Abstract
Background
Peach (Prunus persica L.) is prone to chilling injury as exhibited by inhibition of the ethylene production, failure in softening, and the manifestation of internal browning. The basic leucine zipper (bZIP) transcription factors play an essential role in regulatory networks that control many processes associated with physiological, abiotic and biotic stress responses in fruits. Formerly, the underlying molecular and regulatory mechanism of (bZIP) transcription factors responsive to chilling injury in peach fruit is still elusive.
Methods and results
In the current experiment, the solute peach ‘Zhongyou Peach No. 13’ was used as the test material and cold storage at low temperature (4 °C). It was found that long-term low-temperature storage induced the production of ethylene, the hardness of the pulp decreased, and the low temperature also induced ABA accumulation. The changes of ABA and ethylene in peach fruits during low-temperature storage were clarified. Since the bZIP transcription factor is involved in the regulation of downstream pathways of ABA signals, 47 peach bZIP transcription factor family genes were identified through bioinformatics analysis. Further based on RT-qPCR analysis, 18 PpbZIP genes were discovered to be expressed in refrigerated peach fruits. Among them, the expression of PpbZIP23 and PpbZIP25 was significantly reduced during the refrigeration process, the promoter analysis of these genes found that this region contains the MYC/MYB/ABRES binding element, but not the DRES/CBFS element, indicating that the expression may be regulated by the ABA-dependent cold induction pathway, thereby responding to chilling injury in peach fruit.
Conclusions
Over investigation will provide new insights for further postharvest protocols related to molecular changes during cold storage and will prove a better cope for chilling injury.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Peach (Prunus persica L. Batsch) is more imperative horticultural crop which developed the postharvest chilling injury, when fruits are stored in cold storage for long period [1, 2]. Under cold storage, a series of chilling injury (CI) symptoms like internal browning (IB), membrane fluidity and impairment of softening occur [3]. Formerly, several genes that responsive to cold have been identified and their involvement in cold signaling pathways [4, 5]. In rice, OsbZIP52 belong to bZIP genes which has confirmed his contribution in abiotic stress signaling and acts as negative regulator in cold response. Similarly, GmbZIP1 also enhanced the tolerance to cold, drought and salinity [6, 7]. Moreover, GmbZIP62, GmbZIP44 and GmbZIP78 genes have been proved their functional role as negative regulators in the signaling of ABA and respond to freezing tolerance in transgenic plants of Arabidopsis [8].
Plants face numerous stresses in this natural environment like chilling stress (CS), heat stress (HS) and freezing stress (FS) owing to unfavorable climate fluctuation. bZIP TFs affected the expression of DGD and accumulated the ABA in ‘CM’ fruit under cold storage, thus stimulated the DGDG content and increased the cold resistance of fruit. MYB TFs were engaged in response to LT and regulated the expression of downstream genes in pitted blueberry [9,10,11]. Under low temperature, TaHDZipI-5 indicated its role in cold tolerance during the reproductive stage. It is confirmed that the HD-ZIP I subfamily genes facilitate the resistance mechanism against cold stress by triggering the expression of the cell membrane-related proteins and AFP [12, 13]
Transcription factors (TFs) are more important which play a key role in genes regulation that also control the various decisive biological processes in plants. Characterization related to the function of these TFs is necessary for understanding the biological processes and transcriptional regulatory networks in which they are involved. Previously, at least 64 TFs families in the plant kingdom that have been identified. The basic leucine zipper (bZIP) transcription factor family is one of the most important, biggest and diverse [14]. These are categorized by 40–80 amino acid (bZIP) conserved domains. These domains have two specific structural features. First, there is a leucine zipper region which binds the dimerization motif with DNA [15, 16]. That region is more conserved which contain nearly 16 amino acids and remain with an invariant motif (N-x7-R/K). Second, basic leucine zipper has a motif of dimerization, which is less conserved. It is composed by repetition of heptad leucine residues or other bulky amino acids (hydrophobic).
In several plant genomes, basic leucine zipper (bZIP) domains genes has been comprehensively predicted or identified such as 75 (bZIP) genes has been found in Arabidopsis [17], 89 in Rice [18], 92 in Sorghum [19], 125 in Maize [20], 131 in Castor bean [21], 64 in Cucumber [22], 55 in Grape [23], 50 in Strawberry [24] and 112 in Apple [25]. Such studies provide the understanding related to their evolutionary and functional relationships on herbaceous plant species. Many studies shown that, basic leucine zipper (bZIP) TFs regulates several physiological process which provide resistance against numerous abiotic and biotic stresses, like as osmotic stress, drought and salinity [26,27,28] and low temperatures [29, 30]. They also regulate pathways related to, energy metabolism, nitrogen and carbon [19], sugar signaling, hormones and responses to light and pathogens [31,32,33].These factors are also involved in developmental processes including seed maturation, organ and tissue differentiation [17, 34], cell elongation [35] and embryonic and floral development [36, 37]. To the Authors’ knowledge, there is no systematic analysis of (bZIP) family under chilling injury in peach.
In the current study, we evaluated the physiological changes in the peach fruits under low temperature (4 °C). After that, we reported the 47 (bZIP) genes from the peach genome and examined their phylogenetic relationship, genes characteristics, chromosomal locations, proteins structure, conserved and functional protein motifs, cis-acting elements and their expression patterns induced by cold. Some (bZIP) genes revealed quite response to chilling injury.
Materials and methods
Plant material and postharvest treatment
The peach (Prunus persica L. Batsch) cultivar ‘CN13’ was grown in the experimental orchard that was situated in Zhengzhou Fruit Research Institute, Zhengzhou, China. ‘CN13’ was used as plant material which is a melting flesh (MF) cultivar. Fruits of this cultivar were harvested at maturity stage and stored in chamber at low temperature (4 °C). Fruits were harvested after every 5 days and stored in cold storage. We selected the five self-pollinated 7 years old trees. To conduct this experiment, three independent biological replicates were used, and 20 fruits were collected for each biological replicate at a single time point. Mesocarp tissues were sampled from the cold treated fruits, instantly frozen into the liquid nitrogen and stored at − 80 °C for extraction of RNA and subsequent gene expression analysis.
