Abstract
l-myo-inositol-1-phosphate synthase (MIPS; EC 5.5.1.4) is the key enzyme involved in de novo synthesis of myo-inositol, leading to numerous cellular functions. We isolated an open reading frame of Actinidia deliciosa MIPS (AdMIPS), which is 1,533 bp long and codes for 510 amino acids, with a predicted molecular weight of 56.3 kDa. Sequence analysis revealed its high similarity with MIPS proteins from other organisms. Gene expression and enzyme activity were highest in flower and young fruit. Transcription of AdMIPS was also detected in other tissues. Moderate drought drastically induced expression in the leaves whereas salinity stress induced transcription and enzyme activity in the leaves, phloem, and roots with different degrees. However, a longer period of saline exposure suppressed both expression and enzyme activity in all sampled tissues, indicating that AdMIPS is salt-sensitive.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
l-myo-inositol-1-phosphate synthase (MIPS; EC 5.5.1.4) catalyzes the de novo synthesis of myo-inositol, which is highly conserved in both eukaryotes and bacteria. This is a two-step process. MIPS catalyzes the reaction from d-glucose-6-phosphate to l-myo-inositol-1-phosphate, which is then de-phosphorylated by inositol monophosphatase to release free myo-inositol [1, 2]. The former reaction is regarded as the committed step, making MIPS a rate-limiting enzyme. As the most abundant of the eight isomers of inositols, myo-inositol plays central roles in eukaryotic organisms [3]. Its metabolism, as well as that of derivatives, e.g., inositol phospholipids and inositol hexaphosphate, have vital functions in signal transduction, phosphate storage, membrane formation, stress tolerance, and the synthesis of metabolites such as ascorbic acid [4–7].
Gene cloning and characterization of MIPS have been conducted in animals, plants, yeast, bacteria, and green algae [8, 9]. In plants, the MIPS gene is part of a family in which different members seem to have divergent roles in embryo formation and seed growth [5, 10]. Numerous studies have concentrated on the relationship between plant MIPS and abiotic stress. Smart and Fleming [11] have reported that the transcript levels of MIPS in Spirodela polyrrhiza are induced by abscisic acid (ABA), while Chun et al. [12] have shown that MIPS transcription is down-regulated by salinity during sesame seed germination. Abreu and Aragão [8] have found that transcript levels in yellow passion fruit are differentially regulated by cold and heat and also respond to light stimulus, while research with Jatropha curcas has demonstrated the up-regulation of MIPS transcription and enzyme activity by ABA, drought, and NaCl treatments [13]. A salt-tolerant MIPS gene has been introduced into three genetically different species; transgenic plants exhibit increased inositol production and elevated salt tolerance under salinity stress [14]. Additionally, methylated inositols, such as d-ononitol and d-pinitol, have been associated with tolerances to drought and salinity [15, 16].
Kiwifruit (Actinidia sp.) is a commercially important fruit tree and breeding material all over the world, especially in China and New Zealand. One of its primary carbohydrates is myo-inositol, which accounts for 20–60 % of its total soluble sugar [17]. Drought and salinity are among the environmental stresses that plants most frequently encounter [18]. In some regions of the world, water deficit could be a severe challenge to fruit expansion of kiwifruit trees [19]. It is also known that kiwifruit seedlings accumulate myo-inositol under salinity stress [20]. Because little information is available about the activity of MIPS in that crop, we designed our experiment to focus on the molecular and enzymatic dynamics of MIPS during plant development and in response to stress.
Materials and methods
Plant materials and stress treatments
During the 2011 growing season (13 May to 28 September), fruit samples were collected at 15 day intervals, from 8-year-old vines of kiwifruit (Actinidia deliciosa cv. Qin Mei) at the horticultural experimental field of Northwest Agriculture & Forestry University, Yangling, China. At the mature fruit stage (28 September), shoot tips, phloem, petioles, carpopodia, and young and mature extended leaves were also collected. These samples were used for developmental and tissue-specific analyses of AdMIPS.
