Abstract
The pep4um gene (um04926) of Ustilago maydis encodes a protein related to either vacuolar or lysosomal aspartic proteases. Bioinformatic analysis of the Pep4um protein revealed that it is a soluble protein with a signal peptide suggesting that it likely passes through the secretory pathway, and it has two probable self-activation sites, which are similar to those in Saccharomyces cerevisiae PrA. Moreover, the active site of the Pep4um has the two characteristic aspartic acid residues of aspartyl proteases. The pep4um gene was cloned, expressed in Pichia pastoris and a 54 kDa recombinant protein was observed. Pep4um-rec was confirmed to be an aspartic protease by specifically inhibiting its enzymatic activity with pepstatin A. Pep4um-rec enzymatic activity on acidic hemoglobin was optimal at pH 4.0 and at 40 °C. To the best of our knowledge this is the first report about the heterologous expression of an aspartic protease from a basidiomycete. An in-depth in silico analysis suggests that Pep4um is homolog of the human cathepsin D protein. Thus, the Pep4um-rec protein may be used to test inhibitors of human cathepsin D, an important breast cancer therapeutic target.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aspartic proteases, otherwise known as aspartyl proteases (E.C.3.4.23.X), are enzymes from the pepsin family (A1 family) that usually function in acidic conditions, either outside of the cell or in the lysosome/vacuole compartment. Aspartic proteases are monomeric enzymes with two domains, each one containing an aspartic residue essential for enzymatic activity. These proteases are widely distributed in retroviruses and eukaryotic cells, including mammals, plants, nematodes and fungi [1].
Filamentous fungi and yeast produce two varieties of aspartyl proteases: extracellular and intracellular. Proteins in the secreted aspartyl proteases (SAPs) family, produced by the opportunistic pathogen Candida spp, are a type of phylogenetically related extracellular aspartic proteases that have been studied in-depth. Besides providing nutrition to the cell, SAPs are considered virulence factors [2, 3]. A similar function has been proposed for extracellular aspartic proteases of filamentous fungi such as the phytopathogenic Sclerotinia sclerotiourum, the human opportunistic pathogen Cryptococcus neoformans and the mychophagous Trichoderma harzianum [4,5,6,7].
Yapsins (YAPs) are a second type of extracellular aspartyl proteases that contain a GPI moiety in the C-terminal region. Through this moiety, YAPs bind to the plasma membrane in the fungal cell, apparently implicated in assembly and/or remodeling of the cell wall. Additionally, some of them are virulence factors [8,9,10].
Vacuolar aspartyl protease A (PrA) (E.C.3.4.23.25) is encoded by the PEP4 gene in Saccharomyces cerevisiae. It is synthesized as a preproenzyme and its maturation proceeds in a three-step process. First, a 22-amino-acid signal peptide is removed in the endoplasmic reticulum to form proPrA. Once in the vacuole, 45 amino acids in the proPrA N-terminal section are removed auto-catalytically in a pH-dependent manner, producing an active pseudoPrA that activates proPrB, a vacuolar serine endoprotease. Finally, mature PrB removes 54 amino acid residues from the proPrA N-terminal region yielding matured PrA [11]. Moreover, pseudoPrA triggers a cascade-like activation that drives also carboxypeptidase proCPY and other enzymes [12, 13]. These mature vacuolar enzymes play a pivotal role in cell survival during nitrogen starvation and sporulation process [14].
Like the PrA protease of S. cerevisiae, human lysosomal endoprotease cathepsin D (cath-D) (E.C.3.4.23.5) is synthesized as an inactive preproenzyme, which is maturated through the secretory pathway until it reaches the lysosome. The mature cath-D protein is an essential player in protein turnover. Elevated cath-D levels correlate with tumor malignancy and cancer cell survival [15, 16].
The phytopathogenic fungus Ustilago maydis, the causal agent of corn smut, has a complex life cycle in which the mating of two haploid yeasts leads the formation of a dikaryotic infective mycelium [17]. The yeast-mycelium transition can be generated in vitro when haploid cells growing at pH 7.0 are transferred to pH 3.0 [18]. The addition of pepstatin A, a specific inhibitor of aspartic proteases, inhibits the intracellular aspartic protease activity and the yeast-mycelium transition induced at pH 3.0 [19]. Moreover, the protein encoded by ORF um04926 is up-regulated after the transition of yeast to the filamentous form, induced by the over-expression of the bE2/bW1 heterodimer, a major transcriptional factor in U. maydis [20]. Recently, it has been shown that this putative aspartic protease is involved in the pathogenesis and dimorphism of U. maydis [21]. In the current study, the protein encoded by the pep4um gene, related to S. cerevisiae PrA and to human cath-D, was expressed in Pichia pastoris and biochemically characterized. Even though other proteases from Basidiomycete fungi has been successfully expressed in recombinant systems [22]. To our knowledge this is the firsts vacuolar aspartic protease from Basidiomycete heterologously expressed.
