Abstract
Convolutional neural network (CNN) is one of the deep structured algorithms widely applied to analyze the ability to visualize and extract the hidden texture features of image datasets. The study aims to automatically extract the self-learned features using an end-to-end learning CNN and compares the results with the conventional state-of-art and traditional computer-aided diagnosis system’s performance. The architecture consists of eight layers: one input layer, three convolutional layers and three sub-sampling layers intercepted with batch normalization, ReLu and max-pooling for salient feature extraction, and one fully connected layer that uses softmax function connected to 3 neurons as output layer, classifying an input image into one of three classes categorized as nodules \(\ge\) 3 mm as benign (low malignancy nodules), malignant (high malignancy nodules), and nodules < 3 mm and non-nodules \(\ge\) 3 mm combined as non-cancerous. For the input layer, lung nodule CT images are acquired from the Lung Image Database Consortium public repository having 1018 cases. Images are pre-processed to uniquely segment the nodule region of interest (NROI) in correspondence to four radiologists’ annotations and markings describing the coordinates and ground-truth values. A two-dimensional set of re-sampled images of size 52 \(\times\) 52 pixels with random translation, rotation, and scaling corresponding to the NROI are generated as input samples. In addition, generative adversarial networks (GANs) are employed to generate additional images with similar characteristics as pulmonary nodules. CNNs are trained using images generated by GAN and are fine-tuned with actual input samples to differentiate and classify the lung nodules based on the classification strategy. The pre-trained and fine-tuned process upon the trained network’s architecture results in aggregate probability scores for nodule detection reducing false positives. A total of 5188 images with an augmented image data store are used to enhance the performance of the network in the study generating high sensitivity scores with good true positives. Our proposed CNN achieved the classification accuracy of 93.9%, an average specificity of 93%, and an average sensitivity of 93.4% with reduced false positives and evaluated the area under the receiver operating characteristic curve with the highest observed value of 0.934 using the GAN generated images.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Cancer is one of the major public health issues spread worldwide leading to deaths with high mortality among both men and women due to noninvasive treatment and unclear clinical examinations. The American Cancer Society in the USA estimated the new projected cancer cases of 234,030 and deaths of 154,050 in 2018 [51]. Among the other types of cancers, lung cancer is one of the leading cancers with high mortality rates [1]. Risk factors causing cancer are due to the consumption of tobacco, smoking, biological and chemical reactions (exposure to radon gas), and environmental conditions (exposure to secondhand smoke). The survival rates are low when compared to many other types of cancers. The difficulty lies in detecting the region of nodules in the soft lung tissues at its early stages.
Radiologists recommend different imaging modalities for detecting pulmonary nodule regions, such as computed tomography (CT), magnetic resonance imaging (MRI), and positron emission tomography (PET) [21]. The most commonly preferred imaging modality is the CT scans by the radiologists. CT scans are popular due to their advantages with respect to cost, availability, and rapid acquisition of scans across complete lung sections. In recent years, the mortality of lung cancer has decreased by around 20% with low-dose CT images as reported by the National Lung Screening Trial [55].
Patients with lung cancer symptoms undergo CT scans to distinguish certain abnormal growth observed in the lungs. More exceptionally, sensitively identifying the little knobs (cancerous cells) is a trivial task as the nodules may be attached to vessels or the walls of the chest or false positively considered as irregularly shaped due to noise. The pulmonary nodules are irregularly shaped growth in the lungs with its diameter measured up to 3 mm in the chest region [20]. Also, the pulmonary nodules are categorized based on its shape (round, irregular shaped), size (small or large), location (vascular or pleural regions), texture (solid or non-solid), and so on. As the patient receives the CT scans in a clinical laboratory, the radiologists evaluate and detect the suspicious nodule from the numerous CT images. Based on the possibility of malignancy examined through the nodule information (density, morphology, texture features), their diagnosis is acknowledged with an appropriate treatment plan. The task in identifying the nodules is rigorous. Inappropriate professional experience, distraction, fatigue while capturing scans, etc., may destabilize nodule detection contributing to misinterpretations of false positives with the available data. Therefore, a number of CADx systems were developed to help radiologists process and analyze images automatically and accurately identify the pulmonary lung nodules [36]. But radiologists believe that both detecting and diagnosing the pulmonary lung nodules at the early stages are the key factors for patient’s survival rates. To improve efficiency, CADx systems must be highly sensitive to low false positives, low cost in implementation, low system maintenance, and software security assurance with high levels of automation and must have the ability to detect different types of pulmonary nodules [16, 53].
Recent developed CADx systems are optimized to enhance the performance and interpretation of radiologist’s readings toward medical imaging [4]. Researches have focused on manual feature extraction and classification of lung nodules to distinguish benign from malignant using linear classifiers. However, feature extraction is one of the simplest dimensionality reduction methods widely used in image processing [14]. The features are categorized as texture, density, fractal [42], and morphological features extracted from the whole CT image or from the region of interest (ROI). Texture features include wavelet features [2, 8], histogram of oriented gradient (HOG) [28], gray-level co-occurrence matrix (GLCM) [35], curvelet transform [40], run level features, and local binary pattern (LBP) [56] features. Morphological features include area and circularity [3]. Density features include average intensity, entropy, and standard deviation used for mass detection [61].
The existing CADx systems are in need to design these features as an essential model. But the process is time-consuming and complicated [45]. Moreover, the features are to be correlated to obtain expected performance measures. The feature selection algorithms like the genetic algorithms generate an optimal combination of features, but are limited to high-dimensional features in terms of efficiency [5]. The appropriate extracted features from the ROI are further used for training and testing the data using different classification techniques to eliminate false positives. Some of the classifiers are support vector machine (SVM) [34], AdaBoost, fuzzy logic [38], naive Bayes, k-nearest neighborhood, k-means clustering [17], probabilistic neural networks (PNN) [59], multi-view convolution neural networks (MV-CNN) [48], artificial neural networks (ANN) [29], feed-forward back-propagation neural network (FF-BPNN) [8], Bayesian supervised, decision tree, and regression analysis.
In addition, there are architectures that allow algorithms to automatically learn and extract feature maps at multiple levels of abstraction generating complex functions linking the inputs (input layer) to categories of classes (output layer) directly from the raw image dataset without using any of the above manually handcrafted features. The extracted features at higher layers of hierarchy are determined by the combination of input features at lower layers with appropriate weights and a bias (hidden layers). Each layer consists of hundreds to thousands of neurons as similar to human brain architecture. Different neural network architectures are used in feature learning. Some are categorized as shallow neural networks, deep neural networks, and hybrid neural networks [57, 58]. Learning features from architectures with multiple layers and hierarchical models of input data like deep neural network/hybrid structures are the trend changing concepts topping the big companies like Google, Amazon, Facebook, Microsoft, etc. The study results show that deep neural network algorithms can outperform compared to traditional machine learning concepts [34, 43]. Recent advances in deep learning involve the concept of parallel computing with more accessibility and affordability in using graphics processing units for training huge annotated datasets. Many researchers made progress in training and classifying huge datasets using deep learning algorithms for pattern recognition. This led to substantial advancements in using deep neural network algorithms for medical imaging applications as well [54].