Estimation of physiological parameters and ABA content
To evaluate ethylene production, we incubated the individual peach fruits in 440 ml container for the period of 2 h. After that we withdraw 1 ml of headspace gas and injected into the gas chromatographic machine (Model-GC4 CMPF, Shimadzu, Kyoto, Japan), that equipped with an activated alumina column and flame ionization detector. During this procedure, the detection capacity of minimum ethylene production is 0.01 nl g−1 h−1. By using a penetrometer (model SMT-T-50, Toyo Baldwin, Tokyo, Japan), we measured the flesh firmness (N) at four equatorial regions of peeled flesh, that was fitted with an 8 mm plunger. Ethylene production and flesh firmness of treated fruits were monitored after every 5 days of interval during throughout storage period [38]. ABA extraction and content were determined by using UHPLC-ESI-MS/MS as described by [39]. Quantification was done by considering recovery rates for each sample by means of a deuterium-labeled internal standard.
Bioinformatic analysis of (bZIP) TFs family in peach
Phylogenetic analysis and classification of bZIP genes
We found the protein sequence of (bZIP) TFs genes from strawberry, these sequences were downloaded from GenBank (http://www.ncbi.nlm.nih.gov/Genbank/) and used to inquire the protein blast against the Prunus persica v2.1. Total 47 (bZIP) TFs genes were isolated through transcriptomic screening and those genes strongly changed their expression levels (log2 > 2) and subjected to further analysis. By using the Genome Database of Prunus persica v2.1, the protein sequences of (bZIP) TFs genes in peach were downloaded. In order to construct the phylogenetic trees of (bZIP) superfamily proteins in strawberry and peach, we utilized software (MEGA 5) in combination with Clustal W tool. Phylogenetic trees of (bZIP) genes of peach belong to different classes also constructed. During this process, we used the method of neighbor joining along with replicates of 1000 bootstrap [40].
Genes structure, motif analysis, chromosomal distribution and cis-acting elements
Gene Structure Display Server (GSDS) is a web-based bioinformatics tool (http://gsds.gao-lab.org/Gsds_about.php) that was used to construct (bZIP) TFs genes structure that including exons (CDS), intron numbers and upstream/downstream regions [41]. The motif location of (bZIP) TFs proteins was obtained by using online tool MEME (http://meme.nbcr.net/meme/), along with following parameters, we set the largest number of discovered and conserved motifs which was 10, and width of this motif was kept 30 to 50 aa. Settings of other remaining parameters were put as default. In order to determine the specific chromosomal positions of (bZIP) TFs genes of peach, we used BlastP search of the sequences of (Prunus persica. L) against the PHYTOZOME database (thttp://www.phytozome.net/peach.php) and set the settings as default. All the PpbZIP genes were mapped individually onto the basic eight peach chromosomes to show their physical position. These genes were plotted according to their ascending order, from the short-arm to the long-arm telomere and finally displayed by using Map Chart software 2.32. Cis-elements in the promoter region of bZIP genes determined by using the electronic webtool, New PLACE database (https://www.dna.affrc.go.jp/PLACE/?action=new place).
Extraction of RNA and first-strand cDNA synthesis
In order to extract the RNA from cold treated fruits of peach, we used the total RNA kit (Sangon, Shanghai, China) and followed the manufacturer’s instructions. After that, we observed the RNA degradation and contamination by running (1%) agarose gels. Moreover, according to the manufacturer’s instructions, an Agilent 2100 Bioanalyzer (Agilent Technologies, CA, USA) and Nano Drop ND-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA) were used to determine the quantity and quality of RNA. To carry out (qRT-PCR), we synthesized the cDNA from RNA by using the following M-MLV Reverse Transcriptase kit (Promega, USA). cDNA was further diluted to 20 ng ul-1323 described by [40].
Real-time quantitative reverse transcriptase-PCR analysis
qRT-PCR analysis was performed to investigate the (bZIP) TFs genes expression patterns induced by cold treatment in peach fruits We analyzed whole the data according to 2−ΔΔCt Method [42]. Three biological independent replicates were used to perform gene expression analysis of all the genes. For the reference gene, we used actin [43]. While the primers that was used during the (qRT-PCR) analysis are listed in (Supplementary file 1). The primers that was used for (qRT-PCR), design based on the nucleotide sequence of individual gene by using Primer 3. The range of the entire PCR products size was from 80 to 200 bp. The PCR amplification specificity was observed based on dissociation curves that was monitored throughout qPCR analysis by using a Roche Light Cycler 480 II (Roche, Switzerland).
Results
Physiological changes and deviation in ABA content under LTC (low temperature conditioning)
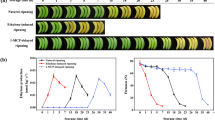
Measurable ethylene and flesh firmness were noticed in low temperature treated fruits throughout the period of cold storage. The production of ethylene after cold treatment was detected, which was at the level of (0.75 nl g−1 h−1) after 5 d and rapidly increased after 15 d in cold storage. Finally, reached at highest level of (57.11 nl g−1 h−1) after 40 d of cold treatment. Hence, significant level of ethylene induction was detected in cold storage fruits as compared to control. Flesh firmness in control fruits remained at highest level (47.65 N), which decreased rapidly in low temperature treated fruits and reached at lowest level (14.94 N) after 40 d of cold storage. Hence, these results confirmed that LTC (low temperature conditioning) at 4 °C significantly increased the induction of ethylene and rapidly decreased the rate of flesh firmness. ABA content sharply decreased from (10 to 40 d) of cold storage as compared to room temperature (Fig. 1).