Materials and methods
Strains and plasmids
The U. maydis FB1 (a1b1) strain used herein was kindly provided by Dr. Flora Banuett (UCSF). The Escherichia coli–P. pastoris shuttle vector pPICZA (Easy Select Pichia Expression Kit, Invitrogen, CA, USA) was employed for cloning and expression of the pep4um gene in P. pastoris X-33. The E. coli strain DH10b was used for plasmid construct and propagation.
Bioinformatic analysis
The pep4um (um04926) nucleotide sequence was identified by BLASTp analysis in the database of the U. maydis genome project, available at http://mips.gsf.de/genre/proj/ustilago. Deduced amino acidic sequences from genes encoding for vacuolar aspartyl proteases in other yeasts and filamentous fungi were used. [ID protein numbers; S. cerevisiae PEP4 (YPL154C), Candida albicans APR1 (orf19.1891) and Neurospora crassa PEP4 (AAA79878)]. The structural analyses of the gene and the translated protein were carried out on the software programs: isoelectric point (pI), helical membranous regions, molecular signatures and subcellular location were predicted by utilizing the ScanProsite database and PSORTII, respectively (http://www.expasy.org). Multiple alignments of nucleotide and deduced protein sequences, and dendrogram were performed on MEGA 6.0 [23] by using Neighbor-Joining algorithms computed with the Poisson correction and Bootstrap analysis (1000 replicates).
Prediction of the tertiary structure of the Pep4um protein
The three-dimensional structure of U. maydis Pep4um and human cath-D were deduced by homology modeling with the SWISS-MODEL Workspace (http://www.expasy.org). Amino acid sequences of these organisms used to search templates were XP_011391245 and NP_001900, respectively. The best templates were selected based on sequence identity and coverage probability. The crystal structures of S. cerevisiae PrA (PDB ID: 1 fmx.1.A) [24], and human complex cath-D-pepstatin A (PDB ID: 1lyb.1.B and .C) [25] were used to model Pep4um and cath-D, respectively. Structure alignment, superposition and root-mean-square (rms) deviation were performed with SuperPose V 1.0 (http://wishart.biology.ualberta.ca/SuperPose/). Superposition was edited on UCSF Chimera software [26].
Heterologous expression of the U. maydis pep4um gene in P. pastoris
The pep4um coding region was amplified by PCR with the following primers: Pep4UMfwd2 5′-CGCGCGAATTCATGAAGCTCAACCTCTCCCTCAC-3′ and Pep4UMrev2 5′-CGCGCGAATTCCTTGGCAGTCGCGAGAC-3′ (EcoRI restriction sites were introduced and are shown in italics). The PCR product was digested with the EcoRI restriction enzyme and purified with QIAquick Gel Extraction (Qiagen, USA), according to the manufacturer’s instructions. The purified product was cloned into the EcoRI site of the pPICZA multiple cloning site to generate the plasmid pPICZA-Pep4um. Construct orientation in frame with the AOX promoter was verified by both PCR and sequencing. Once a clone was selected, P. pastoris was transformed using 10 µg of pPICZA-Pep4um DNA previously linearized with PmeI. Transformant clones were selected according to the manufacturer’s instructions on YPDS zeocin medium (YPD, 100 µg/mL zeocin and 1 M sorbitol). Zeocin-resistant clones were confirmed for construct integration by PCR amplifying of two specific sequences, the first one employing the same primers used for cloning of the pep4um gene, and the second one with the AOX1 5′ forward and 3′ reverse primers supplied in the kit. The wild type P. pastoris X-33 and Cɑ (transformed with the empty plasmid) genomic DNA were utilized as negative controls.
Expression and identification of recombinant Pep4um-rec
Wild type and transformant P. pastoris strains were grown in 50 mL of glycerol-based yeast medium (1.34% YNB without amino acids, 4 × 10−5 % biotin, 10 mM potassium phosphate buffer and 2% glycerol, pH 6.0) at 28 °C until a DO600 = 2.0 was reached. The cells were harvested and resuspended in 100 mL of the same medium supplemented with 0.5% methanol (v/v) instead of glycerol and incubated for 48 h, adding methanol every 24 h. The cells were harvested and washed twice with cold sterile distilled water and disrupted by using glass beads (0.5 mm in diameter) and lysis buffer (50 mM NaH2PO4, 300 mM NaCl and 10 mM imidazole, pH 8). The cell-free extract (CFE) was obtained by centrifugation at 10,000×g for 20 min. Then the extract was ultracentrifuged at 100,000×g for 90 min and the soluble fraction was recovered.
