancy patterns of the ROIs, described in Section 3.2, we propose three similar basic CNN sharing the same fully-connected layer. The ROIs are submitted simultaneously to the evolutionary convolutional neural network for the classification in whether malignant or benign. Fig. 8 shows the proposed architecture.
The individual CNN presents a convolutional layer (C1) followed by rectified linear unit activation and a max-pooling layer (P 1), another convolutional layer (C2) followed by rectified linear unit activation, a max-pooling layer (P 2) followed by a dropout layer to help preventing overfitting, one of the problems that occur during neural network training. The kernel size of C1 and C2 is 5 × 5 and the kernel size of P 1 and P 2 is 2 × 2.
All of the three CNN sharing the same fully-connected layer on the top of network. The MLP presents an input layer followed by rectified linear unit activation and a dropout layer, a hidden layer followed by rectified linear unit activation, at last, an output layer with softmax activation.
The GA optimizes the number of filters in C1 and C2 and the number of neurons in hidden layer. The chromosomes are encoded as a list of three positive integers, called gene, corresponding to the parameters to be optimized. GA operates according to the following algorithm:
-
1.
The chromosomes of initial population are randomly initialized with three genes between 1 and 100. The chosen interval is to avoid a large number of neurons in hidden layer, preventing overfitting.
-
2.
The fitness of a chromosome is evaluated through the results obtained by the network using the proposed parameters. The fitness is defined by Eq. 3. A greater weight is given to the metric sensitivity, in order to get models with a high capacity to malignant nodules correctness.
$$ \mathrm{Fitness}=\left(2\ \mathrm{Sensitivity}\right)+\mathrm{Specificity}+\mathrm{Accuracy} $$(3) -
3.
The population undergoes reproduction until a termination condition is reached. Reproduction consists of three steps:
-
(a)
Selection of chromosomes to reproduce. This research selected two chromosomes using the roulette-wheel selection.
-
(b)
Crossover operator exchanges the genes between the pair of chromosomes selected. This research used the one-point crossover.
-
(c)
Mutation operation replaces randomly a gene in a chromosome. This research used the gaussian mutation.
-
(a)
-
4.
The next population is determined using the elitism method. Chromosomes with highest fitness remain and the new chromosomes are selected randomly. The algorithm starts again from step 2 until the maximum number of epochs is reached.
At the end of evolution, the final population contains the best parameters for the convolutional neural network with respect to the lung nodules diagnosis on CT images using the ROIs. Fig. 9 shows the proposed hybrid model.
3.4 Results evaluation
After the network training, it is necessary to validate the results. This methodology uses metrics commonly used in CADx systems that are widely accepted to analyze the performance of image processing-based systems. These metrics include sensitivity, specificity, accuracy and area under the receiver operating characteristic (ROC) curve (AUC) [10].
Equations 4, 5, 6 calculate the sensitivity, specificity and accuracy, respectively. A ROC curve indicates the true positive rate (sensitivity) as a function of the false positive rate (1 - specificity).
where TP is true positive, FN is false negative, TN is true negative and FP is false positive.
4 Results and discussion
This section presents and discusses the results obtained with the proposed methodology with reference to the lung nodules diagnosis on CT scans. All tests were implemented in Python code using Keras deep learning library [5], running on a machine with an Intel Core i7-5960X processor, 128 GB RAM, GeForce GTX Titan X 12 GB GPU and Linux operating system.
In order to evaluate the proposed hybrid model, each database described in Section 3.2, was divided into two sets: training and test. The training set contains 3043 nodules (1313 malignant and 1730 benign) generating 20,288 slices (12,328 malignant and 7960 benign) and the test set contains 200 nodules (100 malignant and 100 benign) generating 1343 slices (993 malignant and 350 benign). The validation set contains 10% of slices training set, which are used to validate the training model.