In a separate experiment, two-year-old kiwifruit plants were cultivated in individual plastic pots that were of approximately equal weight when the plants were added. Pre-treatment conditions for managing these seedlings followed those previously applied by our laboratory [21]. Drought was induced by withholding irrigation between 12 and 18 August, and before the treatment the maximum soil water capacity was calculated by stoving. Thereafter all pots were weighed daily at 8:00–9:00 a.m. When the soil water capacity reached 55, 45, or 35 % (mild, moderate, or severe drought, respectively), leaf samples were collected. Those from seedlings that continued to receive normal irrigation were used as controls. Immediately after samples from the severe-stress treatment were collected, we re-started the irrigation regimen, and collected more leaf samples after 1 day of this re-watering phase.
Salinity treatments followed time and concentration gradients. Before those experiments began, the seedlings were transferred to a growth room under a 16 h photoperiod (160 μmol m−2 s), 65 % humidity, and a 25 °C/21 °C (day/night) cycle. For 20 day, all plants were irrigated with tap water every 5 day to maintain a soil water capacity of 60–75 %. For the time gradient, 200 mM NaCl was added with the irrigation solution and leaves were collected at days 5, 10, 15, and 20. In addition, the effects of four NaCl concentrations were tested—100, 150, 200, and 250 mM. Each of these solutions was applied only once; thereafter tap water was supplied and any liquid that drained into containers below was poured back into each pot. As our control, some seedlings were irrigated with tap water alone. At the end of this 20 day period, leaf, phloem, and root samples (at least three for one replicate) were collected. All tissues were immediately frozen in liquid nitrogen and stored at −80 °C.
RNA extraction, cDNA synthesis, and cloning of MIPS
Total RNA was extracted from mature fruit by a modified cetyltrimethylammonium bromide (CTAB) method [22]. Prior to reverse-transcription, Rnase-free Dnase I (Invitrogen, USA) was added, per the manufacturer’s instructions, to eliminate DNA contamination. First-strand cDNA was then synthesized with a RevertAid™ First Strand cDNA Synthesis Kit (Fermentas).
The AdMIPS sequence was initially obtained through electronic assembly. Arabidopsis MIPS (GenBank Accession Number U04876) was used as a seed sequence that was aligned with Expression Sequence Tag (EST) sequences [23] of kiwifruit in NCBI. After each round of alignment, we chose five to eight EST sequences that were most similar to the seed sequence. They were assembled with DNAstar (http://www.dnastar.com/) and used as a new seed for more alignments until the acquired sequence could not be elongated further. A sense primer (5′-ATGTTTATCGAGAGCTTTAAG-3′) and antisense primer (5′-TCACTTGTACTCCAAAATC-3′) were designed according to this contig; their product contained the 1,533 bp open reading frame (ORF).
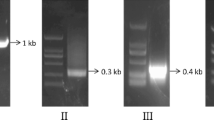
Thermocycling parameters included pre-denaturing at 94 °C for 8 min; then 38 cycles of 94 °C/45 s, 56 °C/45 s, and 72 °C/2 min; followed by a final step at 72 °C for 10 min. The amplified product (~1.5 kb) was purified from the agarose gel and ligased at 4 °C to the pGMET-Easy vector (Promega). The ligation mixture was used to transform Top10 competent cells and the transformants were selected on ampicillin plates. Single colonies with resistance were detected via PCR, and positive samples were sequenced with an ABI 3730 sequencer. The sequence data obtained for AdMIPS were checked against published MIPS sequences from other organisms and were analyzed with standard bioinformatics tools.
Phylogenetic analysis
AdMIPS and protein sequences of MIPS from 20 organisms were obtained from GenBank (www.ncbi.nih.nlm.gov) and aligned using Clustal W [24]. Phylogenetic analysis was conducted with MEGA (Molecular Evolutionary Genetic Analysis) version 5.0 software [25]. A neighbor-joining algorithm was used for constructing the phylogenetic tree, and bootstrap values were computed with 1,000 replicates to evaluate support for the groupings.