The soluble fraction of the strain expressing the recombinant protein was loaded in a ProBondTMNickel-Chelatin Resin (Invitrogen, CA, USA). The elution was performed accordingly to the manufacturer’s instructions using elution buffer supplemented with 250 mM imidazole. However, no recombinant protein was recovered (data not shown). The recombinant Pep4um-rec protein was partially characterized by using the P. pastoris CA1 (pPICZA-Pep4um) soluble fraction, and the activity of a soluble fraction of the Cɑ strain (pPICZA) was subtracted in order to consider background activity. Protein extracts were examined in 10% SDS–PAGE. The protein concentration was estimated by a modified Lowry method (1% SDS).
In order to identify the recombinant protein, the sample was subjected to electrophoresis as aforementioned and a band of the expected size of Pep4um was excised from the SDS–PAGE, treated with trypsin and sent for protein sequence determination by liquid chromatography and nanoelectrospray ionization mass spectrometry (LC–MS) (Thermo-Fischer Co., San Jose, CA, USA) at the Instituto de Biotecnología UNAM proteomics facility.
For the Western blot assay, proteins were transferred from SDS–PAGE to a PVDF membrane (Millipore), blotted with anti-6x His (C-term)-HRP tag antibody (Invitrogen CA, USA), and revealed with diaminobenzidine (Sigma) and 30% hydrogen peroxide.
Endoprotease activity assay
The activity of endoprotease A was evaluated by the acid hemoglobin method [27]. One unit of specific activity was defined as 1 µg of tyrosine-containing peptides released into the supernatant per minute per milligram of protein at 37 °C.
Pep4um-rec activity against other substrates
The denatured hemoglobin used in the standard assay was replaced by albumin, casein and Hide Powder Azure (HPA), a collagen-type substrate. All the substrates were tested at pH 3.5 and 7.0.
Effects of pH and temperature on recombinant Pep4um-rec activity
The optimal pH for protease activity was examined at 37 °C with different buffers sets at the desired pH value. For the pH ranges of 3.0–8.0, 8.0–10.0, and 10.0–11.0, McIlvaine, Tris–HCl and glycine-NaOH 50 mM buffers were used, respectively. pH stability was assessed by overnight preincubation of the enzyme extracts in the appropriate buffer at 4 °C and at different pH values ranging from 2.0 to 10.0, followed in each case by a standard enzyme assay. The optimal temperature for enzyme activity was explored between 5 and 80 °C by a standard enzyme assay. Thermal stability was evaluated by incubating the enzyme solution at 5, 25, 30, 35, 40, 50, 60, 70 and 80 °C for 60 min, and then conducting the standard enzyme assay. The actual activity was always expressed as a percentage of the maximum activity (considered as 100%) obtained at either the optimal pH or temperature.
Effects of protease inhibitors on recombinant Pep4um-rec activity
Potential inhibitors such as pepstatin, leupeptin, Pefabloc, E-64, 1–10 phenanthroline and EDTA-Na2 (Sigma-Aldrich and Roche, Switzerland) were tested at two different concentrations (Table 2). The P. pastoris CA1 soluble extract was preincubated with each potential inhibitor for 30 min at 37 °C, followed by a standard enzyme assay. The actual activity was expressed as a percentage of the enzymatic activity obtained without the inhibitor (considered as 100%). All determinations were performed at least three times.
Results
Identification of aspartic proteases genes of U. maydis
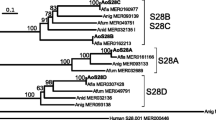
Eight sequences with two aspartic acid residues in the characteristic consensus sites for aspartic proteases were identified in the genome of U. maydis. These were compared to sequences of different aspartic proteases from multiple fungi. Seven of the eight U. maydis predicted proteins are putative extracellular proteases related to both GPI-anchored and soluble types. On the other hand, the predicted protein from the um04926 gene (pep4um) is the only one related to the yeast and filamentous fungi vacuolar aspartic endoproteases, and also to the human lysosomal cathepsin-D (cath-D) (Fig. 1a).