The concept of transfer learning from CNN models is useful for nodule segmentation and classification approaches, encouraging the usage in medical imaging [12, 33]. However, the neural network with hyperparameters is to be explicitly defined for capturing the salient features from heterogeneous volumes of CT images with the same size of input patches. Deeper the network layer increases the power of expression. Training the neural network with more number of layers requires good training data. But the amount of medical data available is limited for image classification according to malignancy suspiciousness due to insufficient professional experience for analysis, time constraints, and ethical issues.
To overcome the drawback, GANs [18] are incorporated in the study generating additional images with similar characteristics as training samples of pulmonary nodules. As a result, the ethical problems are avoided by generating new samples that do not correspond to real cancer patients. The generated new images are meant to compete with actual lung cancer images. Faking the lung nodules using GAN may improve the classification of pulmonary nodules based on malignancy levels.
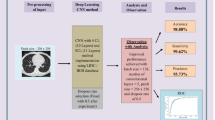
In the study, we incorporate an end-to-end convolutional neural network to automatically learn the features for the classification of pulmonary lung nodules as benign, malignant, or non-cancerous. The NROI is extracted, trained, and tested via deep learning using CNN as shown in Fig. 1. To compare its performance, CNNs are trained using images generated by GAN and are fined-tuned with actual input samples to differentiate and classify the lung nodules based on the classification strategy. In addition, CADx systems are implemented to extract the handcrafted texture, density, and morphological features. The methodology is compared with the state-of-the-art methods and traditional handcrafted methods for performance evaluation.
2 Related work
The lung nodule detection systems with nodule segmentation, feature extraction, and nodule classification have certain challenging tasks to overcome as discussed previously. In nodule segmentation, identifying the exact pulmonary nodule boundaries in CT scans is crucial due to the similar visualization characteristics of candidate nodules and its surroundings close to the ribs, vessels, or walls of the chest. In feature extraction, designing and extracting invariant features from segmented images is time-consuming and complicated if the correlation between features is not properly considered. Reducing false positives plays a vital role in nodule classification, increasing sensitivity rates. Over the past, several researchers have developed methodologies to overcome these challenges. In this section, the works related to the proposed methodology are discussed.
Reference [29] proposed a classification methodology using ANNs using CT images. The lung nodules are segmented using morphological operations, and the features are extracted using statistical descriptors. For classification, feed-forward neural network (FFNN) and FF-BNN are implemented. FF-BNN exhibits better classification compared to FFNN. The methodology exhibits a classification accuracy of 93.33%, the specificity of 100%, and the sensitivity of 91.4%.
Reference [44] implemented an automated CADx system for lung nodule classification. The methodology was evaluated on 3 datasets: (1) sclerotic nodules from 59 patients, (2) lymph nodules from 176 patients, and (3) colonic polyp nodules from 1186 patients. The images were sampled with 2D and 2.5D views through scaling, rotating transformations, and random translations. Deep CNNs (ConvNet) were trained with these views classifying images according to probability scores, resulting in a sensitivity of 70% for sclerotic metastases, 77% for lymph nodules, and 75% for colonic polyps with 3 false positives per patient.
Reference [52] designed an end-to-end machine learning architectures to automatically extract features from CT images for lung cancer diagnosis. The nodules were segmented and re-sampled by rotating nodule slices annotated by four expert radiologist’s markups forming 13,668 samples. Three multichannel ROI-based deep learning algorithms are designed: (1) CNN, (2) deep belief networks (DBN), (3) stacked denoising autoencoder (SDEA). The methodology exhibited an AUC of 0.899 for CNN, 0.884 for DBN, 0.852 for SDEA, and 0.848 for traditional CADx.
Reference [49] designed deep structured algorithms for lung nodule classification using a multi-crop convolutional neural network (MC-CNN). A total of 2618 images from the LIDC-IDRI database having 880 low malignancy nodules and 495 high malignancy nodules were used in the study. The methodology recorded the highest nodule classification accuracy of 87.14%, the specificity of 93%, the sensitivity of 77%, and an AUC score of 0.93.
Reference [13] proposed a classification approach differentiating the patterns of benign and malignant samples using topology-based phylogenetic diversity index on CT images. CNN was used to classify the extracted features. LIDC image dataset comprising 1405 nodules (394 malignant and 1011 benign nodules) was used in the study achieving an accuracy of 92.63%, the specificity of 93.47%, the sensitivity of 90.7%, and the AUC of 0.934.
Reference [37] developed a computer-assisted decision support system for nodule detection and classification according to malignancy stages using internet-of-things (IOTs) on medical datasets obtained from medical body area network (MBAN). Deep fully convolutional neural network (DFCNet) was used for pulmonary nodule classification based on their stages of cancer. The performance of the approach was evaluated on different datasets, resulting in the classification accuracy of 84.58%.
Reference [39] proposed an approach for classifying nodules as primary lung cancer, benign and metastatic nodules using deep CNN. VGG-16 CNN was applied to 1236 patients obtained from Toshiba Medical Systems (TMS) with the cropped volume of interest of \(64\times 64\) pixels. With hyperparameter optimization, the DCNN method resulted in a classification accuracy of 60.7% for an image size of 56, 64.7% for an image size 112, and 68.0% for an image size of 224.
Reference [32] implemented a nodule detection approach to reduce false positives on JSRT image datasets. The nodules were enhanced using unsharp masking. The database images were cropped into \(229\times 229\) pixel sizes with nodules or non-nodules. Ensemble CNNs with layers of 5, 7, and 9 were used to train the input patches separately (CNN1, CNN2, CNN3) with varying inputs of sizes \(12\times 12\), \(32\times 32\), and \(60\times 60\), respectively. The approach resulted in the sensitivity of 94% with 5 FPs per image and 84% with 2 FPs per image differentiating nodules from non-nodules with minimal datasets used.
