Abstract
The nucleotide sugar UDP-glucuronic acid (UDP-GlcA) is the principal precursor for galacturonic acid, xylose, apiose and arabinose residues of the plant cell-wall polymers. UDP-GlcA can be synthesized by two different functional pathways in Arabidopsis involving either UDP-glucose dehydrogenase or inositol oxygenase as the initial enzyme reaction to channel carbohydrates into a pool of UDP sugars used for cell-wall biosynthesis. The genes for the enzyme myo-inositol oxygenase (MIOX) were analyzed in Arabidopsis. They represent a small gene family containing four members. The transcription of all those members indicates a transient and organ-specific gene expression pattern in growing plant tissues as analyzed by RT-PCR and in promoter::GUS reporter gene lines. Two isoforms (MIOX1, MIOX2) are expressed in almost all tissues of the plant, whereas the expression of MIOX4 and MIOX5 is largely restricted to flowers, particularly maturing pollen. T-DNA insertion lines in MIOX genes were isolated; however, single knock-outs show growth phenotypes similar to the wild type. The monosaccharide composition of the cell wall in these mutants is not significantly changed compared to wild type plants. However, the incorporation of 3H-inositol into wall polymers of seedlings is greatly impaired in the mutant lines ΔMIOX1 and ΔMIOX2, which are the only isoforms that are expressed in seedlings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant cells are surrounded by rigid yet flexible cell walls composed of a network of different carbohydrate polymers and glycoproteins. The polymers—except for cellulose and callose—are synthesized from nucleotide sugars in the cisternae of the Golgi apparatus. The polysaccharides are subsequently transported into the apoplast and integrated in the polymer network.
Arabidopsis, as a short-living small plant, has mostly primary cell walls with a high degree of pectic polymers and a low cellulose content (Zablackis et al. 1995). Based on the relative amounts of each polymer in the mature Arabidopsis cell wall and the monosaccharide composition thereof, one can calculate that roughly 50% of the cell-wall biomass (xylose, arabinose, apiose, glucuronic acid, and galacturonic acid) is derived from the precursor UDP-glucuronic acid (UDP-GlcA) through a complex nucleotide sugar interconversion pathway (Gibeaut 2000; Reiter and Vanzin 2001; Seifert 2004). This precursor is converted by an epimerase into UDP-galacturonic acid (Usadel et al. 2004a; Mølhøj et al. 2004), being an important precursor for pectic polymers. Another branch leads from UDP-GlcA after decarboxylation to UDP-xylose (Harper and Bar-Peled 2002; Mølhøj et al. 2003), which can be subsequently epimerized to UDP-arabinose (Burget et al. 2003).
In the last decade, many mutations in the nucleotide sugar interconversion pathway in Arabidopsis have been identified, which often show an altered composition of the cell-wall monomers, morphological changes compared to wild-type plants or structural changes of the cell wall. For instance, the mur1 mutant has a defect in the biosynthesis of GDP-fucose and thus underlines the importance of terminal fucose residues for the rhamnogalacturon side chain and the boron-mediated cross-linking thereof (O‘Neill et al. 2001). Mutants in the NDP-rhamnose biosynthesis show lower pectic mucilage around the seed coat (Usadel et al. 2004b; Western et al. 2004). A mutation in one of the UDP-glucose-4-epimerases of plants, which reduces the available amount of UDP-galactose results in an altered root morphology and less root hairs (Seifert et al. 2002). Most of the mutations are only partially blocked in the respective biosynthesis because isoenzymes partially substitute for the function of the mutated gene. Nevertheless, a reduction of the pool of certain nucleotide sugars has often a dramatic phenotypic effect, which is not compensated and corrected at the level of the polymer synthases.