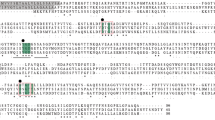
The protein encoded by the U. maydis pep4um gene is an aspartic protease, related to vacuolar and lysosomal proteases. a Neighbor-Joining phylogenetic tree of fungal aspartic proteases; bootstrap values are on branches and protein access numbers are in parenthesis. b Alignment of the predicted amino acid sequences of the U. maydis pep4um gene (Um_um04926) with the S. cerevisiae PrA (Sc_PEP4) and the human cat-D (Hs_CathD). The aspartic residues of the active site and the disulfide bonds are indicated by diamonds and bridges, respectively. The S. cerevisiae PrA has two sites of N-glycosylation, a processing site by the vacuolar serine protease B (PrB), and a self-processing site (shown by asterisks, arrow and arrowhead, respectively). Additionally, grey arrowheads show two probable self-maturation sites in the U. maydis sequence, while the dotted line in N-terminal region denotes its predicted signal peptide and the two probable N-glycosilation sites by asteriks. Two peptides, identified by the sequence of a 54 kDa band from P. pastoris CA1 soluble fraction are designated by a continuous line, underneath the peptide sequence. Residues marked with crosses are involved in the structure maintenance of PrA. Ca—C. albicans, Sc— S. cerevisiae, Bc—B. cinerea, Um—U. maydis, An—A. niger, Ta—T. asperellum, Th—T. harzianum, Cn—C. neoformans, Cb—C. boidinii, Af—A. fumigatus, Nc—N. crassa, Hs—H. sapiens. Sap secreted aspartic protease, Yps yapsins
Characteristics of the vacuolar Pep4um protein of U. maydis
The pep4um gene encodes for a putative protein of 418 amino acids, with a calculated molecular weight of 45.6 kDa and a pI = 4.8. The examination of the Pep4um predicted protein indicated that it contains the two characteristic aspartic protease motifs, VILD125TGSSNLWV and AAID307TGTSLIAM, which are essential for enzymatic activity. As in the S. cerevisiae PrA, Pep4um has: (i) two characteristic N-glycosylation sites (Asn160 and Asn301), (ii) a group of cysteine residues involved in disulfide bond formation (Cys138–Cys143 and Cys341–Cys374), and (iii) a 22-amino acid putative N-terminal signal peptide (Fig. 1b), suggesting that it is a zymogen that transits through the secretory pathway towards the vacuole. In contrast to S. cerevisiae PrA, Pep4um predicted sequence does not contain a PrB-like processing site (Fig. 1b), however it contains two probable auto-maturation sites similar to those found in PrA (Fig. 1b), as well as amino acid regions whose role in its structural stability has been suggested (Fig. 1b). Additionally, amino acid sequences of S. cerevisiae PrA and U. maydis Pep4um are similar to human lysosomal cath-D (with an identity of 40.0 and 36.6%, respectively). However, the Ser80 amino acid in the S3 pocket, which is part of the β hairpin loop (called the “flap”) and plays a role in inhibitor binding, is present in both cath-D and Pep4um proteases.
In order to determine whether these characteristics are structurally conserved in a 3D model, the Pep4um (93–418 region) was deduced by using the uninhibited S. cerevisiae PrA as template. The Pep4um protein structure displayed the characteristic two lobes spaced by a deep cleft of peptidases of the A1 family. In each of these lobes were found the two catalytic aspartic resides. A similar orientation of these residues and the glycosylation site Asn62cathD is shown by the superimposition of the predicted structure of the U. maydis Pep4um on the cath-D structure, modeled using the complex cath-D-pepstatin A, with a rms deviation of 3.55 for 330 Cα (Fig. 2a). Torsion of the Tyr78cathD at the tip of the flap is similar in both cath-D and Pep4um protein structures. Moreover, a structural region similar to the Pro-rich of cath-D is superimposed in the modeling of Pep4um, although these two proteins do not have similar sequences in that region (Fig. 2b). These results show the structural similarity of cath-D and Pep4um and suggest a comparable enzymatic activity.
Predicted tertiary structure of U. maydis Pep4um is similar to the acid aspartic proteases of family A1. a Superimposition of ribbon representations of the predicted Pep4um structure (green) and cath-D (blue). Aspartic catalytic residues are indicated in red, asparagine glycosylation sites in pink, the “flap” region with tyrosine residues in brown and the Cat-D proline loop in yellow. b Schematic representation of the flap region and part of the catalytic site of Pep4um and cath-D. Residues numbers according the modeled sequences are indicated. Pep4um and cath-D 3D structures were modeled as indicated in methods. (Color figure online)
Recombinant Pep4um
In order to test whether pep4um gene encodes for an aspartic protease, this gene was cloned into the pPICZA vector to generate the pPICZA-Pep4um vector and used to transform P. pastoris X-33 (Fig. 3a). The insertion of the recombinant pep4um gene in the genome of the P. pastoris was confirmed by PCR amplification as indicated in the previous section. After confirming gene insertion, the strain was utilized for further characterization of the recombinant Pep4um-rec protein.