Reference [60] designed an automated pulmonary nodule detection scheme using a faster region-based CNN with two regions and a deconvolutional layer to detect the candidate nodules. In order to reduce false positives, all 3 models were trained using 2D CNN in the study. Experiments were conducted on total candidates of 150,414 (339 nodule images and 150,075 non-nodule images) from LUNA 16 datasets for training. Each candidate was labeled with class 0 for non-nodules and 1 for nodule images. The system exhibited a nodule detection sensitivity of 86.42%.
Reference [12] proposed a lung nodule classification methodology using transfer learning and CNN. Several CNN’s like VGG16, VGG19, Xception, InceptionV3, MobileNet, ResNet50, DenseNet169, DenseNet201, InceptionResNetV2, NASNetMobile, and NASNetLarge were built and trained on ImageNet dataset with feature extractors applied on LIDC images. The extracted deep features were classified with SVM, naive Bayes, k-nearest neighbor, multilayer perception, and random forest classifier. Upon experiments, ResNet50-based feature extractor with SVM classifier exhibited the highest AUC of 93.1% among the other evaluated combinations.
Reference [50] developed a CADx system for lung nodule detection using deep CNN based on transfer learning. Images were extracted from the LIDC dataset comprising of 700 nodules and 700 non-nodule samples. The samples were pre-processed and cropped into a \(224\times 244\) pixel rectangle. VGG-16 were used as feature extractors to extract the features from the input patches and were classified using SVM classifiers. The system exhibited a sensitivity of 87.2% with 0.39 FPs per scan and 85.4% with 4 FPs per scan.
Reference [27] proposed an attention-based fully CNN for medical image segmentation. The methodology incorporated a separate attention mechanism into ResNet + SE nets [22] hybrid architecture defined as focusNet. A total of 267 images of re-size \(256\times 256\) were used in the study. An accuracy of 99.32% was recorded by the hybrid architecture with a drawback of lack in responsiveness to sensitivity metrics of the input data. The time complexity is undefined, and the architecture is trained on limited image datasets.
A study on transfer learning by Ref. [33] depicts the performance of deep CNN classifying pulmonary lung nodules as benign or malignant. Images from subjects of 796 patients with biopsy-proven ground-truth values from one institution from 2012 to 2017 were used in the study. The nodule locations were manually traced from high-resolution CT images. The network was trained with prior knowledge of pathology confirmed diagnosis from the CT-guided biopsy values. Transfer learning on initial layers of the network with input datasets resulted in an AUC of 0.70 and a classification accuracy of 71%. Similarly, Ardila et al. [6] predicts the risk of lung cancers using a deep learning algorithm considering the patient’s current and prior CT volumes. The AUC of 94.4% was observed in 1139 validation cases with 11% reduction in false positives and a 5% reduction in false negatives. The methodology optimizes the screening process with prior CT and radiologists testing. Rather than training and testing the network with prior knowledge on nodule locations, which risks the time complexity on huge datasets, we employed CNN with ROI-based feature learning as most of the studies rely on subjective analysis and ratings of malignancy levels annotated by expert radiologists.
Reference [47] developed a lung classification approach for lung cancer diagnosis using DCNN on high-level image representations. The images were acquired from the Kaggle Data Science Bowl 2017 comprising of 63,890 cancer patients and 171,345 non-cancer patients. Images from the dataset were re-sized from \(512\times 512\) to \(120\times 120\) pixels. The first convolution layer in the method uses 50 feature maps with a filter size of \(11\times 11\). The second convolution layer uses 120 feature maps with a filter size of \(5\times 5\). The last layer used 120 feature maps with a filter size of \(3\times 3\). The approach categorizes the candidate nodules as cancerous and non-cancerous. The time complexity is undefined, and the training set of 50% and testing set of 25% were used in the experiments. The classification accuracy of 94.1%, the sensitivity of 0.87, and the specificity of 0.991 were recorded. With a minimum number of filters, feature maps, and augmented dataset with balanced categories of images, our methodology classifies candidate nodules as non-cancerous, benign, and malignant nodules based on their malignancy levels.
All the above methods exhibit promising results related to sensitivity, but few approaches result in high false positives either per scan or per patient or per image basis with minimal datasets used for validation, in turn, affecting the performance of classification accuracy. In addition, one of the major challenges in medical image classification techniques with CNN’s is the difficulty in acquiring datasets with enough samples for training. Most of the methods discussed do not have an equal number of image samples for each class of malignancy categorized, resulting in overfitting. Overall, several approaches demonstrated potential progress in lung nodule detection and classification, but still require significant improvement to overcome the challenging issues like detection of irregularly structured nodules from heterogeneous volumes of lung CT images with varying shape, size and location, differentiating vascular, solitary (non-solid or part-solid), pleural and juxta-pleural nodules with high accuracy and sensitivity, exhibiting robust methodologies applicable across different databases, and reduced time complexity on huge datasets as well.
The proposed work aims to automatically extract the self-learned features using an end-to-end learning CNN for lung cancer diagnosis based on their malignancy suspiciousness. The results are promising in terms of classification accuracy, sensitivity, specificity, and AUC with reduced false positives. The above issues are addressed with ROI-based feature learning with subjective analysis on annotated images from expert radiologists on huge datasets. In summary, Table 1 represents the key differences between the CNN method and state-of-the-art method. The methodology is highlighted with techniques and datasets used, and the results obtained are compared with other works.
3 Methodology
3.1 Data acquisition
Images used in the study are acquired from the Lung Image Database Consortium and Infectious Disease Research Institute (LIDC-IDRI) publically available repository, consisting of diagnostic and lung cancer screening thoracic CT scans with marked-up annotated lesions [7, 11, 24]. A total of 1018 cases from seven academic centers and eight medical imaging companies are collaborated to create the LIDC dataset with the scan slice thickness varying from 1.25 to 3 mm, indicating the malignancy suspiciousness from levels 1 to 5. CT scan images are pre-processed to uniquely segment the NROI in correspondence to four radiologists’ annotations and markings. The XML files associated with the CT scan images have annotations evaluated by four expert radiologists. Each radiologist reviewed and labeled the nodules/lesions to one of the three key categories: nodule greater than or equal to 3 mm, non-nodule larger than 3 mm, and nodule less than 3 mm. The retrieved Digital Imaging and Communications in Medicine (DICOM) images are \(512\times 512\) dimensions in size. Each individual annotations are read from the XML files, and their corresponding locations in DICOM images are traced and cropped. The dataset images are segmented based on these annotations in correspondence to the malignancy levels ranging from level 1 to 5, extracting the nodule area in each slice into a \(52\times 52\) pixel rectangle. The segmented candidate nodules are fitted into a \(52\times 52\) rectangle and are converted into a TIF image format for easier processing [30]. The extracted ROI is rescaled, translated in a range of [− 3 3], and rotated to three different angles (90, 180, 270) forming a training set of images. Figure 2 shows the original CT scan image, extracted nodule ROI rectangle of \(52\times 52\) pixels by four expert radiologists and the corresponding ground-truth values.