Our interest is the biosynthesis of UDP-GlcA. Plants synthesize UDP-GlcA via two independent pathways (Fig. 1). In the more prominent pathway, UDP-glucose is oxidized directly into UDP-GlcA by the enzyme UDP-glucose dehydrogenase (UGD; Tenhaken and Thulke 1996). This pathway is often the only one mentioned in plant biochemistry textbooks. However, from early studies by the Loewus group, it became clear that a second pathway for the biosynthesis of UDP-GlcA exists in plants, which is called the inositol oxygenation pathway (Loewus et al. 1962). In cyanobacteria, fungi, animals and plants, myo-inositol is synthesized from glucose-6-phosphate by the enzyme inositol-1-phosphate synthase. The corresponding genes are known from many organisms including plants (Smart and Fleming 1993; Loewus and Murthy 2000). A specific myo-inositol-1-phosphate phosphatase generates the free myo-inositol. Within the inositol oxygenation pathway, myo-inositol is converted into free GlcA by oxygenative cleavage. Subsequently, GlcA is activated via GlcA-1-phosphate into UDP-GlcA (Loewus and Murthy 2000). The enzymatic activities necessary for the activation of GlcA were shown to be present in different plant tissues (Roberts 1971; Leibowitz et al. 1977).
The key enzyme of the inositol oxygenation pathway is myo-inositol oxygenase (MIOX; E.C. 1.13.99.1). The enzyme uses molecular oxygen and catalyzes the oxygenative cleavage of myo-inositol to GlcA, which makes the overall reaction thermodynamically irreversible. The enzyme MIOX is therefore a good candidate to control the flux of carbohydrates through this pathway. Until recently, not much was known about the enzyme MIOX, because it is difficult to measure in plant extracts and because no molecular sequences were available. By purifying the enzyme from pig kidney (Arner et al. 2001) and from the yeast Cryptococcus lactativorus (Kanter et al. 2003), peptide sequences were obtained which allowed the subsequent cloning of the corresponding genes . These sequences now provide molecular tools to analyze the role of the MIOX pathway for nucleotide sugar biosynthesis. Using a similar approach, Lorence et al. (2004) investigated the role of MIOX for ascorbate biosynthesis in transgenic Arabidopsis plants. They isolated the Miox4 gene by a homology-based PCR approach and found a flower-specific gene expression for the Miox4 gene. A recombinant Miox catalyzed the conversion of myo-inositol into glucuronic acid. The ectopic expression of the Miox4 gene in transgenic Arabidopsis results in plants with elevated levels of ascorbic acid, suggesting a role for glucuronic acid in the vitamin C biosynthesis in plants (Lowrence et al. 2004).
Myo-inositol is also central to several other pathways in plants. Inositol phosphates are essential components of cellular signal transduction pathways (Majerus 1992). The hexaphosphate of inositol, named phytate, is the most important phosphate storage component in seeds (Raboy 2003). Furthermore, inositol can be incorporated into membrane lipids. Some plant species like legumes synthesize the raffinose oligosaccharide series, which requires galactinol, made from UDP-galactose and inositol, as substrate for elongation. In some plants, inositol is mono- or di-methylated to produce compatible solutes as osmoprotectant compounds under drought–stress conditions (Vernon and Bohnert 1992). Thus, myo-inositol biosynthesis and conversion must be highly controlled and regulated to meet the requirements of the different pathways.
Here, we report on the Arabidopsis gene family for inositol oxygenases, which contribute to the pool of nucleotide sugars for the synthesis of plant cell walls.
Materials and methods
Plant material
Arabidopsis seeds, Col-0 and T-DNA-insertion lines of the SALK collection were obtained from the Arabidopsis stock center (NASC, Nottingham, UK). T-DNA-insertion lines of the GABI collection were obtained from GABI-KAT (Cologne, Germany).