Physiological changes in peach fruits at the time of harvest under low temperature; Ethylene production (nl/g−1 h−1) and flesh firmness (N). Fruits were transferred to 4 °C for 5d to 40d of cold storage. Data was collected after 5d intervals and represented as Mean ± SD of three biological replications. Determination of ABA contents at Room temperature (RT) and Low temperature (4 °C). ABA content sharply decreased from (10 to 40 d) of cold storage as compared to room temperature
Phylogenetic relationship of peach and strawberry (bZIP) genes
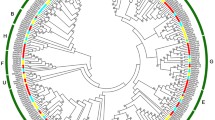
In different species, the dynamic method to understand the evolutionary history, origin and function of un-characterized genes is the comparison of the genomes. Since, a well-known study has been done on strawberry (bZIP) genes. The function of these genes also well-characterized. We used the full-length protein sequences of PpbZIP and mrnabZIP genes to construct an unrooted neighbor-joining (NJ) tree, which permitted the identification of 47 (bZIP) genes in peach. Ppbzip1/mrna01680, Ppbzip13/mrna13716, Ppbzip27/mrna18928 and Ppbzip29/mrna29546 had high homology of protein sequence in same clade, while Ppbzip2, Ppbzip24, Ppbzip31 and Ppbzip36 showed the similarity in another clade. Ppbzip14 and Ppbzip15 had high similarity in protein sequence with mrna04504, mrna29546, mrna11019 and mrna00517 in same clade. Ppbzip3/mrna29159, Ppbzip46/mrna23487, Ppbzip45/mrna03633 had homology that belonging to another clade. Ppbzip4, Ppbzip8, Ppbzip11, Ppbzip28 and Ppbzip38 had similarity with mrna30280, mrna09110 and mrna02177 in one clade. Similarly, Ppbzip5, Ppbzip40 and Ppbzip41 shared the same clade with mrna21344, mrna28103 and mrna17796 respectively. Ppbzip6 and Ppbzip47 revealed the homology with mrna30252, mrna08186 and mrna31322. While another clade had high similarity of Ppbzip7, Ppbzip22, Ppbzip30, Ppbzip42, Ppbzip43 with mrna21882, mrna23487, mrna32022, mrna11979, mrna07554. Ppbzip19/mrna21797, Ppbzip23/mrna03778, Ppbzip26/mrna11837, Ppbzip9/mrna31621, Ppbzip12/mrna08757, Ppbzip10/mrna22776, Ppbzip32/mrna31621, Ppbzip23/mrna03778, Ppbzip17/mrna11666, Ppbzip44/mrna32629 tended to cluster together in distinct clades. While, Ppbzip18/mrna21832, Ppbzip20/mrna15193, Ppbzip21/mrna14942, Ppbzip16/mrna02284, Ppbzip33/mrna21882, Ppbzip34/mrna14220, Ppbzip37/mrna28250, Ppbzip25/mrna09110 and Ppbzip39/mrna00393 had high homology of protein sequence in shared clades of phylogenetic tree (Supplementary Fig. 1). Phylogenetic tree of peach (bZIP) TFs was also constructed and classified the genes into further (I-VIII) sub-groups. Each PpbZIP gene belong to different groups including leucine zipper, CAMP response, G-Box, TGA, MYB, ABA insensitive, H-loop and HY5 proteins. Each group was assigned a different color according to well-known members in peach specie (Fig. 2).
Phylogenetic tree of peach (bZIP) proteins. The proteins were classified into eight distinct clusters. Each PpbZIP gene belong to different groups including leucine zipper (bZIP1-bZIP13), CAMP response (bZIP14-bZIP26), G-Box (bZIP27-bZIP30), TGA (bZIP31-bZIP35), MYB and ABA insensitive (bZIP36-bZIP40), H-loop and HY5 proteins (bZIP41-bZIP47). Each group was assigned a different color according to well-known members in peach specie. (Color figure online)
Genomic characterization of (bZIP) genes in peach
All the PpbZIP proteins characteristics were deliberated and listed in (Table 1), which including genome length, CDS length, protein length, exon number, molecular weight and theoretical pI. Genomic length of PpbZIPs from 441 (PpbZIP 17) to 13,282 (PpbZIP 34), CDS length varied from 429 (PpbZIP 16) to 2298 (PpbZIP 12). While protein length of PpbZIP had wide-ranging from 142 (PpbZIP 16) to 1266 (PpbZIP 37) aa with an average 364 aa. Similarly, range of exon number of all PpbZIP from 1 to 12. Meanwhile, the predicted molecular weights of PpbZIP proteins range from 2101.45 (PpbZIP 7) to 81,968.37 (PpbZIP 12) Da, with pI from 4.69 (PpbZIP 3) to 9.82 (PpbZIP 37).
Exon–intron organization, conserved protein motifs, chromosomal location and cis-regulatory elements of (bZIP) genes in peach
For better understanding of genomic structures evolution, exon–intron organization of 47 (bZIP) genes were examined. Schematic diagrams of the (bZIP) genes of peach were constructed by using the online tool GSDS utility that was presented in (Fig. 3). The members of (bZIP) TFs family have no. of introns, exons, and CDS sequence. 38 out of 47 PpbZIP genes have variable number of exons and nine genes have no intron in their ORF region. This indicated that exon–intron organization of PpbZIPs family and protein structure could be important during gene evolution process. Motif distribution remained helpful to insight the function and divergence of the PpbZIPs proteins, we captured the overall 10 conserved protein motifs by using online MEME software that was annotated with the Inter Pro database. All peach (bZIP) TFs genes have the basic domain of (leucine zipper) in motif 1 (Fig. 4). These results suggested that proteins contain analogous amino acid sequence. Further, these results also supported the phylogenetic analyses of PpbZIPs. There are total eight chromosomes in peach, all PpbZIP genes were physically mapped on these chromosomes. Among these, chromosome 1 contains the maximum no of PpbZIP (19.14%), while, the minimum number of genes (2.12%) were dispersed on chromosome 4. Certain physical regions with a relatively higher accumulation of gene clusters revealed by distribution pattern of the PpbZIP genes on individual chromosome. For example, PpbZIP genes which were located on chromosomes 2, 5, 6 and chromosomes 8 appear to be assembled at upper and lower end of the arms, respectively. The accurate position of every PpbZIP on the chromosome of peach was displayed (Fig. 5). Promoter sequence analysis of bZIP genes revealed ABA responsive cis-regulatory elements (MYB, MYC and ABRE recognition sites) and some other stress elements (Table 2). ABREs binding element contained conserved 5 base pair signal sequence (ACGTG), which had immense capacity to regulate the gene expression.