In the study, 5188 ROI samples with an augmented image data store are used as a training set as suggested by [52] eliminating the intermediate samples having level 3 malignancy, level 1 and 2 samples combined to form the benign samples (low malignancy nodules), level 4 and level 5 samples combined to form malignant samples (high malignancy nodules), and nodules less than 3 mm and non-nodules greater than 3 mm combined forming non-cancerous samples, with each sample containing 2704 pixels. Most of the methods discussed in related works have eliminated the nodules with malignancy 3 level and nodules having ambiguous ids. Few methods have categories like nodules and non-nodules for classification. Few methods have categories like low malignancy nodules (LMNs) and high malignancy nodules(HMNs) as shown in Table 1. For an accurate comparison, similar datasets and training metrics are followed in our study as well.
System specification—The proposed methodology is run on Matlab 2018b version on a desktop machine with a memory of 8 GB, 12(4C and 8G) core AMD A10 processor and an Nvidia GeForce GTX 960 GPU enabled to analyze the results of the proposed method.
3.2 Pre-processing
The raw CT scan images are pre-processed to improve the quality and more often reduce the noisy artifacts that occurred during the image acquisition process. In addition, few algorithms incorporate pre-processing as an important component to enhance the detection ability, reduce noisy artifacts, and optimize the input information for further processing. Some of the pre-processing steps adopted by many researchers are the contrast stretching [25], discrete wavelet transforms [2, 15], Hough transform [41], thresholding [19], morphological operations [46], unsharp mask technique [32], smoothing and median filtering [9, 26]. In the study, we used the schema suggested by [2] using discrete wavelet transforms as a pre-processing step to enhance the input CT images. The images are decomposed into 4 frequency sub-bands at different scales using the low-pass and high-pass filters by applying Daubechies filters. Daubechies filters achieve the perfect reconstruction of the original signal when compared to other filters. These filters help in identifying the sudden changes in intensities in detail with respect to the original image. The LL band represents information about light illumination. The LH, HL, HH bands represent the intensity values of edges. In order to enhance the intensity values of edges, unsharp energy mask (UEM) is employed on LH, HL, HH bands. Finally, all 4 frequency sub-bands are reconstructed using inverse DWT, resulting in the enhanced original image.
The mathematical representation of high-pass filters and low-pass filters are defined, respectively, as
where \(Y_{\mathrm{high}}[n]\)—output of high-pass filters and \(Y_{\mathrm{low}}[n]\)—output of low-pass filters.
The two-level high-pass and low-pass wavelet decomposition can be expressed in a combined way reconstructing the high-contrast CT image and representing low energy of each band as e(b)
e(b) is the low energy of each sub-band and is calculated as
and the final cost of low level energy (UEM) is expressed as
3.3 Method
The study involves the design and implementation of a convolutional neural network architecture for learning the feature maps of images for lung nodule classification as benign, malignant, or non-cancerous. In addition, GANs are employed to generate additional images with similar characteristics as pulmonary nodules. CNN’s are trained using images generated by GAN and are fined-tuned with actual input samples to differentiate and classify the lung nodules based on the classification strategy. For comparison, the traditional handcrafted features are also extracted from the categories—texture, density, and morphological features to classify the pulmonary nodules using SVM classifiers.
3.3.1 Convolutional neural network
One of the deep neural network algorithms used in our study is the convolutional neural network based on LeCun’s model. CNNs are the most powerful, commonly used neural networks designed for applications having inputs with an inherent two-dimensional structure like images. CNN’s are end-to-end learning algorithms having several convolutional layers and sub-sampling layers, followed by a fully connected layer with each layer having a topographic structure [31]. The augmented ROI images with the size of \(52\times 52\) pixels mentioned above are fed as the input to the input layer. The size of the sub-patch/region passed as input is referred to as “receptive field,” that is, the region space that a particular CNN’s feature is being extracted for (the input from the previous layer).
Figure 3 shows the architecture of the proposed CNN algorithm. The architecture contains a total of 8 layers: The first and last layer forms the input and the output layer, respectively, and the layers 2, 4, and 6 are the convolutional layers, and 3, 5, and 7 are the sub-sampling layers intercepted with max-pooling, ReLu, and batch normalization for salient feature extraction, and the last layer before the output layer is the fully connected layer that uses softmax function connected to 3 neurons as output layer, classifying an input image into one of three classes categorized. Convolution helps to extract the salient features from the input patches preserving the spatial relationships among pixels. In particular, the design details are as follows: The second layer has 12 filters of size \(5\times 5\) (feature maps) connected to the input image, followed by a max-pooling layer of \(2\times 2\). The pooling layer performs down-sampling along the width and height of the convolved image. Max-pooling reduces the computational cost as the dimensionality of the features maps reduces and helps the neural network remain unchanged to any translations, distortions, and small transformations on the sampled input patches. The number of output feature maps in each dimension (width, height) can be calculated using Eq. 6. The fourth layer has 8 feature maps connected to the previous layer (small region connected to previous layers \(12\times 8 = 96\)) through 96 \(5\times 5\) filters. There exists one more max-pooling layer, followed by the sixth layer having 6 feature maps, and 48 \(5\times 5\) filters were used from the previous layer (\(8\times 6 = 48\)). The last layer before the output layer (eight layers), which is the fully connected layer, has the input shrunk to \(3\times 3\) matrices using softmax nonlinear functions having 3 output neurons that fall into one of the 3 categories of classes benign, malignant, or non-cancerous nodules. Fully connected implies that every neuron in one layer is connected to the same location at the other layers. As a result, each neuron receives input as linear combinations from its corresponding neurons with a set of input weights and bias in the previous layer as shown in Eq. 7. Finally, the output layer provides the strength of the network prediction for each possible category of classes. The output of each layer in the CNN architectures was normalized, whitened to enhance the contrast before it was sent to the next layer [23].