Evolutionary convolutional neural networks were trained using SGD with a batch size of 30 ROIs of each database, submitted simultaneously, over 30 epochs. Learning rate parameter determines how much the weights can change in response to an observed error on the training set. The value of the learning rate should be sufficiently large to allow a fast learning process but small enough to guarantee its e effectiveness [41]. In order to find the global minimum, we used a small rate of 0.01. The loss function used was the cross entropy [36]. The GA parameters used in the training are assigned based on Table 2.
Table 3 presents the results obtained for all chromosomes of the final population, including the sensitivity (SEN), specificity (SPE), accuracy (ACC) and AUC, followed by the standard deviations. These results were obtained using the 1343 slices (993 malignant and 350 benign).
The ideal CADx system must presents a good balance among the sensitivity, specificity and accuracy. Hence, a good methodology must be capable of successfully classifying both malignant and benign nodules. Based on this, chromosome 1 obtained the best result of the proposed methodology with a final error of 0.2808 taking about 9.5 min computation time.
Despite that, all chromosomes obtained good results for every metric, exceeding 92%. The final population presented low standard deviations in all the metrics between the chromosomes, indicating the methodology’s robustness. Fig. 10 shows the final errors obtained by all chromosomes. The mean computation time of a chromosome was of about 11 min.
Besides these results, we tested the performance of the chromosome 1 to classify the volume of lung nodules. The classification is still based on slices, however, the metric is evaluated considering all slices of a nodule. The criterion for the lung nodule classification consisting in, if at least a slice of the nodule is classified as malignant, the nodule is considered malignant. Confusion matrix shows the true and false rates between the actual cases and predicted cases. Table 4 presents the confusion matrix obtained, resulting in a sensitivity of 98%, specificity of 91% and accuracy of 94.5%. These results were obtained using the 200 nodules (100 malignant and 100 benign) of test set.
4.1 Comparison with other related works
Table 5 presents a comparison between the results found on this research and the related works. It is important to reiterate that to perform a trustworthy comparison with these previous researches, it should be necessary to use the same image database, same training and test sets, and same settings for the classifiers, including the architecture, weight initialization, among other parameters.
Comparing the best result obtained in this research with the related works, only that of Cheng et al. [4] presents accuracy better than our research, with the difference of less than 1%, however, our research presents a better sensitivity, of about 2.26% of difference. In term of CADx system, the sensitivity is the metric more important, because it shows the model performance to classify correctly malignant nodules, allowing the fast medical intervention. Besides, our research used large samples of LIDC-IDRI database (21,631 slices). Only Sun et al. [39] used an amount of samples superior to ours, consisting in 174,412 samples. Nonetheless, the accuracy presents a low value. In brief, the proposed deep learning-based CADx system presents good results, exceeding 94%, in the classification of both malignant and benign nodules, even applied in large samples.
5 Conclusion
Lung cancer is considered to be the main cause of cancer death worldwide. It evidences the importance of developing research aimed at precocious diagnosis, providing a better treatment to patients. Thereof, this research has proposed a methodology to classify lung nodules in whether malignant or benign on CT images using an evolutionary convolutional neural network, a deep learning technique jointly with the genetic algorithm.
The results obtained demonstrate the promising performance of the proposed hybrid model. All results exceeded 91%. Besides, the proposed deep learning-based CADx system avoids the need of the feature extraction and selection steps. It reduces the computational complexity of the CADx system without compromising the performance.
Although the database used in this research was robust and ensured a high diversity of the nodules, additional tests with other databases are necessary to improve the proposed methodology and to make it more robust and generic. Finally, the methodology presented can integrate a CADx system to classify lung nodules in whether malignant or benign, making, therefore, the analysis of exams by radiologists more efficient and less exhaustive.