Distribution of 47 (bZIP) genes onto eight peach chromosomes. Chromosome 1 contains the maximum no of PpbZIPs (19.14%), while, the minimum number of genes (2.12%) were dispersed on chromosome 4. Physical locations for each PpbZIP gene belong to different groups including leucine zipper (bZIP1-bZIP13), CAMP response (bZIP14-bZIP26), G-Box (bZIP27-bZIP30), TGA (bZIP31-bZIP35), MYB and ABA insensitive (bZIP36-bZIP40), H-loop and HY5 proteins (bZIP41-bZIP47) on peach chromosomes (numbered 1–8) represented by graphical (scaled)
Expression of (PpbZIP) TFs genes induced by cold
To investigate the chilling responsive (bZIP) TFs family members, we performed the qRT-PCR analyses. We selected 18 distinct (bZIP) family members and assessed their expression levels during cold storage period. These results exposed that seven (bZIPs) genes were downregulated, two genes were upregulated while all other genes exhibited differential expression during cold storage period. Among them, expression of the Ppbzip20 (Prupe.1G374400), Ppbzip16 (Prupe.8G267100), Ppbzip23 (Prupe.1G455300), Ppbzip42 (Prupe.8G165400), Ppbzip44 (Prupe.8G026300), Ppbzip38 (Prupe.7G112200) and Ppbzip25 (Prupe.1G434500) revealed gradual reduction with progressing cold treatment. On the contrary, Ppbzip7 (Prupe.5G211200) was measured as up regulated during cold storage period. While Ppbzip1 (Prupe.1G369300), Ppbzip2 (Prupe.1G575800), Ppbzip3 (Prupe.5G003000), Ppbzip5 (Prupe.2G263700), Ppbzip11 (Prupe.8G232800), Ppbzip12 (Prupe.2G020400), Ppbzip13 (Prupe.5G027000), Ppbzip27 (Prupe.2G182800), Ppbzip33 (Prupe.7G037900), and Ppbzip29 (Prupe.7G150700) displayed diverse expression levels, first decreased to 20 d, then increased to 25 d and finally decreased till 45 d of cold treatment as shown in (Fig. 6). qRT-PCR results revealed that, the peach (bZIP) transcription factors were remarkably affected which exposed the altering expression levels in response to chilling temperature.
Cellular process induced by cold acclimation and molecular mechanism involved in regulation of genes responsive to chilling injury
Low temperature exposures generated serious stresses, which overcome by triggering the cascade of plant actions that cause alterations in expression of genes. Consequently, induced the modification in biochemical and some physiological processes which enhanced the stress tolerance. The process is called cold acclimation or chilling. Cold acclimation or chilling induced several process such as plant membrane modification (molecular or physiological), the activation of scavenger systems of (ROS) by increasing the ROS level, alteration of cold regulating and transcription factors (TFs) gene expression, increasing the level of cytosolic Ca2+, increasing the level of sugars and proline, increased the level of ABA and photosynthesis affected by these biochemical changes (Supplementary Fig. 2).
Plants response to cold (low temperature) goes through a certain process that initiated from cell recognition (low temperature sensor) and pass through signaling pathway, which induced the cold response genes. Low temperature sensors including membranes, elements of cytoskeletal, phytochromes, chromatin, specific proteins, DNA/RNA and sugars perceived signals and mediated the ABA dependent and ABA independent signaling pathways. In ABA independent pathway, CBF/DREB1 transcription factors binds with the regulatory signal sequence in the promoter of CBF (C-repeating binding factors), DREs (drought responsive elements) and LTREs (low temperature responsive elements) in cold regulating genes, which regulate the transcription and response to cold or chilling. While, in ABA dependent pathway, MYB/MYC/bZIPs families transcriptions factors bind with regulatory signal sequence in the promoter of ABREs or AREBs (ABA responsive elements) in cold response genes and improved the cold tolerance (Supplementary Fig. 2). Thus, ABA dependent and independent signaling pathways, regulatory components and their interactions help us to understand the regulatory mechanism of chilling responsive genes.
Cold storage induced the physiological disorder CI in peaches by membrane modification (physiological), changes in the expression of genes and protein synthesis (molecular). Several transcription factor TFs families such as DREB, MYB, ERF, WRKY, bHLH, ZF and bZIP are induced as response to cold stress which either developed the cold acclimation or stabilize other cold associated physiological disorders. Stability of the membrane and metabolic responses which controlled the chilling injury under cold storage, regulated by bZIP genes which contain ABRE or AREBs binding factors, which had immense capacity to up or downregulate the expression of genes through ABA dependent signaling pathway. Thus, increased the tolerance against chilling injury in peach fruit.
Discussion
Low temperature alleviates CI (chilling injury) and permits the normal fruit softening in peach
Cold storage is one of the most valuable postharvest technology which we used to extent the shelf life of the fresh horticultural produce. Formerly, cold storage reduces the most cell metabolic activities. Thus, fruit ripening and plants senescence become delay [44]. Low temperature treatment produces many physiological disorders in chilling sensitive fruits. These disorders including dry or wooly texture, tissue browning and abnormality in cell metabolism like membrane permeability related disorders which cause chilling injury [45]. Previously, the fruits stored at 2.5 °C developed 98% of LTB (low temperature breakdown) symptoms [46]. During this study, we observed no LTB symptoms during cold storage in peach. Under LTC (low temperature conditions), fruits indicated the increased in ethylene production, along with losing of original flesh firmness by alteration of cell wall structure. Peach fruits stored at 4 °C rapidly decreased the softening rate. These results are consistent with previous findings in many species that reported by [1, 47,48,49,50]. Therefore, Low temperature (4 °C) seemed valuable measure to alleviate the chilling injury and permitted the normal process of peach fruit softening during the period of cold storage. Moreover, Endogenous ABA concentrations gradually decreased in peach fruits during post-harvest at 4 °C. Thus, cold treatment led to an inhibition of over-ripening. These results are similar in sweet cherries [51]. Low temperature decreased the ABA concentration along with ethylene which inhibited the development of pulp browning. Consequently, taste was enhanced in peach [52]. While in kiwifruit, LTC alleviated the chilling injury with improving the activities of antioxidant enzyme and maintaining higher levels of endogenous ABA [53].
Evolutionary process of (PpbZIP) transcription factors family
There are substantial evidences that genes of (bZIP) family are the key regulators of several crucial growth, developmental, ripening and numerous physiological processes of plants [54], several biological, cellular processes as well as response to biotic and abiotic stresses [55]. Although, there are many plant species in which the (bZIP) TFs family has been reported. While in peach fruit, the genome-wide documentation of (bZIPs) TFs family has not been formerly reported. In this study, 47 (bZIP) TFs genes of this family were detected in the genome of peach (Prunus persica). Based on their evolutionary relationship of phylogenetic, they were classified into eight sub-groups, which was consistent with maize [20], Arabidopsis [17], strawberry [24], B. distachyon [56] and Apple [25]. The phylogenetic analysis accomplished that was supported by both structural analyses (Exon–intron and conserved motif prediction).