Real-time PCR analysis of MIPS
RNA was extracted from plant tissues as described above. Reverse transcription was performed with 1 μg of RNA and a PrimeScript® RT reagent Kit (Takara). All of the reverse transcripts were adjusted with double-distilled water to a concentration of 150 ng uL−1. A sense primer (5′-TCTCTCGGTCCCTCAAACTT-3′) and antisense primer (5′-CCCACATACGGCACATACTT-3′) were designed with Primer Premier, version 5.0, software (Palo Alto, CA). The RT-PCR was carried out by Bio-Rad iQ5 thermocycler in triplicate and the RT-PCR program included pre-denaturing at 94 °C for 3 min; then 45 cycles of denaturing at 94 °C for 30 s, annealing at 60 °C for 20 s, and elongation at 72 °C for 30 s. For the first cycle, we used a melt curve to ensure that the primers could not form dimers. Gene expression for each sample was normalized against house-keeping gene Actin with sense primer (5′-GCTTACAGAGGCACCACTCAACC-3′) and antisense primer (5′-CCGGAATCCAGCACAATACCAG-3′) and analyzed with Bio-Rad iQ5 optical system software.
Crude enzyme extraction and study of AdMIPS activity
Samples from the flowers, leaves, phloem, and roots were ground in a tenfold volume of extraction buffer (Tris–HCl; pH 7.5) containing 10 mM NH4Cl, 10 mM β-mercaptoethanol, and 2 mM phenylmethylsulfonyl fluoride that was supplemented with 1 % Triton X-100 and 4 % polyvinylpolypyrrolidone. The homogenates were centrifuged for 30 min at 4 °C and 10,000 g. The supernatant was collected and stored at 4 °C, with one portion designated for detecting enzyme activity and the other for determining the soluble protein content. Enzyme activity was monitored according to the method of Barnett et al. [26] and was presented as nmol inositol-1-phosphate formed in each milligram of soluble protein within 1 h of reaction. The amount of soluble protein was measured as described by Bradford [27].
Results
Cloning and characterization of MIPS gene in Actinidia deliciosa
Through electronic assembly, we created a 1,756 bp contig and cloned a complete ORF sequence of MIPS from Actinidia deliciosa. This ORF contains 1,533 bp, coding for a protein with 510 predicted amino acids (Fig. 1). We have deposited this 1,533 bp sequence into GeneBank under Accession Number: JX122766. A comparison with protein information from other organisms revealed its high similarity with enzyme in eukaryotes; alignment was more divergent with prokaryotes, as previously described [28, 29]. When aligned with other plant MIPS protein sequences, four stretches of ‘highly conserved’ amino acids were also found in the AdMIPS sequence—GWGGNNG, LWTANTER, NGSPQNTFVPGL, and SYNHLGNNDG (Fig. 2). This is presumed to be essential for substrate-binding to the MIPS enzyme, perhaps pointing toward a conserved ‘core structure’ for catalytic activity through evolution [28]. A phylogenetic tree for MIPS from different species produced clustering into four groups—plant, animal, alga, and bacterium (Fig. 3). Terrestrial and aquatic plants had clearly been segregated into two groups early whereas the divergence of dicotyledons and monocotyledons within terrestrial plants occurred later in the evolutionary process. Interestingly, Triticum aestivum clustered with dicotyledons rather than with other monocotyledons such as Zea mays. AdMIPS was most closely related to that from Citrus paradisi, a tropical fruit.
Alignment of MIPS amino acid sequences from different plants. Boxed residues indicate four conserved stretches. GenBank Accession Numbers for nucleotides include: Arabidopsis thaliana, U04876; Sesamum indicum, AF284065; Phaseolus vulgaris, U38920; Nicotiana tabacum, AB009881; Citrus paradisi, Z32632; Actinidia deliciosa, JX122766; Mesembryanthemum crystallinum, U32511; Oryza sativa, AB012107; Spirodela polyrhiza, Z11693
Phylogenetic analysis of MIPS proteins from different organisms. AdMIPS in phylogenetic tree is marked by triangle. GenBank Accession Numbers for nucleotides include: Arabidopsis thaliana, U04876; Phaseolus vulgaris, U38920; Triticum aestivum, AF120146; Nicotiana tabacum, AB009881; Citrus paradisi, Z32632; Actinidia deliciosa, JX122766; Mesembryanthemum crystallinum, U32511; Sesamum indicum, AF284065; Zea mays, AF056326; Hordeum vulgare, AF056325; Oryza sativa, AB012107; Spirodela polyrhiza, Z11693; Xenopus laevis, BC077437; Mus musculus, AF288525; Rattus norvegicus, AABR03100304; Bos taurus, BC111160; Homo sapiens, AF207640; Macaca fascicularis, AB168239; Candida albicans, L22737; Candida glabrata, CR380955; Saccharomyces cerevisiae, L23520; Mycobacterium smegmatis, CP000480; Mycobacterium tuberculosis, BX842572
MIPS gene expression and enzyme activity of A. deliciosa fruits at different developmental stages
Transcript levels for AdMIPS were highest at days 0 and 15 after flowering, being three to four times greater than at day 30. Thereafter, expression declined gradually, almost to zero (Fig. 4a).