The entire training process of CNN is depicted in Fig. 4. A learning rate of 0.1 was used, the number of epochs to train was set to 50, the batch size was set to 100, and the sub-sampling rate was set constantly to 2. An adaptive moment estimation optimizer (Adam) is used in the research to optimize the performance of the neural network. Figure 5 shows the learning curve of the deep learning scheme using Adam optimizer. We also experimented using stochastic gradient descent with momentum (Sgdm) to optimize the performance of the network, but observed good results with Adam when compared to the Sgdm optimizer. The prediction results from the test data provide aggregate probability scores for nodule detection, reducing false positives while generating high sensitivity scores. Figure 6 shows the feature maps extracted from (a) first convolutional layer using Adam optimizer and (b) first convolutional layer using Sgdm optimizer, through the proposed CNN architecture. The learning rate of the Sgdm optimizer is shown in Fig. 7.
were \(F_{\mathrm{out}}\)—number of output feature maps, \(F_\mathrm{in}\)—number of input feature maps, k—kernel, s—stride, p—zero padding.
where \(v_{k}\)—\(k{\text {th}}\) output neuron, \(W_{kn}\)—weight connecting the input neurons \(u_{k}\) with \(v_{k}\), \(u_{k}\)—\(n{\text {th}}\) input neuron, \(b_{k}\)—bias term with \(v_{k}\).
3.3.2 Generating pulmonary nodules using GAN
Figure 8 depicts the architecture of GAN to generate new image datasets for performance evaluation. GAN comprises of two networks—generator and discriminator that train together. The structure of the generator is composed of 4 convolution layers. The generator network generates images from an array of 100 random numbers drawn from the input data and outputs an image of size \(64\times 64\) pixels. The convolution layer with the filter size of \(5\times 5\) is followed by sub-sampling layers intercepted with max-pooling, ReLu, and batch normalization. Since the output image from the generator is of size \(64\times 64\) pixels, to match with the input ROI of size \(52\times 52\) pixels, we performed down-sampling to fit the new nodule images into the ROI rectangle of the same size without any loss in information. The discriminator is composed of 4 convolution layers with the filter size of \(5\times 5\) and helps determine whether the given input image is fake or real. The GAN is trained using the Jensen–Shannon divergence expressing the distance between the probability scores. A learning rate of 0.01 was used, and the number of epochs to train was set to 500. An Adam optimizer is used in the research to optimize the performance of the neural network. In the study, 5000 images with pulmonary nodules are generated to conduct experiments.
3.3.3 Traditional handcrafted features
To have a comparison, the traditional handcrafted feature maps were extracted from the pre-processed samples. Traditional CADx systems with statistical, morphological, and texture-based descriptors discussed in the literature are extracted for nodule classification in the methodology. For extracting texture features, the images are decomposed into 4 frequency sub-bands at different scales using the low-pass and high-pass filters by applying Daubechies filters with level 2 decomposition. In the study, 20 features were extracted from the NROI based on three categories as described in Table 2. The pulmonary nodules are trained using SVM classifiers to distinguish the nodules as benign or malignant based on the suspicious malignancy level features extracted. SVM is straightforward supervised learning algorithms used for classifying categories of classes. The re-sampled \(52\times 52\) rectangle images from the LIDC datasets were analyzed by the existing SVM classifiers of the Matlab tool. SVM classifier uses a radial basis function as the kernel to standardize the input data for classifying the lung nodules. A range of values was calculated from the benign and malignant features for classifying the images according to levels of malignancy based on their corresponding probability scores. Using the posterior probability scores, the standard area under the receiver operating characteristic curve (AUC) is plotted.
4 Results
The main objective of the study is to evaluate if the CNN methodology can achieve better performance metrics in classifying the nodules as normal, benign, or malignant in comparison with the conventional feature extraction and classification methods. To compare the results of the CNN method with conventional state-of-art methods and traditional handcrafted methods, the re-sampled ROI extracted images were tested with the above-mentioned methods. We also conducted a detailed analysis of modeling the input data based on the nodule malignancy suspiciousness according to their uncertainty.
The CNN method was evaluated on 4 LIDC datasets as shown in Table 3. The classification of candidate nodules with the DS1 dataset was easy compared to the other 2 datasets. Datasets DS2 and DS3 having intermediate samples (malignancy level 3) made the CNN architecture to perform less when compared to the results obtained without these intermediate nodules. Therefore, the intermediate malignancy level 3 nodules were neither categorized as benign (LMNs) or malignant (HMNs) due to the ambiguity in differentiating the nodules. Hence, the intermediate malignancy level 3 samples were eliminated in the study to distinguish the pulmonary nodules better. We additionally tested DS4 considering these intermediate samples as separate categories forming 4 groups of classes. The resultant classification accuracy was 87.91% compared to DS1.
Later, to have a separate training and testing set of images with an equal number of image samples for each class of malignancy, tenfold cross-validation was applied to generate the training and testing folds. The experiment was carried in 2 phases: Phase 1: A training set of samples with GAN generated images (5000 samples) and Phase 2: A training set of samples with actual ROI images. Image sets from the training folds were used for training, while the ROIs from the testing folds were used for testing purposes. For the classification of candidate nodules, CNN was trained with GAN generated and actual ROI samples, respectively. Table 4 shows the classification accuracy of both the training datasets with a testing combination of input samples to see if the algorithm results in consistent behavior. From Table 4, CNN trained with GAN generated samples exhibited good classification accuracy compared to actual ROI pre-training. As the number of generated samples increases, a decrease in classification accuracy was observed. But with actual ROI trained CNN, a behavior with classification accuracy all above 91.9% was observed. In addition, we also conducted experiments with pre-trained models as depicted in Fig. 9. Among these methods, CNN with GAN generated images outperformed and ResNet50 produced better classification accuracy compared to other pre-trained models.
A systematic evaluation was conduct against different input parameters including the convolution filter sizes, learning rates, and the number of layers in the architecture. The performance of the algorithm was evaluated by calculating the ROI-based accuracy and AUC scores, and the testing results were significantly efficient. Table 5 shows the tested combinations of input parameters for the CNN architecture.
The CNN architecture with Adam training optimizer was tested upon different configurations of candidate parameters with performance values all above 88.5%, with a maximum classification accuracy of 93.46% as shown in Table 6 and AUC score of 0.9316. We also evaluated if there is any drastic change in performance measurements when the number of neurons varies in the hidden layers. But the variations in performance were quite stable and less than 0.4% for certain kernel size in correspondence to a particular epoch value. Finally, based on the experiments, we fixed the hidden layer’s neurons as [12, 8, 6] with the kernel size of [5, 5, 5] due to their relative stability while continuing our experiments. In addition, the experiments were repeated to train CNN using Sgdm optimizer having the same input size, the number of layers, and same convolution filters to evaluate its performance as shown in Table 7. As a result, the methodology achieved good promising results with Adam optimizer compared to Sgdm as shown in Fig. 10. The classification accuracy for all the tested combinations of input parameters using Sgdm optimizer was all above 88.1%, with a maximum value of 91.9% as highlighted bold.