The genes structure analysis indicated that PpbZIPs comprised no. of exons that varying range from 1 to 12 with the similar exon–intron structure organizations. PpbZIP gene structure features have also been observed in various species, such as grapes [57], Apple [25], Strawberry [24] and sorghum [56]. All the PpbZIPs contain the typical (bZIP) domains that suggested by conserved motif analysis. Conserved protein motifs like features have been noticed in some former plant species like as cassava and grapes [57, 58]. Both structural analyses indicated that all PpbZIP genes had similar exon–intron structure organizations and conserved protein motifs which suggesting that peach (bZIPs) TFs had a very close relationship through their gene evolution process and had similar function due to analogous conserved domains.
bZIP genes response to chilling injury in Peach
In fruits, cold temperature responses involve numerous crosstalk with the process of senescence and ripening at physiological, cellular, biochemical and molecular level [59]. Peach fruit is sensitive to chilling, which causes the symptoms of CI like IB along with failure of fruit softening process and prohibition of ethylene production [3, 60].There are some evidence about (bZIP) proteins that has been suggested that they were widely involved during signaling and abiotic/biotic stimuli responses. [18, 61, 62]. But, there is limited information related to (bZIP) family and their involvement on chilling responses in peach fruit. In order to investigate the (bZIP) genes of peach and their potential involvement in cold responses. We observed the expression of most important genes induced cold. Expression patterns of genes can provide crucial evidences for gene function. During this study, Group I (basic leucine zipper) including Ppbzip1, Ppbzip2, Ppbzip3, Ppbzip5, Ppbzip11, Ppbzip12 and Ppbzip13 had differential expression while Ppbzip7 upregulated and respond to cold stress. These results are consistent with previous findings reported in rice, in which large number of bZIP genes including OsbZIP73, OsbZIP38, OsbZIP87 and OsbZIP52 regulated positively or negatively by cold temperature [29, 30, 63]. Furthermore, Ppbzip20, Ppbzip16, Ppbzip23, Ppbzip25 and Ppbzip38 transcription factors belong to Group II (Camp response binding Factor) and Group V (ABA and MYB insensitive protein) respectively, were downregulated with progressive cold treatment and showed an agreement with previous investigation which revealed that OsMYBS3 and OsMYB3R2 induced under cold stress in rice and suppressed during well-known cold signaling pathway [64]. While, Group III (G-Box binding factors) and Group IV (TGA transcription factors) contained Ppbzip27, Ppbzip29 and Ppbzip33 transcription factors which respond to cold stress and had different expressions during cold storage period respectively. These findings were similar with inquiry in rice which indicated that bZIP factors including GCC-box and TGA-like elements through various regulatory enrichment clusters involved in chilling response during oxidative signaling (ROS) pathway [65]. Transcript level of Ppbzip42 and Ppbzip44 decreased under chilling period that transcription factors belong to Group VI (H-loop) and Group VII (HY5). Such results matched with previous findings in Arabidopsis [66] and tomato [67] which indicated that SlHY5 in a Phytochrome expressed under cold stress and inhabit the tomato growth and also induce cold tolerance. While, MdHY5 also modulated cold tolerance positively in Apple through CBF- pathways [68].
Additionally, in transcriptional regulatory network, DREBs/CBFs binding and regulatory factors play an essential role to regulate the ABA independent pathway. Such as, in wheat, TaCBF14 and TaCBF15 were involved in cold signaling pathway and enhanced the cold tolerance in transgenic plants [69]. While in fruits species, [70] reported the five CBFs genes in peach and overexpression of PpCBF1gene in apple enhanced the level of cold tolerance. While, AREBs/ABREs binding elements facilitates to regulate the ABA dependent pathway and respond to cold regulating genes under chilling stress [71]. The expression of genes in ABA dependent pathway is regulated by MYC, MYB and bZIP (ABRE or AREBs binding factors) transcription factors. For example, OsbZIP52 and OsbZIP38 genes in rice responded to drought and chilling stress through signaling pathways of cold [29]. Both dependent and independent cold acclimation pathways facilitate to regulate the chilling responsive genes. While, ABA plays a function synergistically under cold signal [72]. bZIP TFs has been proven to contribute in ABA-dependent signaling pathway in some plants in response to several abiotic stress (cold, drought and salinity) like Rice [73], Arabidopsis thaliana [74], Barley [75] and Wheat [76]. In Arabidopsis, AtbZIP1 transcription factor regulates the ABA signaling by binding factor ABREs, which alter the expression of chilling responsive genes and provide the tolerance against chilling stress [77]. Chilling and ABA inducible bZIP gene, ABF1 binding factor has been cloned in Arabidopsis. Zinc finger protein (C2H2‐type) triggers bZIP transcription factor which regulates the expression of chilling responsive genes by ABRE elements. Overexpression of these protein from soybean to Arabidopsis provides the tolerance against chilling [78]. CsbZIP18 was a gene of tea plant that overexpressed in Arabidopsis, which played a role as negative regulator and decreased the freezing tolerance via ABA dependent signaling pathway under cold or chilling stress [79]. In soybean, SGBF1 belongs to bZIP subfamily (G‐Box‐binding factor 1) which induced the expression of chilling responsive geneses in an ABA dependent pathway and enhanced the DNA‐binding activity during cold acclimation and respond to chilling related physiological disorders [80]. Overall, it ponders that bZIP transcription factor genes induced as response to cold stress, which either developed the cold acclimation or alleviated other cold associated physiological disorder (chilling injury).
Conclusions
In conclusion, physiological and molecular aspects respond to chilling injury in peach fruit during the period of cold storage were monitored. Peach fruit is susceptible to CI as exhibited by inhibition of the ethylene production. Low temperature (4 °C) increased the production of ethylene and rapidly decreased the rate of flesh firmness with no LTB (Low temperature breakdown). Even so, ABA content sharply decreased by low temperature which led to inhibition of over ripening. In PpbZIP family, Ppbzip23 and Ppbzip25 are candidate genes which triggered under cold stress by ABA dependent cold signaling pathway. These verdicts revealed that these bZIP genes response to cold stress and reduced the CI, which permitted the normal process of peach fruit softening under low temperature. In near future, we will clarify the function of chilling responsive (bZIP) genes through molecular cloning. Consequently, these identified genes can be used as a road map to validate with other cultivars of peach that respond to chilling injury, which will prove the first step for postharvest technological applications.