Identification of the Arabidopsis MIOX genes
The protein sequence of the inositol oxygenase gene from Cryptococcus lactativorus, that we have recently cloned by PCR, using data from the de novo sequencing of the purified enzyme (Kanter et al. 2003), was used to search the Arabidopsis genome for homologous sequences with the BLASTP program (http://www.ncbi.nlm.nih.gov/BLAST/). The protein sequences were aligned with the CLUSTAL software and a bootstrapped neighbor-joining tree was visualized with TreeView (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). The gene model for each MIOX gene was verified by amplifying the cDNA for each isoform from reverse-transcribed RNA. The following primers were used: MIOX1: 5′-ATCCATGGCTATTCTCATAGATCGCCACTCTG-3′; 5′-ACCATTTTAGCTTGGACGGA-3′; MIOX2: 5′-TGGATCCATTCCGAGCAAGATGACTATTCT-3′; 5′-ATAAGCTTGGGGGAAAAAATTACAAATGAACAC-3′; MIOX4: 5′-ATCCATGGCGATCTCTGTTGAGAAGCCG-3′; 5′-TCACCACCTCAAGTTTTCCG-3′; MIOX5: 5′-ATGGATCCAGATGAATATCTCCGTCGAAAACC-3′; 5′-ATAAGCTTATGCCGTTTTCACCACCTCAGA-3′.
RNA isolation and RT-PCR
RNA from different plant tissues of Arabidopsis Col-0 plants was isolated by the acid phenol/guanidinium thiocyanate method according to Chomczynski and Sacchi (1987). First-strand cDNA from 3 μg of total RNA was synthesized with an MMuLV-reverse transcriptase (MBI-Fermentas) according to the supplier’s protocol. The primer pair for APT1 (5′-TCCCAGAATCGCTAAGATTGCC-3′; 5′-CCTTTCCCTTAAGCTCTG-3′) was used to amplify a 478-bp fragment. The APT1 gene is considered to be constitutively expressed in Arabidopsis (Moffat et al. 1994).
The conditions for the RT-PCR of MIOX-isoforms were as follows: MIOX1: 5′-TCTCATAGATCGCCACTCTG-3′; 5′-CAGGATCACTCTCGTCGATA-3′ (331 bp); MIOX2: 5′-GAAAGTGATAGAGGAGAGGGATAA-3′; 5′-AGTGAGATGGAGCCAATCTT-3′ (396 bp); MIOX4: 5′-TCTCTGTTGAGAAGCCGATTT-3′; 5′-GCTCGTCAAGATCTGGATCA-3′ (365 bp); MIOX5: 5′-GGACTCGACAACGCAGAA-3′; 5′-GGATTGCTTCCGCAGATTA-3′ (361 bp). The PCR program for all isoforms was as follows: 94°C for 3 min; 5× (94°C for 15 s, 61–1°C/cycle for 30 s (touch down), 72°C for 1 min); 30× (94°C for 15 s; 56°C for 30 s; 72°C for 1 min); 72°C for 3 min. Products were separated on a 1.5% TAE-agarose gel, stained with ethidium bromide and photographed.
RNA for real-time PCR experiments was extracted from different tissues from greenhouse grown Arabidopsis thaliana Col-0 plants as reported earlier (Logeman et al. 1987). Residual genomic DNA was removed by treatment with RNAse-free DNAse (Roche, Mannheim, Germany) and approximately 2 μg of total RNA was reverse transcribed with an MMuLV reverse transcriptase (Qbiogene, Heidelberg, Gemany) using an oligo (dT) primer according to the manufacturer’s suggestion, thus generating 21 μl of first-strand cDNA. Real-time RT-PCR was performed using 10 μl of a 1/30 (v/v) dilution of the first-strand cDNA reaction and SyberGreen master mix (Applied Biosystems, Darmstadt, Germany) in a reaction volume of 20 μl on a GeneAmp 5700 Sequence Detection System (PE-Applied Biosystems, Darmstadt, Germany) using conditions described earlier (Czechowski et al. 2004). The following primers were used: MIOX1: 5′- CACACCAACTCTTTTGGTCGC-3′; 5′-GTACGATTTAGCTTCTCGTATTCTTC-3′; MIOX2: 5′- TGATATGAATTTCTTGGGCCATT-3′; 5′- ATCTTGTTAAGTTTTCCATACTCTTTCC-3′; MIOX4: 5′- GAGATGAATGCATTTGGCCG-3′; 5′- TTTATCTAATTTTCCATATTCAGCCC-3′; MIOX5: 5′- GAGATGAACGCATTTGGTCGT-3′; 5′- CTTGTCCAATTTTCCATACTCACTT-3′. Each primer pair amplified a single product, as indicated by the curve of the melting temperature of the amplicons. The resulting CT values were normalized to the average of the CT values of the two standard transcripts; adenine-phosphoribosyltransferase (At1g27450) and Actin2 (At3g18780) using primers described previously (Czechowski et al. 2004).