Changes in enzyme activity followed a trend similar to that for gene expression, with activities on days 0 and 15 being five to six times higher than measured on day 30. Afterward, activities remained relatively stable, increasing only slightly to just less than 2 nmol h−1 mg−1 at day 105 before dropping to the level recorded at day 30 (Fig. 4b).
Expression of AdMIPS in different tissues of kiwifruit
AdMIPS transcription was detected in all six tissue types. Compared with levels found for the house-keeping gene actin, expression of AdMIPS was higher in the petiole than in any other tissue. Whereas shoot tips and mature leaves showed relatively higher expression, much less was detected in the young leaves, carpopodium, and phloem (Fig. 5).
Expression profile and enzyme activity of MIPS in A. deliciosa plants subjected to drought or salinity
Relative expression by AdMIPS was about 3 times (mildly drought-stressed plants) and 25 times higher (moderately drought-stressed plants) than the control respectively. Under severe drought, the transcription level decreased by approximately two-thirds of the highest amount, then, after 1 day of re-watering, declined to the level measured in the control (Fig. 6a). Enzyme activity of MIPS did not follow a similar trend but, instead, showed no significant difference among stress severities. After 1 day of re-watering, enzyme activity continued to decrease significantly to just above 2 nmol h−1 mg−1. (Fig. 6b).
a Expression of AdMIPS in leaves under increasingly severe drought stress and re-watering. b Enzyme activity of AdMIPS in leaves during drought and re-watering treatments. For a and b, D3, D5, D6 represent drought treatment for 3, 5, and 6 days (mild, moderate, and severe levels) respectively. D7 is 1 day after re-watering. c Time courses for AdMIPS expression in leaves under 200 mM NaCl treatment. d Enzyme activity of AdMIPS in leaves under 200 mM NaCl stress. e Expression of AdMIPS in leaves, phloem, and roots after 20 day of treatment with various NaCl concentrations. f Enzyme activity of AdMIPS in leaves, phloem, and roots after 20 day of treatment with various NaCl concentrations. For e and f, S0, S100, S150, S200, S250 represent salt concentration of 0, 100, 150, 200 and 250 mM respectively. Significant differences from control values are indicated by * (for leaves) and + (for roots); * and +, P < 0.05; ** and ++, P < 0.01. Data are mean ± SE of 3 independent extracts
Under 200 mM NaCl treatment, transcription levels were 1.5 and 3.5 times higher than the control at days 5 and 10 after treatment, respectively. Afterward, expression declined sharply to one third of the control level at day 15 and to less than one-quarter of the control at day 20 (Fig. 6c). MIPS activity showed similar changes in response to salinity stress, rising gradually at days 5 and 10 before being repressed to levels below those of the control at days 15 and 20 (Fig. 6d).
Under control conditions, AdMIPS expression was identical between leaves and phloem. However, the expression in leaves was induced by 150 mM NaCl, repressed by 100, 200 and 250 mM NaCl; whereas that in phloem was decreased only by 250 mM NaCl. Root tissues from control plants showed the lowest expression; all four tested concentrations of NaCl led to an increase in AdMIPS transcription there (Fig. 6e). For the MIPS activity of these tissues, a converse tendency in leaves and roots was observed. Except at 150 mM NaCl, enzyme activity in the leaves was increased at all other concentrations whereas the highest level of enzyme activity in the roots was found in the control. The AdMIPS activity was slightly induced in the phloem by 100 mM NaCl (Fig. 6f).