Data availability
All supporting data is available within the text and supplementary files. Further queries can be directed to corresponding author.
References
Wang K, Yin XR, Zhang B, Grierson D, Xu CJ, Chen KS (2017) Transcriptomic and metabolic analyses provide new insights into chilling injury in peach fruit. Plant Cell Environ 40(8):1531–1551
Wang L, Shan T, Xie B, Ling C, Shao S, Jin P, Zheng Y (2019) Glycine betaine reduces chilling injury in peach fruit by enhancing phenolic and sugar metabolisms. Food Chem 272:530–538
Lurie S, Crisosto CH (2005) Chilling injury in peach and nectarine. Postharvest Biol Technol 37(3):195–208
Park S, Lee CM, Doherty CJ, Gilmour SJ, Kim Y, Thomashow MF (2015) Regulation of the Arabidopsis CBF regulon by a complex low-temperature regulatory network. Plant J 82(2):193–207
Zhu J, Dong CH, Zhu JK (2007) Interplay between cold-responsive gene regulation, metabolism and RNA processing during plant cold acclimation. Curr Opin Plant Biol 10(3):290–295
Gao SQ, Chen M, Xu ZS, Zhao CP, Li L, Xu HJ, Tang YM, Zhao X, Ma YZ (2011) The soybean GmbZIP1 transcription factor enhances multiple abiotic stress tolerances in transgenic plants. Plant Mol Biol 75(6):537–553
Kim JB, Kang JY, Kim SY (2004) Over-expression of a transcription factor regulating ABA-responsive gene expression confers multiple stress tolerance. Plant Biotechnol J 2(5):459–466
Liao Y, Zou HF, Wei W, Hao YJ, Tian AG, Huang J, Liu YF, Zhang JS, Chen SY (2008) Soybean GmbZIP44, GmbZIP62 and GmbZIP78 genes function as negative regulator of ABA signaling and confer salt and freezing tolerance in transgenic Arabidopsis. Planta 228(2):225–240
Zhang Y, Raza A, Huang H, Su W, Luo D, Zeng L, Ding X, Cheng Y, Liu Z, Li Q (2022) Analysis of Lhcb gene family in rapeseed (Brassica napus L.) identifies a novel member “BnLhcb3. 4” modulating cold tolerance. Environ Exp Bot 198:104848
Raza A, Tabassum J, Kudapa H, Varshney RK (2021) Can omics deliver temperature resilient ready-to-grow crops? Crit Rev Biotechnol 41(8):1209–1232
Wang Z, Wang Y, Li A, Miao Y, Deng L, Wang H, Meng J, Liu H, Niu L, Pan L (2022) Transcriptome, lipidome, and phytohormone analyses elucidate the mechanism of chilling injury during postharvest cold storage in different cold-sensitive peach fruit. Available at SSRN 4085272
Sharif R, Raza A, Chen P, Li Y, El-Ballat EM, Rauf A, Hano C, El-Esawi MA (2021) HD-ZIP gene family: potential roles in improving plant growth and regulating stress-responsive mechanisms in plants. Genes 12(8):1256
Zahra N, Shaukat K, Hafeez MB, Raza A, Hussain S, Chaudhary MT, Akram MZ, Kakavand SN, Saddiq MS, Wahid A (2021) Physiological and molecular responses to high, chilling, and freezing temperature in plant growth and production consequences and mitigation possibilities. Harsh Environment and Plant Resilience. Springer, Cham, pp 235–290
Pérez-Rodríguez P, Riano-Pachon DM, Corrêa LGG, Rensing SA, Kersten B, Mueller-Roeber B (2010) PlnTFDB: updated content and new features of the plant transcription factor database. Nucleic Acids Res 38(suppl_1):D822–D827
Talanian RV, McKnight CJ, Kim PS (1990) Sequence-specific DNA binding by a short peptide dimer. Science 249(4970):769–771
Hurst HC (1995) Transcription factors 1: bZIP proteins. Protein Profile 2(2):101–168
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7(3):106–111
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146(2):333–350
Wang J, Zhou J, Zhang B, Vanitha J, Ramachandran S, Jiang SY (2011) Genome-wide expansion and expression divergence of the basic leucine zipper transcription factors in higher plants with an emphasis on Sorghum F. J Integr Plant Biol 53(3):212–231
Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D (2012) Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19(6):463–476
Jin Z, Xu W, Liu A (2014) Genomic surveys and expression analysis of bZIP gene family in castor bean (Ricinus communis L.). Planta 239(2):299–312
Baloglu MC, Eldem V, Hajyzadeh M, Unver T (2014) Genome-wide analysis of the bZIP transcription factors in cucumber. PLoS ONE 9(4):e96014
Gao M, Zhang H, Guo C, Cheng C, Guo R, Mao L, Fei Z, Wang X (2014) Evolutionary and expression analyses of basic zipper transcription factors in the highly homozygous model grape PN40024 (Vitis vinifera L.). Plant Mol Biol Report 32(5):1085–1102
Wang XL, Chen X, Yang TB, Cheng Q, Cheng ZM (2017) Genome-wide identification of bZIP family genes involved in drought and heat stresses in strawberry (Fragaria vesca L). Int J Genom. https://doi.org/10.1155/2017/3981031
Zhao J, Guo R, Guo C, Hou H, Wang X, Gao H (2016) Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front Plant Sci 7:376
Weltmeier F, Ehlert A, Mayer CS, Dietrich K, Wang X, Schütze K, Alonso R, Harter K, Vicente-Carbajosa J, Dröge-Laser W (2006) Combinatorial control of Arabidopsis proline dehydrogenase transcription by specific heterodimerisation of bZIP transcription factors. EMBO J 25(13):3133–3143
Xiang Y, Tang N, Du H, Ye H, Xiong L (2008) Characterization of OsbZIP23 as a key player of the basic leucine zipper transcription factor family for conferring abscisic acid sensitivity and salinity and drought tolerance in rice. Plant Physiol 148(4):1938–1952
Ying S, Zhang D-F, Fu J, Shi Y-S, Song Y-C, Wang T-Y, Li Y (2012) Cloning and characterization of a maize bZIP transcription factor, ZmbZIP72, confers drought and salt tolerance in transgenic Arabidopsis. Planta 235(2):253–266
Liu C, Wu Y, Wang X (2012) bZIP transcription factor OsbZIP52/RISBZ5: a potential negative regulator of cold and drought stress response in rice. Planta 235(6):1157–1169
Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T (2005) LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol 46(10):1623–1634
Nieva C, Busk PK, Domínguez-Puigjaner E, Lumbreras V, Testillano PS, Risueño MC, Pagès M (2005) Isolation and functional characterisation of two new bZIP maize regulators of the ABA responsive gene rab28. Plant Mol Biol 58(6):899–914
Thurow C, Schiermeyer A, Krawczyk S, Butterbrodt T, Nickolov K, Gatz C (2005) Tobacco bZIP transcription factor TGA2. 2 and related factor TGA2. 1 have distinct roles in plant defense responses and plant development. Plant J 44(1):100–113
Ulm R, Baumann A, Oravecz A, Máté Z, Ádám É, Oakeley EJ, Schäfer E, Nagy F (2004) Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci U S A 101(5):1397–1402
Silveira AB, Gauer L, Tomaz JP, Cardoso PR, Carmello-Guerreiro S, Vincentz M (2007) The Arabidopsis AtbZIP9 protein fused to the VP16 transcriptional activation domain alters leaf and vascular development. Plant Sci 172(6):1148–1156
Yin Y, Zhu Q, Dai S, Lamb C, Beachy RN (1997) RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J 16(17):5247–5259
Guan Y, Ren H, Xie H, Ma Z, Chen F (2009) Identification and characterization of bZIP-type transcription factors involved in carrot (Daucus carota L.) somatic embryogenesis. Plant J 60(2):207–217
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66(6):675–683
Zeng W, Tan B, Deng L, Wang Y, Aslam MM, Wang X, Qiao C, Wang Z, Feng J (2020) Identification and expression analysis of abscisic acid signal transduction genes during peach fruit ripening. Sci Hortic 270:109402
Wang X, Zeng W, Ding Y, Wang Y, Niu L, Yao J-L, Pan L, Lu Z, Cui G, Li G (2019) PpERF3 positively regulates ABA biosynthesis by activating PpNCED2/3 transcription during fruit ripening in peach. Hortic Res. https://doi.org/10.1038/s41438-018-0094-2
Aslam MM, Deng L, Wang X, Wang Y, Pan L, Liu H, Niu L, Lu Z, Cui G, Zeng W (2019) Expression patterns of genes involved in sugar metabolism and accumulation during peach fruit development and ripening. Sci Hortic 257:108633
Zhang C, Shangguan L, Ma R, Sun X, Tao R, Guo L, Korir N, Yu M (2012) Genome-wide analysis of the AP2/ERF superfamily in peach (Prunus persica L). Genet Mol Res 11(4):4789–4809
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408
Tatsuki M, Nakajima N, Fujii H, Shimada T, Nakano M, Hayashi KI, Hayama H, Yoshioka H, Nakamura Y (2013) Increased levels of IAA are required for system 2 ethylene synthesis causing fruit softening in peach (Prunus persica L. Batsch). J Exp Bot 64(4):1049–1059
McGlasson W, Scott KJ, Mendoza D Jr (1979) The refrigerated storage of tropical and subtropical products. Int J Refrig 2(6):199–206
Hardeburg RE (1986) The commercial storage of fruits, vegetables, and florist and nursery stocks. USDA Agriculture Handbook, vol 66, pp 11–12
Lallu N (1995) Low temperature breakdown in kiwifruit. In: III International Symposium on Kiwifruit (444): pp 579–586
Mworia EG, Yoshikawa T, Salikon N, Oda C, Fukuda T, Suezawa K, Asiche WO, Ushijima K, Nakano R, Kubo Y (2011) Effect of MA storage and 1-MCP on storability and quality of ‘Sanuki Gold’ kiwifruit harvested at two different maturity stages. J Jpn Soc Hortic Sci 80(3):372–377
Koukounaras A, Sfakiotakis E (2007) Effect of 1-MCP prestorage treatment on ethylene and CO2 production and quality of ‘Hayward’kiwifruit during shelf-life after short, medium and long term cold storage. Postharvest Biol Technol 46(2):174–180
Jin P, Wang K, Shang H, Tong J, Zheng Y (2009) Low-temperature conditioning combined with methyl jasmonate treatment reduces chilling injury of peach fruit. J Sci Food Agric 89(10):1690–1696
Ritenour MA, Crisosto CH, Garner DT, Cheng GW, Zoffoli JP (1999) Temperature, length of cold storage and maturity influence the ripening rate of ethylene-preconditioned kiwifruit. Postharvest Biol Technol 15(2):107–115
Tijero V, Teribia N, Muñoz P, Munné-Bosch S (2016) Implication of abscisic acid on ripening and quality in sweet cherries: differential effects during pre-and post-harvest. Front Plant Sci 7:602
Li P, Dai S, Zhao B, Zhang Y, Liao K, Xu F, Du C, Leng P (2014) Effect of low temperatures on pulp browning and endogenous abscisic acid and ethylene concentrations in peach (Prunus persica L.) fruit during post-harvest storage. J Hortic Sci Biotechnol 89(6):686–692
Yang Q, Zhang Z, Rao J, Wang Y, Sun Z, Ma Q, Dong X (2013) Low-temperature conditioning induces chilling tolerance in ‘Hayward’ kiwifruit by enhancing antioxidant enzyme activity and regulating en-dogenous hormones levels. J Sci Food Agric 93(15):3691–3699
Alves MS, Dadalto SP, Gonçalves AB, De Souza GB, Barros VA, Fietto LG (2013) Plant bZIP transcription factors responsive to pathogens: a review. Int J Mol Sci 14(4):7815–7828
Sornaraj P, Luang S, Lopato S, Hrmova M (2016) Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim Biophys Acta (BBA)-General Subj 1860(1):46–56
Liu X, Chu Z (2015) Genome-wide evolutionary characterization and analysis of bZIP transcription factors and their expression profiles in response to multiple abiotic stresses in Brachypodium distachyon. BMC Genom 16(1):227
Liu J, Chen N, Chen F, Cai B, Dal Santo S, Tornielli GB, Pezzotti M, Cheng ZMM (2014) Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera L). BMC Genom 15(1):281
Hu W, Zuo J, Hou X, Yan Y, Wei Y, Liu J, Li M, Xu B, Jin Z (2015) The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front Plant Sci 6:742