References
Armato SG III, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA et al (2011) The lung image database consortium (lidc) and image database resource initiative (idri): a completed reference database of lung nodules on ct scans. Med Phys 38(2):915–931
Boughrara H, Chtourou M, Amar CB, Chen L (2016) Facial expression recognition based on a mlp neural network using constructive training algorithm. Multimedia Tools and Applications 75(2):709–731
Chen H, Zhang J, Xu Y, Chen B, Zhang K (2012) Performance comparison of artificial neural network and logistic regression model for differentiating lung nodules on ct scans. Expert Syst Appl 39(13):11503–11509
Cheng JZ, Ni D, Chou YH, Qin J, Tiu CM, Chang YC, Huang CS, Shen D, Chen CM (2016) Computer-aided diagnosis with deep learning architecture: Applications to breast lesions in us images and pulmonary nodules in ct scans Scientific reports:6
Chollet F (2015) Keras. https://github.com/fchollet/keras
da Silva GLF, de Carvalho Filho AO, Silva AC, de Paiva AC, Gattass M (2016) Taxonomic indexes for differentiating malignancy of lung nodules on ct images. Research on Biomedical Engineering 32(3):263–272
Dandil E, Cakiroglu M, Eksi Z, Ozkan M, Kurt OK, Canan A (2014) Arti cial neural network-based classification system for lung nodules on computed tomography scans. In: Soft Computing and Pattern Recognition (SoCPaR), 2014 6th International Conference of, pp. 382–386. IEEE
de Carvalho Filho AO, de Sampaio WB, Silva AC, de Paiva AC, Nunes RA, Gattass M (2014) Automatic detection of solitary lung nodules using quality threshold clustering, genetic algorithm and diversity index. Artificial intelligence in medicine 60(3):165–177
Dolejs M (2007) Detection of pulmonary nodules from ct scans. Ph.D. thesis, Citeseer
Duda RO, Hart PE et al (1973) Pattern classification and scene analysis, vol 3. Wiley, New York
Eberhart RC, Kennedy J et al. (1995) A new optimizer using particle swarm theory. In: Proceedings of the sixth international symposium on micro machine and human science, vol. 1, pp. 39–43. New York
El-Baz A, Suri JS (2011) Lung imaging and computer aided diagnosis. CRC Press, Boca Raton
Elizabeth D, Nehemiah H, Raj CR, Kannan A (2012) Computer-aided diagnosis of lung cancer based on analysis of the significant slice of chest computed tomography image. IET Image Process 6(6):697–705
Gupta B, Tiwari S (2014) Lung cancer detection using curvelet transform and neural network. International Journal of Computer Applications 86(1)
Hansell DM, Bankier AA, MacMahon H, McLoud TC, Muller NL, Remy J (2008) Fleischner society: glossary of terms for thoracic imaging 1. Radiology 246(3):697–722
Hecht-Nielsen R (1989) Theory of the backpropagation neural network. In: Neural Networks, 1989. IJCNN., International Joint Conference on, pp. 593–605. IEEE
Holland JH (1975) Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. U Michigan Press, Ann Arbor
Hong WK, Tsao AS (2007) Lung carcinoma: Tumots of the lungs. merck manual professional edition. Online
Hua KL, Hsu CH, Hidayati SC, Cheng WH, Chen YJ (2015) Computer-aided classification of lung nodules on computed tomography images via deep learning technique. OncoTargets and therapy 8:2015–2022
Huang PW, Lin PL, Lee CH, Kuo C (2013) A classification system of lung nodules in ct images based on fractional brownian motion model. In: System Science and Engineering (ICSSE), 2013 International Conference on, pp. 37–40. IEEE
INCA (2016) Instituto nacional do câncer, tipos de câncer: Pulmão http://www2.inca.gov.br/wps/wcm/connect/tiposdecancer/site/home/pulmao. Accessed: 29 July 2016
Kang K, Wang X (2014) Fully convolutional neural networks for crowd segmentation. arXiv preprint arXiv:1411.4464
Krewer H, Geiger B, Hall LO, Goldgof DB, Gu Y, Tockman M, Gillies RJ (2013) Effect of texture features in computer aided diagnosis of pulmonary nodules in low-dose computed tomography. In: Systems, Man, and Cybernetics (SMC), 2013 I.E. International Conference on, pp. 3887–3891. IEEE