Discussion
AdMIPS is a highly conserved enzyme
Previous studies of the nucleotide sequences and protein structures of myo-inositol-1-phosphate synthase have provided supporting evidence that MIPS is a highly conserved enzyme throughout the eukaryotic and prokaryotic phyla [28]. Although those groups tend to segregate into two divergent branches of the phylogenetic tree, they show high similarity in amino acid sequences. The ‘core catalytic domains’ present in both groups suggest two critical functions. First, MIPS catalyzes the de novo synthesis of myo-inositol, which is a precursor to several important metabolites, e.g., phytic acid, raffinose, ascorbic acid, and phosphoinositides. Second, it can act directly as an osmoprotectant for cellular functions [3].
Young fruit and reproductive tissue are major sources of AdMIPS
Many research groups have examined the relationship between seeds and MIPS because of the abundance of inositol hexaphosphate (phytic acid), the storage form of phosphate [30]. MIPS genes have been shown to be differentially expressed in Arabidopsis embryos at different developmental stages [5]. However, little information is available about MIPS expression in fruit trees. We detected MIPS gene expression and enzyme activity at several developmental stages in kiwifruit, especially the young fruit and flowers, which are major sources of AdMIPS. These findings are corroborated by previous reports [17, 31, 32]. This high amount of myo-inositol in young fruit may be associated with the maintenance of turgor during the cell expansion phase [31]. Additionally, high detection in the flowers might indicate a reproductive role for MIPS in kiwifruit. At the mature fruit stage, transcripts of AdMIPS were found in all tissue types that we collected, suggesting that MIPS expression is ubiquitous. Thus, the coordination of inositol metabolism and cellular growth can be achieved through such differential regulation [1, 33]. Several species, such as Zea mays [34], Glycine max [35], and Arabidopsis thaliana [36] contain more than one MIPS, and various isoforms have been localized to different organs in Arabidopsis, also indicating tissue-specific roles [5].
AdMIPS is regulated by drought or salt stress
MIPS can be induced upon exposure to drought, salinity, freezing, darkness, or treatment with ABA, demonstrating its important function in response to environmental stresses [8, 12, 37]. Here, under drought or a high NaCl concentration, gene expression and enzyme activity of AdMIPS were induced differentially. Despite a transient increase, both then declined gradually, indicating that AdMIPS could be suppressed at both the transcript and enzyme levels as the stress period lengthened. MIPS from a salt-tolerant rice has shown enhanced mRNA expression and retained a constant level of enzyme activity even under high salt concentrations. By contrast, the transcription of MIPS is not induced under saline conditions in Arabidopsis thaliana, suggesting an essential difference in response between halophytes and glycophytes [15, 38]. Myo-inositol accumulation was detectable before kiwifruit seedlings showed any signs of salt stress [20]. Our experiment confirms this result at transcription and enzyme levels. Actinidia deliciosa is known not salt- or drought-tolerant. However, the increase in transcript, enzyme and myo-inositol levels may still suggest some protection to plant. Myo-inositol may have functions such as osmotic protection, scavenging oxygen radicals or storage of unused carbohydrate under osmotic stress [3, 20]. In other plant species, MIPS transcription or its enzyme activity were also induced by salt, drought or heat stress within 24 h [8, 13], indicating the importance of myo-inositol at the early stage of stress. When exposed to salinity stress, the higher enzyme activity in roots versus leaves/phloem might suggest that green tissues are rapidly impaired while roots may continue to elongate as they take up water from deeper soil layers [18].
To summarize, we have cloned a complete ORF of MIPS from A. deliciosa, and analyzed its transcription and enzyme activity in terms of fruit development, tissue specificity, and responsiveness to abiotic stresses. AdMIPS is an evolutionarily conserved enzyme, showing high similarity with MIPS from other plant sources. It might serve as a key element in young fruit given its high expression and level of enzyme activity. As reported with other glycophytes, AdMIPS can be induced by drought or salinity at both the mRNA and enzyme levels [8, 12]. Engineering of enzymes that catalyze the production of osmoprotectants is a useful tool for improving stress tolerance in plants [39]. In addition, MIPS tends to present as multigene families in plants, with each member appearing to be tissue-specific and having different functions [5]. To gain greater insight into the mechanism of myo-inositol metabolism in kiwifruit, further research should focus on the overexpression and downregulation of AdMIPS gene(s).