Sevillano L, Sanchez-Ballesta MT, Romojaro F, Flores FB (2009) Physiological, hormonal and molecular mechanisms regulating chilling injury in horticultural species. Postharvest technologies applied to reduce its impact. J Sci Food Agric 89(4):555–573
Brummell DA, Dal Cin V, Crisosto CH, Labavitch JM (2004) Cell wall metabolism during maturation, ripening and senescence of peach fruit. J Exp Bot 55(405):2029–2039
Hsieh TH, Li CW, Ruey-Chih S, Cheng CP (2010) A tomato bZIP transcription factor, SlAREB, is involved in water deficit and salt stress response. Planta 231(6):1459–1473
Kesarwani M, Yoo J, Dong X (2007) Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol 144(1):336–346
Liu C, Ou S, Mao B, Tang J, Wang W, Wang H, Cao S, Schläppi MR, Zhao B, Xiao G (2018) Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat Commun 9(1):1–12
Zhang T, Zhao X, Wang W, Pan Y, Huang L, Liu X, Zong Y, Zhu L, Yang D, Fu B (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS ONE 7(8):e43274
Yun K-Y, Park MR, Mohanty B, Herath V, Xu F, Mauleon R, Wijaya E, Bajic VB, Bruskiewich R, delosReyes BG (2010) Transcriptional regulatory network triggered by oxidative signals configures the early response mechanisms of japonica rice to chilling stress. BMC Plant Biol 10(1):16
Chen D, Xu G, Tang W, Jing Y, Ji Q, Fei Z, Lin R (2013) Antagonistic basic helix-loop-helix/bZIP transcription factors form transcriptional modules that integrate light and reactive oxygen species signaling in Arabidopsis. Plant Cell 25(5):1657–1673
Wang F, Wu N, Zhang L, Ahammed GJ, Chen X, Xiang X, Zhou J, Xia X, Shi K, Yu J (2018) Light signaling-dependent regulation of photoinhibition and photoprotection in tomato. Plant Physiol 176(2):1311–1326
An JP, Yao JF, Wang XN, You CX, Wang XF, Hao YJ (2017) MdHY5 positively regulates cold tolerance via CBF-dependent and CBF-independent pathways in apple. J Plant Physiol 218:275–281
Soltész A, Smedley M, Vashegyi I, Galiba G, Harwood W, Vágújfalvi A (2013) Transgenic barley lines prove the involvement of TaCBF14 and TaCBF15 in the cold acclimation process and in frost tolerance. J Exp Bot 64(7):1849–1862
Wisniewski M, Norelli J, Artlip T (2015) Overexpression of a peach CBF gene in apple: a model for understanding the integration of growth, dormancy, and cold hardiness in woody plants. Front Plant Sci 6:85
Nakashima K, Ito Y, Yamaguchi-Shinozaki K (2009) Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol 149(1):88–95
Xiong L, Ishitani M, Zhu J-K (1999) Interaction of osmotic stress, temperature, and abscisic acid in the regulation of gene expression in Arabidopsis. Plant Physiol 119(1):205–212
Lu G, Gao C, Zheng X, Han B (2009) Identification of OsbZIP72 as a positive regulator of ABA response and drought tolerance in rice. Planta 229(3):605–615
Fujita Y, Fujita M, Satoh R, Maruyama K, Parvez MM, Seki M, Hiratsu K, Ohme-Takagi M, Shinozaki K, Yamaguchi-Shinozaki K (2005) AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17(12):3470–3488
Casaretto J, Ho TH (2003) The transcription factors HvABI5 and HvVP1 are required for the abscisic acid induction of gene expression in barley aleurone cells. Plant Cell 15(1):271–284
Kobayashi F, Maeta E, Terashima A, Takumi S (2008) Positive role of a wheat HvABI5 ortholog in abiotic stress response of seedlings. Physiol Plant 134(1):74–86
Xiao-Li S, Li Y, Hua C, Xi B, Wei J, Zuo-Jun J, Yan-Ming Z (2011) Arabidopsis bZIP1 transcription factor binding to ABRE cis-element regulates abscisic acid signal transduction. Acta Agron Sin 37(4):612–619
Choi H-i, Hong J-h, Ha J-o, Kang J-y, Kim SY (2000) A family of ABA-responsive element binding factors. J Biol Chem 275(3):1723–1730
Yao L, Hao X, Cao H, Ding C, Yang Y, Wang L, Wang X (2020) ABA-dependent bZIP transcription factor, CsbZIP18, from Camellia sinensis negatively regulates freezing tolerance in Arabidopsis. Plant Cell Rep 39(4):553–565
Kim JC, Lee SH, Cheong YH, Yoo CM, Lee SI, Chun HJ, Yun DJ, Hong JC, Lee SY, Lim CO (2001) A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J 25(3):247–259
Acknowledgements
This work was supported by the National Natural Science Foundation of China [No. 31872085], the National Key Research and Development Program [2018YFD1000200], and the Agricultural Science and Technology Innovation Program (ASTIP) [CAAS-ASTIP-2019-ZFRI]. Authors are highly grateful for providing financial assistance for this research.
Funding
This work was supported by the National Natural Science Foundation of China [No. 31872085], the National Key Research and Development Program [2018YFD1000200], and the Agricultural Science and Technology Innovation Program (ASTIP) [CAAS-ASTIP-2019-ZFRI].
Author information
Authors and Affiliations
Contributions
WZ conceived this project and designed all the experiment. MMA, LD, YW performed the experiments. JM analyzed data. ZW supervised the experiments MMA and WZ wrote the article, LP, LN, ZL and GC help to revise the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
We declare that none of the work contained in this manuscript is published in any language or current under consideration at any other journal; there is no conflict of interest to declare. All authors have contributed to, read, and approved this submitted manuscript in its current form.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aslam, M.M., Deng, L., Meng, J. et al. Characterization and expression analysis of basic leucine zipper (bZIP) transcription factors responsive to chilling injury in peach fruit. Mol Biol Rep 50, 361–376 (2023). https://doi.org/10.1007/s11033-022-08035-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-022-08035-3