The recombinant Pep4um-rec encoded by the U. maydis pep4um gene is an intracellular soluble endoprotease. a Cartoon of the pPICZA-Pep4um plasmid and its recombination in P. pastoris AOX1 gene. b Specific activity against denatured hemoglobin of the cell-free extract (CFE), soluble (Sol) and membrane (Mem) fractions from the transformants CA1 (carrying the pPICZA-Pep4um plasmid) and Cɑ carrying the empty plasmid (pPICZA), which were grown in methanol during 48 h. Error bars show ± SD. c Molecular weight determination of the Pep4um-rec recombinant protein. CA1 and Cɑ soluble fractions were examined by SDS–PAGE. A 54 kDa band (arrow) in the CA1extract was sequenced by LC–MS and corresponded to the deduced amino acid sequence of Pep4um. d A unique band of 54 kDa (arrow) was evidenced in the CA1 soluble fraction by Western blot analysis by using an anti-His tag antibody
Pichia pastoris strains transformed with plasmids pPICZA and pPICZA-Pep4um, were grown in medium containing methanol as the sole carbon source to elicit recombinant protein expression. The cells were harvested and then disrupted to obtain the CFE, from which the soluble and the membrane fractions were separated, as before mentioned. The P. pastoris transformant CA1 soluble fraction presented nearly a fourfold higher activity after 48 h of induction when compared to P. pastoris Cɑ (carrying empty plasmid pPICZA) (Fig. 3b).
The soluble fractions from the CA1 and the control Cɑ strains were subjected to SDS–PAGE and a band of approximately 54 kDa was evident in the CA1 but not in the Cɑ soluble extract (Fig. 3c). This band was excised and sequenced by LC–MS. Two sequenced peptides matched the predicted protein sequence of the Pep4um gene. One of them corresponded to the first motif of the aspartic protease and the other was part of the last amino acid region of the sequence (Fig. 1b). A Western blot analysis with an anti-His antibody also revealed a band of 54 kDa in the CA1 clone soluble fraction (Fig. 3d).
Pep4um-rec is an endoprotease, with optimal activity at pH 4.0 and 40 °C
Pichia pastoris CA1 and Cɑ soluble fractions were tested in neutral and acidic conditions to determine their activity towards four different substrates hemoglobin, albumin, casein and HPA, as indicated in previously. Then the activity of the Cɑ strain was subtracted from that of CA1, the highest Pep4um-rec specific activity was obtained in acidic conditions using hemoglobin as substrate. In neutral conditions, no recombinant activity was detected towards albumin or HPA (Table 1). The recombinant enzyme was stable in an acidic pH ranging from 4.0 to 6.0. Optimal activity was found at pH 4.0 with hemoglobin as substrate (Fig. 4a). Moreover, Pep4um-rec was stable for 60 min in a range of 25–35 °C, the optimal temperature being 40 °C (Fig. 4b).
Effect of pH and temperature on recombinant Pep4um-rec activity. The CA1 soluble fraction was tested at the indicated pHs and temperatures a (i) McIlvaine (pH 3.0–7.0), (ii) Tris–HCl (pH 8.0–9.0) and (iii) glycine–NaOH (pH 9.0–10.0) buffers were used in this assay for the indicated pH. The continuous line designates optimum pH and the dotted line pH stability. b The continuous line denotes optimum temperature and the dotted line temperature stability
Additionally, the effect of six protease inhibitors on Pep4um-rec activity was evaluated. Whereas enzyme activity was inhibited by pepstatin A, a specific aspartic protease inhibitor, no inhibitory effects were observed with the cysteine (leupeptin), serine (E64), or metalloprotease (pefabloc, EDTA or 1–10 phenanthroline) protease inhibitors (Table 2). Overall, these evidences show that Pep4um is an aspartic protease.
Discussion
Ustilago maydis genome has at least eight genes encoding for putative aspartyl proteases, seven of them are grouped with extracellular aspartyl proteases of other yeasts and filamentous fungi. Meanwhile, the Pep4um (um04926) is the only one grouped with vacuolar and lysosomal aspartyl proteases. The vacuolar/lysosomal location of the Pep4um protein has been demonstrated by our group [21], suggesting a phylogenetic relation of Pep4um with the S. cerevisiae PrA and human lysosomal cath-D.
Like PrA and cath-D, the Pep4um vacuolar protease could be synthesized as a zymogen, since it displays two putative auto activation sites as S. cerevisiae PrA, however no PrB-like processing site was found in Pep4um sequence. It is known that S. cerevisiae strain mutant lacking PrB protease, is capable to produce an active pseudo-PrA by self-activation [13]. On the other hand, it has been suggested that C. glabrata PrA might has a role in the autophagy process [28, 29].
The overexpression of pep4um in P. pastoris allowed us to get an intracellular recombinant protein of 54 kDa, confirmed as a Pep4um protein by peptide sequencing. This is in agreement with the molecular masses described for vacuolar intracellular aspartic proteases from other fungi such as S. cerevisiae PrA, H. polymorpha and N. crassa PEP4 (41.7, 45.4 and 40 kDa, respectively) [30,31,32].