Kumar D, Wong A, Clausi DA (2015) Lung nodule classification using deep features in ct images. In: Computer and Robot Vision (CRV), 2015 12th Conference on, pp. 133–138. IEEE
Kuruvilla J, Gunavathi K (2014) Lung cancer classification using neural networks for ct images. Comput Methods Prog Biomed 113(1):202–209
LeCun Y, Boser B, Denker JS, Henderson D, Howard RE, Hubbard W, Jackel LD (1989) Backpropagation applied to handwritten zip code recognition. Neural Comput 1(4):541–551
LeCun Y, Kavukcuoglu K, Farabet C et al (2010) Convolutional networks and applications in vision. ISCAS, In, pp 253–256
Lederlin M, Revel MP, Khalil A, Ferretti G, Milleron B, Laurent F (2013) Management strategy of pulmonary nodule in 2013. Diagnostic and interventional imaging 94(11):1081–1094
Leef JL, Klein JS (2002) The solitary pulmonary nodule. Radiol Clin N Am 40(1):123–143
Liu Y, Yin B, Yu J, Wang Z (2016) Image classification based on convolutional neural networks with cross-level strategy. Multimedia Tools and Applications 1–15
Nascimento LB, de Paiva AC, Silva AC (2012) Lung nodules classification in ct images using shannon and Simpson diversity indices and svm. Machine Learning and Data Mining in Pattern Recognition. Springer, In, pp 454–466
Ngo TA, Carneiro G (2015) Lung segmentation in chest radiographs using distance regularized level set and deep-structured learning and inference. In: Image Processing (ICIP), 2015 I.E. International Conference on, IEEE, pp. 2140–2143
Orozco HM, Villegas OOV, Dominguez HdJO, Sanchez VGC (2013) Lung nodule classi cation in ct thorax images using support vector machines. In: Artificial Intelligence (MICAI), 2013 12th Mexican International Conference on. IEEE, pp. 277–283
Otsu N (1975) A threshold selection method from gray-level histograms. Automatica 11(285–296):23–27
Parveen SS, Kavitha C (2014) Classification of lung cancer nodules using svm kernels. International Journal of Computer Applications 95(25)
Rubinstein RY, Kroese DP (2013) The cross-entropy method: a unified approach to combinatorial optimization. Monte-Carlo simulation and machine learning, Springer Science & Business Media
Schaer JD, Caruana RA, Eshelman LJ (1990) Using genetic search to exploit the emergent behavior of neural networks. Physica D: Nonlinear Phenomena 42(1–3):244–248
Sun T, Zhang R, Wang J, Li X, Guo X (2013) Computer-aided diagnosis for early-stage lung cancer based on longitudinal and balanced data. PLoS One 8(5):e63559
Sun W, Zheng B, Qian W (2016) Computer aided lung cancer diagnosis with deep learning algorithms. SPIE Medical Imaging. International Society for Optics and Photonics, In, pp 97850Z–97850Z
WHO (2012) Globocan 2012: World cancer statistics. http://globocan.iarc.fr/Pages/fact sheets cancer.aspx?cancer = lung. Accessed 04 Aug 2016
Wilson DR, Martinez TR (2001) The need for small learning rates on large problems. In: Neural Networks, 2001. Proceedings. IJCNN'01. International Joint Conference on, vol. 1. IEEE, pp. 115–119
Wu Z, Zhao K, Wu X, Lan X, Meng H (2015) Acoustic to articulatory mapping with deep neural network. Multimedia Tools and Applications 74(22):9889–9907
Zhang W, Li R, Deng H, Wang L, Lin W, Ji S, Shen D (2015) Deep convolutional neural networks for multi-modality isointense infant brain image segmentation. NeuroImage 108:214–224
Acknowledgements
The authors acknowledge CAPES and CNPq for their financial support. The authors acknowledge the National Cancer Institute and the Foundation for the National Institutes of Health for their critical role in the creation of the free, publicly available LIDC-IDRI database used in this research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
da Silva, G.L.F., da Silva Neto, O.P., Silva, A.C. et al. Lung nodules diagnosis based on evolutionary convolutional neural network. Multimed Tools Appl 76, 19039–19055 (2017). https://doi.org/10.1007/s11042-017-4480-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-017-4480-9