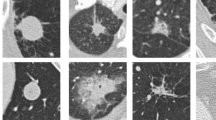
To compare the performance metrics of automatically generated salient features and the traditional handcrafted features, we tested the methods with the same ROI input samples. The classification accuracy of 85.36% and 77.5% was recorded by traditional CADx systems using the actual and GAN generated images, respectively. Overall, the automatically extracted features from LIDC datasets by deep learning procedures using CNN outperformed the conventional state-of-the-art methods as shown in Table 8 as well as traditional handcrafted methods as shown in Table 9. In most cases, traditional CADx methods evaluate nodules by considering the “size” as a characteristic feature to distinguish between benign or malignant but not taking into consideration large variations of nodule patterns. However, dependence on the nodule size led to misclassification of small candidate nodules as benign and large candidate nodules as malignant in some cases. Few ROI images as depicted in Fig. 11 exhibit good results by extracting their salient features manually, but are uncertain to the different patterns of nodules due to the sensitivity observed with small variations in the features.
In addition, some malignancy nodules may be false positively classified as non-nodules by radiologists in their reviews with similar morphological appearances due to their irregular shapes closer to the ribs, vessels, or walls of the chest or presence of noise. Therefore, critically analyzing and classifying malignancy nodules play an important role. Few cases observed in the study with the above-discussed criterion are depicted in Fig. 12. The methodology was successful in classifying these nodules as true nodules differentiating their intensity values. A consistent improvement in the performance of the CNN was observed by automatically learning the salient feature for differentiating vascular, solitary (non-solid or part-solid), pleural and juxta-pleural nodules with varying size, shape, and location compared to the conventional state-of-the-art methods. Non-cancerous lung nodule samples are generated from the benchmark (LIDC) datasets based on marked-up annotations by expert radiologists and are very well classified as a separate category in the methodology with reduced false positives without affecting the classification accuracy. The approach exhibits better results in classifying the candidate nodules as normal, benign, or malignant nodules when compared to other implementation approaches in Sect. 2. Figure 13 shows the confusion matrix of the proposed CNN with possible outcomes as true positive (TP), true negative (TN), false positive (FP), and false negative (FN) generated upon the tested data with average specificity and sensitivity of 92.8% and 93%, respectively, calculated using Eqs. 8–10. As a performance metric, the area under the receiver operating characteristic curve (AUC) was plotted as shown in Fig. 14 with the highest value of 0.9316. Also, the time complexity in training the CNN was 1.33 min on GPU mode, indicating an efficient computation time when compared to computation time more than hours on CPU mode.
5 Discussions
In the study, we proposed a deep structured algorithm to automatically extract the self-learned featured using an end-to-end learning CNN in diagnosing lung cancer CT images. A well-tuned CNN algorithm with GAN exhibited promising results when compared to actual ROI samples in terms of classification accuracy (93.9%), differentiating the benign, malignant, and non-cancerous lung nodules with low false positives. Some of the images generated using GAN may be dissimilar to actual lung nodules, blurred or even different from its resolution compared to the original images, henceforth are easily spotted by the human eye. Increasing the number of GAN generated samples decreased the classification accuracy of pulmonary nodules with respect to malignancy levels. As a result, CNN trained with the actual ROI dataset having images with real non-cancerous, benign, or malignant images categorized separately for classification exhibits almost the same results compared to GAN generated images while training. In the study, non-cancerous images segmented manually from the XML file of the LIDC dataset using radiologists annotation play a vital role similar to using fake lung images for performance evaluation. Since non-cancerous images are included in the actual ROI dataset, introducing fake lung nodules had no major effect on the classification accuracy.
In addition, the algorithm was compared with pre-trained models and traditional handcrafted methods as discussed in Sect. 4. A challenging task with traditional CADx systems was identifying and designing a set of features relevant to image datasets. The procedures in identifying features are time-consuming and may not guarantee good results if correlations between features were not properly considered. In the study, a set of features were manually computed as described in Sect. 3.3.2, resulting in the classification accuracy of 85.36% on a similar dataset, but are still uncertain to different image datasets due to the sensitivity observed with small variations in the features. However, deep learning methods can handle huge datasets and automatically generate computational features potential to the existing problem, sustainable and reliable to any changing situations.
As the deep learning algorithm performs an end-to-end learning procedure, the only input passed is the re-sampled ROI images. However, pre-processing the image datasets plays a vital role in our study enhancing the nodules for early detection of lung cancers. The prerequisites for the deep structured scheme include input data of the same size and a feasible procedure for pre-processing all the images in the datasets. The candidate nodules segmented differs in sizes having information concerning to nodule’s shape and its surroundings. Using the deep learning scheme, the information around the nodules structure along with its shapes and sizes was extracted efficiently at the same time. As a result, if the segmented ROI has a nodule of large size as input to train and test the network, it may result in increased dimensions of data, leading to more number of entries to the network. To avoid this criterion, the entire nodule with its information should be embedded in the ROI rectangle, if not down-sampling is performed. In case the nodules exceed the ROI rectangle size, we down-sampled the large nodules to fit into the ROI rectangle of size \(52\times 52\) pixels. Not all nodules are down-sampled as it incurs information loss. Meanwhile, the smaller nodules are retained without any loss in information making each nodule fit into ROI rectangle of the same size. Nodule size details must be taken into consideration as an important feature for pre-processing the data. This makes the deep structured scheme preserve the nodule size information for processing huge datasets and can be applied to other datasets in medical imaging for better computational efficiency.
6 Conclusion
In this paper, we visualized the convolutional neural network architecture with GAN generated and original image datasets and compared the performance metrics with the traditional CADx system’s texture, density, and morphological behavior. CNN with GAN generated images achieved good promising results in handling the challenging problem of malignancy classification at early stages with the highest classification accuracy among the other methods. Deep learning the features potentially increased the ability in capturing the salient information of the nodules with the limited number of layered structures in the method.
The following observations were made as to future works
-
1.
Although the results of the preliminary study are encouraging, we tested the images only to a limited number of deep learning layers. CNNs need to be further tested using deep layered structures on larger datasets and incorporate the extracted featured into the traditional state-of-the-art methods to further improve the performance metrics for lung cancer diagnosis.
-
2.
The optimal size of the input patch for deep learning algorithms is to be further investigated.
-
3.
Also features from 3D input data are to be extracted to train the CNN even though it could incur more network complexity.
-
4.
Although the ROI-based feature learning with CNN exhibits promising results, advancement in feature extractors from ROI segmented rectangle with an attention mechanism incorporated in CNN architecture can be further investigated.
-
5.
To further investigate, high-quality realistic generated fake lung nodules samples can be generated in addition to the current implementation using generative adversarial networks [10] as future work to train radiologists for learning discriminative features for educational purposes and improve the diagnostic decision making on cancer images.