Both the CFE and the soluble fraction obtained from the CA1 strain expressing Pep4um-rec digest preferentially hemoglobin in acidic conditions. A similar effect has been shown for S. cerevisie PrA, C. boidinii PEP4, N. crassa PEP4 and the previously purified intracellular aspartic protease PumAi from U. maydis. Also, human cath-D that is more active on denatured hemoglobin than on native proteins [30,31,32,33,34]. Pep4um-rec is stable at a pH between 4 and 6 and at temperatures ranging from 25 to 35 °C, having an optimum activity at pH 4.0 and at 40 °C. This is similar to what has been found for PrA and PumAi [30, 34]. Contrary to the activity observed with the soluble fraction and CFE, which are also able to digest casein, albumin and collagen, no protease activity was exhibited in the membrane fraction, suggesting that Pep4um-rec is a soluble protein. In addition, the endoprotease activity of the P. pastoris CA1 soluble fraction towards acid hemoglobin was completely inhibited by adding 25 µM pepstatin A, while no effect was detected in general with the other inhibitors tested, except with EDTA and 1–10 phenantroline, thus Pep4um-rec could be affected by metal ions such as other aspartic proteases [35]. Thus, our prediction that the product of the pep4um gene is an aspartic protease was confirmed.
The predicted tertiary structure of U. maydis Pep4um, using as the search model a non-ligated PrA protease, is similar to that of other acid aspartic proteases. When it is superimposed on cath-D-pepstatin A complex, they share important similarities such as the “flap” structure at the Tyr 78, Gly 79 and Ser 80 residues located at the tip, that have an important function in substrate and inhibitor specificity [25]. In contrast, for S. cerevisiae PrA the rotation of the Tyr 75 of the “flap” is different, occupying the S1 binding pocket of the non-inhibited protein [24]. Also Ser 80 residue is absent. Despite these structural differences, a phylogenetic analysis demonstrated that PrA is more related to cath-D than to other mammalian and fungal aspartic endoproteases [36]. However, U. maydis was proposed as a model for understanding what happens in mammalian cellular processes, since U. maydis and humans share disease-related proteins not observed in S. cerevisiae [37].
Aditionally U. maydis is proposed a suitable model for heterologous expression of proteins [38] and the knowledge of the characteristics of proteases whose main role is the post-translational modification is important to get high efficiencies of the recombinant proteins [39]. In conclusion, we found that Pep4um is an aspartic endoprotease active and stable under acidic conditions and a homolog of PrA and lysosomal cath-D. We propose that this version of recombinant Pep4um can serve as an alternative model for screening and testing human cath-D inhibitors and chemotherapeutic agents, and for studying processes such as autophagy, protein turnover and programmed cell death in the basidiomycete U. maydis.
References
Tang J, Wong RNS (1987) Evolution in the structure and function of aspartic proteases. J Cell Biochem 33:53–63. https://doi.org/10.1002/jcb.240330106
Loaiza-Loeza S, Parra-Ortega B, Cancino-Díaz JC, Illades-Aguiar B, Hernández-Rodríguez CH, Villa-Tanaca L (2009) Differential expression of Candida dubliniensis-secreted aspartyl proteinase genes (CdSAP1-4) under different physiological conditions and during infection of a keratinocyte culture. FEMS Immunol Med Microbiol 56:212–222. https://doi.org/10.1111/j.1574-695X.2009.00570.x
Parra-Ortega B, Cruz-Torres H, Villa-Tanaca L, Hernández-Rodríguez C (2009) Phylogeny and evolution of the aspartyl protease family from clinically relevant Candida species. Memórias do Instituto Oswaldo Cruz 104:505–512. https://doi.org/10.1590/S0074-02762009000300018
Delgado-Jarana J, Rincón AM, Benítez T (2002) Aspartyl protease from Trichoderma harzianum CECT 2413: cloning and characterization. Microbiology 148:1305–1315. https://doi.org/10.1099/00221287-148-5-1305
Mandujano-González V, Villa-Tanaca L, Anducho-Reyes MA, Mercado-Flores Y (2016) Secreted fungal aspartic proteases: a review. Revista Iberoamericana de Micología 33:76–82. https://doi.org/10.1016/j.riam.2015.10.003