References
Majumder AL, Johnson MD, Henry SA (1997) l-myo-Inositol-1-phosphate synthase. Biochim Biophys Acta (BBA)-Lipids and Lipid. Metabolism 1348(1–2):245–256
Loewus FA, Loewus MW (1983) Myo-inositol: its biosynthesis and metabolism. Annu Rev Plant Physiol 34(1):137–161
Loewus FA, Murthy PPN (2000) Myo-Inositol metabolism in plants. Plant Sci 150(1):1–19
Lorence A, Chevone BI, Mendes P, Nessler CL (2004) Myo-Inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134(3):1200–1205
Donahue JL, Alford SR, Torabinejad J, Kerwin RE, Nourbakhsh A, Ray WK, Hernick M, Huang X, Lyons BM, Hein PP (2010) The Arabidopsis thaliana myo-inositol 1-phosphate synthase1 gene is required for myo-inositol synthesis and suppression of cell death. Plant Cell 22(3):888–903
Kanter U, Usadel B, Guerineau F, Li Y, Pauly M, Tenhaken R (2005) The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221(2):243–254
Raboy V (2003) Myo-Inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64(6):1033–1043
Abreu EFM, Aragão FJL (2007) Isolation and characterization of a myo-inositol-1-phosphate synthase gene from yellow passion fruit (Passiflora edulis f. flavicarpa) expressed during seed development and environmental stress. Ann Bot 99(2):285–292
RayChaudhuri A, Hait NC, DasGupta S, Bhaduri TJ, Deb R, Majumder AL (1997) l-myo-lnositol 1-phosphate synthase from plant sources (characteristics of the chloroplastic and cytosolic enzymes). Plant Physiol 115(2):727–736
Kaur H, Shukla RK, Yadav G, Chattopadhyay D, Majee M (2008) Two divergent genes encoding l-myo-inositol 1-phosphate synthase1 (CaMIPS1) and 2 (CaMIPS2) are differentially expressed in chickpea. Plant Cell Environ 31(11):1701–1716
Smart CC, Fleming AJ (1993) A plant gene with homology to d-myo-inositol-3-phosphate synthase is rapidly and spatially up-regulated during an abscisic acid-induced morphogenic response in Spirodela polyrrhiza. Plant J 4(2):279–293
Chun JA, Jin UH, Lee JW, Yi YB, Hyung NI, Kang MH, Pyee JH, Suh M, Kang CW, Seo HY (2003) Isolation and characterization of a myo-inositol 1-phosphate synthase cDNA from developing sesame (Sesamum indicum L.) seeds: functional and differential expression, and salt-induced transcription during germination. Planta 216(5):874–880
Wang Y, Huang J, Gou CB, Dai X, Chen F, Wei W (2011) Cloning and characterization of a differentially expressed cDNA encoding myo-inositol-1-phosphate synthase involved in response to abiotic stress in Jatropha curcas. Plant Cell Tissue Organ 106(2):269–277
Das-Chatterjee A, Goswami L, Maitra S, Dastidar K, Ray S, Majumder A (2006) Introgression of a novel salt-tolerant l-myo-inositol 1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka (PcINO1) confers salt tolerance to evolutionary diverse organisms. FEBS Lett 580(16):3980
Nelson DE, Rammesmayer G, Bohnert HJ (1998) Regulation of cell-specific inositol metabolism and transport in plant salinity tolerance. Plant Cell 10(5):753–764
Patra B, Ray S, Richter A, Majumder AL (2010) Enhanced salt tolerance of transgenic tobacco plants by co-expression of PcINO1 and McIMT1 is accompanied by increased level of myo-inositol and methylated inositol. Protoplasma 245(1):143–152
Bieleski RL, Clark CJ, Klages KU (1997) Identification of myo-inositol as a major carbohydrate in kiwifruit, Actinidia deliciosa. Phytochemistry 46(1):51–55
Pardo JM (2010) Biotechnology of water and salinity stress tolerance. Curr Opin Biotechnol 21(2):185–196
Judd M, McAneney K, Wilson K (1989) Influence of water stress on kiwifruit growth. Irrigation Sci 10(4):303–311