References
American Lung Association Lung Cancer Fact Sheet (2019) https://www.lung.org/lung-health-and-diseases/lung-disease-lookup/lung-cancer/resource-library/lung-cancer-fact-sheet.html
Abbas Q (2017) Segmentation of differential structures on computed tomography images for diagnosis lung-related diseases. Biomed Signal Process Control 33:325–334
Al-Fahoum AS, Jaber EB, Al-Jarrah MA (2014) Automated detection of lung cancer using statistical and morphological image processing techniques. J Biomed Graph Comput 4(2):33
Amer HM, Abou-Chadi FE, Kishk SS, Obayya MI (2018) A computer-aided early detection system of pulmonary nodules in CT scan images. In: Proceedings of the 7th international conference on software and information engineering. ACM, pp 81–86
Arabasadi Z, Alizadehsani R, Roshanzamir M, Moosaei H, Yarifard AA (2017) Computer aided decision making for heart disease detection using hybrid neural network-genetic algorithm. Comput Methods Programs Biomed 141:19–26
Ardila D, Kiraly AP, Bharadwaj S, Choi B, Reicher JJ, Peng L, Tse D, Etemadi M, Ye W, Corrado G et al (2019) End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat Med 25(6):954
Armato SG III, McLennan G, Bidaut L, McNitt-Gray MF, Meyer CR, Reeves AP, Zhao B, Aberle DR, Henschke CI, Hoffman EA et al (2011) The lung image database consortium (LIDC) and image database resource initiative (IDRI): a completed reference database of lung nodules on CT scans. Med Phys 38(2):915–931
Arulmurugan R, Anandakumar H (2018) Early detection of lung cancer using wavelet feature descriptor and feed forward back propagation neural networks classifier. In: Computational vision and bio inspired computing. Springer, Berlin, pp 103–110
Bhuvaneswari P, Therese AB (2015) Detection of cancer in lung with k-NN classification using genetic algorithm. Proc Mater Sci 10:433–440
Chuquicusma MJ, Hussein S, Burt J, Bagci U (2018) How to fool radiologists with generative adversarial networks? A visual Turing test for lung cancer diagnosis. In: 2018 IEEE 15th international symposium on biomedical imaging (ISBI 2018). IEEE, pp 240–244
Clark K, Vendt B, Smith K, Freymann J, Kirby J, Koppel P, Moore S, Phillips S, Maffitt D, Pringle M et al (2013) The cancer imaging archive (TCIA): maintaining and operating a public information repository. J Digit Imaging 26(6):1045–1057
da Nóbrega RVM, Rebouças Filho PP, Rodrigues MB, da Silva SP, Júnior CMD, de Albuquerque VHC (2018) Lung nodule malignancy classification in chest computed tomography images using transfer learning and convolutional neural networks. Neural Comput Appl. https://doi.org/10.1007/s00521-018-3895-1
de Carvalho Filho AO, Silva AC, de Paiva AC, Nunes RA, Gattass M (2018) Classification of patterns of benignity and malignancy based on CT using topology-based phylogenetic diversity index and convolutional neural network. Pattern Recognit 81:200–212
El-Sherbiny B, Nabil N, El-Naby S.H, Emad Y, Ayman N, Mohiy T, AbdelRaouf A (2018) BLB (brain/lung cancer detection and segmentation and breast dense calculation). In: Deep and representation learning (IWDRL), 2018 first international workshop on. IEEE, pp 41–47
Fernandes SL, Gurupur VP, Lin H, Martis RJ (2017) A novel fusion approach for early lung cancer detection using computer aided diagnosis techniques. J Med Imaging Health Inf 7(8):1841–1850
Firmino M, Morais AH, Mendoça RM, Dantas MR, Hekis HR, Valentim R (2014) Computer-aided detection system for lung cancer in computed tomography scans: review and future prospects. Biomed Eng Online 13(1):41
Ghosh S, Dubey SK (2013) Comparative analysis of k-means and fuzzy c-means algorithms. Int J Adv Comput Sci Appl 4(4):35–39
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B, Warde-Farley D, Ozair S, Courville A, Bengio Y (2014) Generative adversarial nets. In: Advances in neural information processing systems, pp 2672–2680
Han H, Li L, Han F, Song B, Moore W, Liang Z (2015) Fast and adaptive detection of pulmonary nodules in thoracic CT images using a hierarchical vector quantization scheme. IEEE J Biomed Health Inf 19(2):648–659
Hansell DM, Bankier AA, MacMahon H, McLoud TC, Muller NL, Remy J (2008) Fleischner society: glossary of terms for thoracic imaging. Radiology 246(3):697–722
Hochhegger B, Zanon M, Altmayer S, Pacini GS, Balbinot F, Francisco MZ, Dalla Costa R, Watte G, Santos MK, Barros MC et al (2018) Advances in imaging and automated quantification of malignant pulmonary diseases: a state-of-the-art review. Lung 196(6):633–642
Hu J, Shen L, Sun G (2018) Squeeze-and-excitation networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 7132–7141
Hyvärinen A, Oja E (2000) Independent component analysis: algorithms and applications. Neural Netw 13(4–5):411–430
Jacobs C, van Rikxoort EM, Murphy K, Prokop M, Schaefer-Prokop CM, van Ginneken B (2016) Computer-aided detection of pulmonary nodules: a comparative study using the public LIDC/IDRI database. Eur Radiol 26(7):2139–2147
Javaid M, Javid M, Rehman MZU, Shah SIA (2016) A novel approach to CAD system for the detection of lung nodules in CT images. Comput Methods Programs Biomed 135:125–139
Kalpana V, Rajini G (2016) Segmentation of lung lesion nodules using dicom with structuring elements and noise—a comparative study. In: Electrical, computer and electronics engineering (UPCON), 2016 IEEE Uttar Pradesh Section international conference on. IEEE, pp 252–257
Kaul C, Manandhar S, Pears N (2019) Focusnet: an attention-based fully convolutional network for medical image segmentation. arXiv preprint arXiv:1902.03091
Korkmaz S.A, Akçiçek A, Bínol H, Korkmaz M.F (2017) Recognition of the stomach cancer images with probabilistic hog feature vector histograms by using hog features. In: Intelligent systems and informatics (SISY), 2017 IEEE 15th international symposium on. IEEE, pp 000339–000342
Kuruvilla J, Gunavathi K (2014) Lung cancer classification using neural networks for CT images. Comput Methods Programs Biomed 113(1):202–209
Lampert TA, Stumpf A, Gançarski P (2016) An empirical study into annotator agreement, ground truth estimation, and algorithm evaluation. IEEE Trans Image Process 25(6):2557–2572