Pinti M, Orsi CF, Gibellini L, Esposito R, Cossarizza A, Blasi E, Peppoloni S, Mussini C (2007) Identification and characterization of an aspartyl protease from Cryptococcus neoformans. FEBS Lett 581:3882–3886. https://doi.org/10.1016/j.febslet.2007.07.006
Poussereau N, Gente S, Rascle C, Billon-Grand G, Fèvre M (2001) aspS encoding an unusual aspartyl protease from Sclerotinia sclerotiorum is expressed during phytopathogenesis. FEMS Microbiol Lett 194:27–32. https://doi.org/10.1016/S0378-1097(00)00500-0
Cortés-Acosta E, Ibarra JA, Ramírez-Saad H, Vargas-Mendoza CF, Villa-Tanaca L, Hernández-Rodríguez C (2017) Polymorphism in the regulatory regions of genes CgYPS1 and CgYPS7 encoding for yapsins in Candida glabrata is associated to changes in expression levels. FEMS Yeast Res. https://doi.org/10.1093/femsyr/fox077
Gagnon-Arsenault I, Tremblay J, Bourbonnais Y (2006) Fungal yapsins and cell wall: a unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res 6:966–978. https://doi.org/10.1111/j.1567-1364.2006.00129.x
Parra-Ortega B, Villa-Tanaca ML, Hernández-Rodríguez CH (2011) Evolution of GPI—aspartyl proteinases (Yapsines) of Candida spp. In: Friedberg F (ed) Gene duplication. InTech, Rijeka, Croatia, pp 289–314. https://doi.org/10.5772/23456
Rothman JH, Hunter CP, Valls LA (1986) Overproduction-induced mislocalization of a yeast vacuolar protein allows isolation of its structural gene. Proc Natl Acad Sci USA 83:3248–3252
Ammerer G, Hunter CP, Rothman JH, Saari GC, Valls LA, Stevens TH (1986) PEP4 gene of Saccharomyces cerevisiae encodes proteinase A. Mol Cell Biol 6:2490–2499
van den Hazel HB, Kielland-brandt MC, Winther JR (1992) Autoactivation of proteinase A initiates activation of yeast vacuolar zymogens. Eur J Biochem 207:277–283
Jones EW (1991) Three proteolytic systems in the yeast. J Biol Chem 266:7963–7966
Carmona-Gutiérrez D, Bauer MA, Ring J, Knauer H, Eisenberg T, Büttner S, Ruckenstuhl C, Reisenbichler A, Magnes C, Rechberger GN, Birner-Gruenberger R, Jungwirth H, Fröhlich K-U, Sinner F, Kroemer G, Madeo F (2011) The propeptide of yeast cathepsin D inhibits programmed necrosis. Cell Death Dis 2:e161. https://doi.org/10.1038/cddis.2011.43
Laurent-Matha V, Derocq D, Prébois C, Katunuma N, Liaudet-Coopman E (2006) Processing of human cathepsin D is independent of its catalytic function and auto-activation: involvement of cathepsins L and B. J Biochem 139:363–371. https://doi.org/10.1093/jb/mvj037
Banuett F (1995) Genetics of Ustilago maydis, a fungal pathogen that induces tumors in maize. Annu Rev Genet 29:179–208. https://doi.org/10.1146/annurev.ge.29.120195.001143
Ruiz-Herrera J, Leon CG, Guevara-Olvera L, Carabez-Trejo A (1995) Yeast-mycelial dimorphism of haploid and diploid strains of Ustilago maydis. Microbiology 141:695–703. https://doi.org/10.1099/13500872-141-3-695
Mercado-Flores Y, Guerra-Sanchez G, Villa-Tanaca L, Hernandez-Rodriguez C (2003) Purification and characterization of an extracellular non-aspartyl acid protease (pumAe) from Ustilago maydis. Curr Microbiol 47:408–411. https://doi.org/10.1007/s00284-003-4047-z
Böhmer M, Colby T, Böhmer C, Bräutigam A (2007) Proteomic analysis of dimorphic transition in the phytopathogenic fungus Ustilago maydis. Proteomics 7:675–685. https://doi.org/10.1002/pmic.200600900
Soberanes-Gutiérrez CV, Juárez-Montiel M, Olguín-rodríguez O, Hernández-Rodríguez C, Ruiz-herrera J, Villa-Tanaca L (2015) The pep4 gene encoding proteinase A is involved in dimorphism and pathogenesis of Ustilago maydis. Mol Plant Pathol 16:837–846. https://doi.org/10.1111/mpp.12240
Juárez-Montiel M, Ibarra JA, Chávez-Camarillo G, Hernández-Rodríguez C, Villa-Tanaca L (2014) Molecular cloning and heterologous expression in Pichia pastoris of X-prolyl-dipeptidyl aminopeptidase from basidiomycete Ustilago maydis. Appl Biochem Biotechnol 172:2530–2539. https://doi.org/10.1007/s12010-013-0682-4
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Gustchina A, Li M, Phylip LH, Lees WE, Kay J, Wlodawer A (2002) An unusual orientation for Tyr75 in the active site of the aspartic proteinase from Saccharomyces cerevisiae. Biochem Biophys Res Commun 295:1020–1026. https://doi.org/10.1016/S0006-291X(02)00742-8