Klages K, Boldingh H, Smith G (1999) Accumulation of myo-inositol in Actinidia seedlings subjected to salt stress. Ann Bot 84(4):521–527
Wang Y, Ma F, Li M, Liang D, Zou J (2011) Physiological responses of kiwifruit plants to exogenous ABA under drought conditions. Plant Growth Regul 64(1):63–74
Chang S, Puryear J, Cairney J (1993) A simple and efficient method for isolating RNA from pine trees. Plant Mol Biol Rep 11(2):113–116
Crowhurst RN, Gleave AP, MacRae EA, Ampomah-Dwamena C, Atkinson RG, Beuning LL, Bulley SM, Chagne D, Marsh KB, Matich AJ (2008) Analysis of expressed sequence tags from Actinidia: applications of a cross species EST database for gene discovery in the areas of flavor, health, color and ripening. BMC genomics 9(1):351
Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Barnett J, Brice R, Corina D (1970) A colorimetric determination of inositol monophosphates as an assay for d-glucose 6-phosphate-1l-myoinositol 1-phosphate cyclase. Biochem J 119(2):183
Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Majumder AL, Chatterjee A, Ghosh Dastidar K, Majee M (2003) Diversification and evolution of l-myo-inositol 1-phosphate synthase. FEBS Lett 553(1–2):3–10
Chen L, Zhou C, Yang H, Roberts MF (2000) Inositol-1-phosphate synthase from Archaeoglobus fulgidus is a class II aldolase. Biochemistry 39(40):12415–12423
Raboy V (2009) Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci 177(4):281–296
Boldingh H, Smith G, Klages K (2000) Seasonal concentrations of non-structural carbohydrates of five Actinidia species in fruit, leaf and fine root tissue. Ann Bot 85(4):469–476
Klages K, Donnison H, Boldingh H, MacRae E (1998) Myo-Inositol is the major sugar in Actinidia arguta during early fruit development. Funct Plant Biol 25(1):61–68
Ishitani M, Majumder AL, Bornhouser A, Michalowski CB, Jensen RG, Bohnert HJ (1996) Coordinate transcriptional induction of myo-inositol metabolism during environmental stress. Plant J 9(4):537–548
Larson S, Raboy V (1999) Linkage mapping of maize and barley myo-inositol 1-phosphate synthase DNA sequences: correspondence with a low phytic acid mutation. Theor Appl Genet 99(1):27–36
Hegeman CE, Good LL, Grabau EA (2001) Expression of d-myo-inositol-3-phosphate synthase in soybean. Implications for phytic acid biosynthesis. Plant Physiol 125(4):1941–1948
Johnson MD, Sussex IM (1995) 1l-myo-Inositol 1-Phosphate Synthase from Arabidopsis thaliana. Plant Physiol 107(2):613–619
Keller R, Brearley CA, Trethewey RN, Müller-Röber B (1998) Reduced inositol content and altered morphology in transgenic potato plants inhibited for 1d myo-inositol 3-phosphate synthase. Plant J 16(4):403–410
Majee M, Maitra S, Dastidar KG, Pattnaik S, Chatterjee A, Hait NC, Das KP, Majumder AL (2004) A novel salt-tolerant l-myo-inositol-1-phosphate synthase from Porteresia coarctata (Roxb.) Tateoka, a halophytic wild rice. J Biol Chem 279(27):28539
Vinocur B, Altman A (2005) Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotech 16(2):123–132
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30871700). The authors are grateful to Priscilla Licht for help in revising our English composition.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Cui, M., Liang, D. & Ma, F. Molecular cloning and characterization of a cDNA encoding kiwifruit l-myo-inositol-1-phosphate synthase, a key gene of inositol formation. Mol Biol Rep 40, 697–705 (2013). https://doi.org/10.1007/s11033-012-2110-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-012-2110-1