LeCun Y, Bottou L, Bengio Y, Haffner P (1998) Gradient-based learning applied to document recognition. Proc IEEE 86(11):2278–2324
Li C, Zhu G, Wu X, Wang Y (2018) False-positive reduction on lung nodules detection in chest radiographs by ensemble of convolutional neural networks. IEEE Access 6:16060–16067
Lindsay W, Wang J, Sachs N, Barbosa E, Gee J (2018) Transfer learning approach to predict biopsy-confirmed malignancy of lung nodules from imaging data: a pilot study. In: Image analysis for moving organ, breast, and thoracic images. Springer, Berlin, pp 295–301
Lu L, Yapeng L, Hongyuan Z (2018) Benign and malignant solitary pulmonary nodules classification based on CNN and SVM. In: Proceedings of the international conference on machine vision and applications. ACM, pp 46–50
Makaju S, Prasad P, Alsadoon A, Singh A, Elchouemi A (2018) Lung cancer detection using CT scan images. Proc Comput Sci 125:107–114
Manikandan T, Bharathi N (2016) A survey on computer-aided diagnosis systems for lung cancer detection. Int Res J Eng Technol 3(5):1562–70
Masood A, Sheng B, Li P, Hou X, Wei X, Qin J, Feng D (2018) Computer-assisted decision support system in pulmonary cancer detection and stage classification on CT images. J Biomed Inf 79:117–128
Nguyen T, Khosravi A, Creighton D, Nahavandi S (2015) Classification of healthcare data using genetic fuzzy logic system and wavelets. Expert Syst Appl 42(4):2184–2197
Nishio M, Sugiyama O, Yakami M, Ueno S, Kubo T, Kuroda T, Togashi K (2018) Computer-aided diagnosis of lung nodule classification between benign nodule, primary lung cancer, and metastatic lung cancer at different image size using deep convolutional neural network with transfer learning. PLoS ONE 13(7):e0200721
Obayya M, Ghandour M (2015) Lung cancer classification using curvelet transform and neural network with radial basis function. Int J Comput Appl 120(13):33–37
Orozco HM, Villegas OOV, Sánchez VGC, Domínguez HdJO, Alfaro MdJN (2015) Automated system for lung nodules classification based on wavelet feature descriptor and support vector machine. Biomed Eng Online 14(1):9
Ozekes S, Osman O (2010) Computerized lung nodule detection using 3D feature extraction and learning based algorithms. J Med Syst 34(2):185–194
Rastegari M, Ordonez V, Redmon J, Farhadi A (2016) Xnor-net: imagenet classification using binary convolutional neural networks. In: European conference on computer vision. Springer, Berlin, pp 525–542
Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers RM (2015) Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 35(5):1170–1181
Roth HR, Lu L, Liu J, Yao J, Seff A, Cherry K, Kim L, Summers RM (2016) Improving computer-aided detection using convolutional neural networks and random view aggregation. IEEE Trans Med Imaging 35(5):1170–1181
Saad M.N, Muda Z, Ashaari N.S, Hamid H.A (2014) Image segmentation for lung region in chest X-ray images using edge detection and morphology. In: Control system, computing and engineering (ICCSCE), 2014 IEEE international conference on. IEEE, pp 46–51
Serj M.F, Lavi B, Hoff G, Valls D.P (2018) A deep convolutional neural network for lung cancer diagnostic. arXiv preprint arXiv:1804.08170
Setio AAA, Ciompi F, Litjens G, Gerke P, Jacobs C, Van Riel SJ, Wille MMW, Naqibullah M, Sánchez CI, van Ginneken B (2016) Pulmonary nodule detection in CT images: false positive reduction using multi-view convolutional networks. IEEE Trans Med Imaging 35(5):1160–1169
Shen W, Zhou M, Yang F, Yu D, Dong D, Yang C, Zang Y, Tian J (2017) Multi-crop convolutional neural networks for lung nodule malignancy suspiciousness classification. Pattern Recognit 61:663–673
Shi Z, Hao H, Zhao M, Feng Y, He L, Wang Y, Suzuki K (2019) A deep cnn based transfer learning method for false positive reduction. Multimed Tools Appl 78(1):1017–1033
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics. CA Cancer J Clin 68(1):7–30. https://doi.org/10.3322/caac.21442
Sun W, Zheng B, Qian W (2017) Automatic feature learning using multichannel roi based on deep structured algorithms for computerized lung cancer diagnosis. Comput Biol Med 89:530–539
Takahashi R, Kajikawa Y (2017) Computer-aided diagnosis: a survey with bibliometric analysis. Int J Med Inf 101:58–67
Tan J, Huo Y, Liang Z, Li L (2017) Apply convolutional neural network to lung nodule detection: recent progress and challenges. In: International conference on smart health. Springer, Berlin, pp 214–222
Team NLSTR (2011) Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 365(5):395–409
Wan S, Lee HC, Huang X, Xu T, Xu T, Zeng X, Zhang Z, Sheikine Y, Connolly JL, Fujimoto JG et al (2017) Integrated local binary pattern texture features for classification of breast tissue imaged by optical coherence microscopy. Med Image Anal 38:104–116
Wikipedia Contributors (2018) Cellular neural network—Wikipedia, the free encyclopedia. https://en.wikipedia.org/w/index.php?title=Cellular_neural_network&oldid=869201596. Accessed 31 Dec 2018
Wikipedia contributors (2018) Deep learning— Wikipedia, the free encyclopedia. https://en.wikipedia.org/w/index.php?title=Deep_learning&oldid=875207371. Accessed 31 Dec 2018
Woźniak M, Połap D, Capizzi G, Sciuto GL, Kośmider L, Frankiewicz K (2018) Small lung nodules detection based on local variance analysis and probabilistic neural network. Comput Methods Programs Biomed 161:173–180
Xie H, Yang D, Sun N, Chen Z, Zhang Y (2019) Automated pulmonary nodule detection in CT images using deep convolutional neural networks. Pattern Recognit 85:109–119
Zeng JY, Ye HH, Yang SX, Jin RC, Huang QL, Wei YC, Huang SG, Wang BQ, Ye JZ, Qin JY (2015) Clinical application of a novel computer-aided detection system based on three-dimensional CT images on pulmonary nodule. International J Clin Exp Med 8(9):16077
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to report this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Suresh, S., Mohan, S. ROI-based feature learning for efficient true positive prediction using convolutional neural network for lung cancer diagnosis. Neural Comput & Applic 32, 15989–16009 (2020). https://doi.org/10.1007/s00521-020-04787-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00521-020-04787-w