Baldwin ET, Bhat TN, Gulnik S, Hosur MV, Sowder RC, Cachau RE, Collins J, Silva AM, Erickson JW (1993) Crystal structures of native and inhibited forms of human cathepsin D: implications for lysosomal targeting and drug design. Proc Natl Acad Sci USA 90:6796–6800. https://doi.org/10.1073/pnas.90.14.6796
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Jones EW (1977) Proteinase mutants of Saccharomyces cerevisiae. Genetics 85:23–33
Cortez-Sánchez J, Cortés-Acosta E, Cueto-Hernández V, Reyes-Maldonado E, Hernández-Rodríguez C, Villa-Tanaca L, Ibarra JA (2018) Activity and expression of Candida glabrata vacuolar proteases in autophagy-like conditions. FEMS Yeast Res. https://doi.org/10.1093/femsyr/foy006
Sepúlveda-González E, Parra-Ortega B, Betancourt-Cervantes Y, Hernández-Rodríguez C, Xicohtencatl-Cortés J, Villa-Tanaca L (2016) Vacuolar proteases from Candida glabrata: acid aspartic protease PrA, neutral serine protease PrB and serine carboxypeptidase CpY, nitrogen source influence expression level. Revista Iberoamericana de Micología 33:26–33. https://doi.org/10.1016/j.riam.2014.10.005
Bae J, Sohn J, Rhee S, Choi E (2005) Cloning and characterization of the Hansenula polymorpha PEP4 gene encoding proteinase A. Yeast 22:13–19. https://doi.org/10.1002/yea.1193
Meussdoerffer F, Tortora P, Holzer H (1980) Purification and properties of proteinase A from yeast. J Biol Chem 255:12087–12093
Vázquez-Laslop N, Tenney K, Bowman BJ (1996) Characterization of a vacuolar protease in Neurospora crassa and the use of gene RIPing to generate protease-deficient strains. J Biol Chem 271:21944–21949. https://doi.org/10.1074/jbc.271.36.21944
Komeda TK, Akai YS, Ato NK, Ondo KK (2002) Construction of protease-deficient Candida boidinii strains useful for recombinant protein production: cloning and disruption of proteinase A gene (PEP4) and proteinase B gene (PRB1). Biosci Biotechnol Biochem 66, 628–631
Mercado-Flores Y, Trejo-Aguilar A, Ramirez-Zavala B, Villa-Tanaca L, Hernandez-Rodriguez C (2005) Purification and characterization of an intracellular aspartyl acid proteinase (pumAi) from Ustilago maydis. Can J Microbiol 51:171–175. https://doi.org/10.1139/w04-125
Hsiao NW, Chen Y, Kuan YC, Lee YC, Lee SK, Chan HH, Kao CH (2014) Purification and characterization of an aspartic protease from the Rhizopus oryzae protease extract, Peptidase R. Electron J Biotechnol 17:89–94
Aguilar CF, Cronin NB, Badasso M, Dreyer T, Newman MP, Cooper JB, Hoover DJ, Wood SP, Johnson MS, Blundell TL (1997) The three-dimensional structure at 2.4 A resolution of glycosylated proteinase A from the lysosome-like vacuole of Saccharomyces cerevisiae. J Mol Biol 267:899–915
Münsterkötter M, Steinberg G (2007) The fungus Ustilago maydis and humans share disease-related proteins that are not found in Saccharomyces cerevisiae. BMC Genom 8:473–482. https://doi.org/10.1186/1471-2164-8-473
Monreal-Escalante E, Navarro-Tovar G, León-Gallo A, Juárez-Montiel M, Becerra-Flora A, Jiménez-Bremont JF, Rosales-Mendoza S (2016) The corn smut-made cholera oral vaccine is thermostable and induces long-lasting immunity in mouse. J Biotechnol 234:1–6. https://doi.org/10.1016/j.jbiotec.2016.04.047
Broekhuijsen MP, Mattern IE, Contreras R, Kinghorn JR, van den Hondel CA (1993) Secretion of heterologous proteins by Aspergillus niger: production of active human interleukin-6 in a protease-deficient mutant by KEX2-like processing of a glucoamylase-hIL6 fusion protein. J Biotechnol 31:135–145. https://doi.org/10.1016/0168-1656(93)90156-H
Acknowledgements
JAI, CHHR, and LVT received support from COFAA-IPN, EDI-IPN and SNI-CONACyT. JAI and LVT were hired through the “Programa Institucional de Contratación de Personal Académico de Excelencia IPN”.
Funding
This work was supported by SIP-IPN [20181873, 20171553, 20180647, 20182010]. MJM, PTM, VSG were recipients of fellowships from CONACyT and BEIFI-IPN.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Juárez-Montiel, M., Tesillo-Moreno, P., Cruz-Angeles, A. et al. Heterologous expression and characterization of the aspartic endoprotease Pep4um from Ustilago maydis, a homolog of the human Chatepsin D, an important breast cancer therapeutic target. Mol Biol Rep 45, 1155–1163 (2018). https://doi.org/10.1007/s11033-018-4267-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-018-4267-8