Reporter-gene constructs
The promotor sequences of the ArabidopsisMIOX2 and MIOX4 genes were amplified by PCR, using the primers MIOX2 5′-ATAAGCTTTTTTTGTCGTTTCCTTAAGTATG-3′; 5′-TAGGATCCTCGGAATTTGATACGTAATTTAGAT-3′ and MIOX4 5′- ATAAGCTTCACCAACTCAAGTCAAGCAGCA-3′; 5′-AAGGATCCGAGAAGAAACGAAACGAAACGAA-3′ and a PCR program with the steps 94°C 3 min; 35× (94°C 15 s, 60°C 30 s, 72°C 1 min); 72°C 5 min. The PCR products were cut with HindIII amd BamHI, gel purified and cloned in front of the GUS gene of vector pBI101. After verifying the construct by DNA sequencing, the plasmids were transferred into Agrobacterium tumefaciens GV3101 (pMP90). Arabidopsis plants were transformed by the floral dip method (Clough and Bent 1998).
Seedlings of transgenic Arabidopsis reporter gene lines were grown sterile on MS-agar plates (0.5×strength, 0.2% phytagel) with either 0.5% sucrose or 0.5% inositol. Plates were grown at 23°C in the light (fluorescent bulbs ~80 μE m−2 s−1) or in the dark to analyze etiolated seedlings. Seedlings, carrying the MIOX2::GUS or the MIOX4::GUS constructs, were stained for GUS activity using the protocol of Jefferson (1987). The plant organs were stained for 4–16 h, followed by an ethanol series extraction (20%, 50%, 70%) to remove chlorophyll. Plants were photographed with a Leica stereo microscope, equipped with a digital camera. Photos were assembled in Adobe PhotoshopCS.
Inositol labeling of Arabidopsis seedlings
Surface sterilized seeds of Arabidopsis wild-type plants or T-DNA insertion lines were germinated in a film (0.5 ml each in 12-well plates) of liquid MS medium (Duchefa M0245) supplemented with 0.1 g l−1 sucrose and 3H-inositol (74 kBq; specific activity 555 GBq mMol−1) in the dark at 23°C. After 7 days, ten seedlings were removed from the germination fluid and washed to remove unbound inositol and subsequently homogenized in 70% ethanol in a bead beater with glass beads. The extraction was repeated three times until less than 2% of the insoluble radioactivity was determined in the liquid extract, followed by two extractions with chloroform/methanol (1:1). The polymer-bound radioactivity was determined by liquid scintillation counting. In some experiments, the dried crude cell wall was hydrolyzed in 2 M of TFA and sugars were recovered by lyophilization and separated on a Polygram SIL G/UV254 thin-layer chromatography plate (Macherey Nagel, Düren, Germany) as described earlier (Seitz et al. 2000).
Isolation of homozygous insertional mutants
Seeds for putative insertional mutants in some MIOX genes were obtained from the SALK (ΔMIOX2: SALK_040608, ΔMIOX5: SALK_112535) and the GABI-KAT collection (ΔMIOX1: 450d10, ΔMIOX2: 161e06). Homozygous insertional mutants were isolated from these using a PCR-based scheme with the recommended T-DNA-specific primers and gene-specific primers spanning the putative insertion site (ΔMIOX1: 5′-CTCTACAGCAAAAGCAAAGTC-3′; 5′-CACAAACGAAACAACACTCTG-3′; ΔMIOX2 (SALK) 5′-CATTTTCAGATCTTGGCAAGGTTC-3′; 5′-CCAGCGTGAGGAAGGGTCG-3′, ΔMIOX2 (GABI) 5′-ATTCCTTCAGGTTTGTTTAATGG-3′; 5′-GAATTTGAGGCTCATCAAGATC-3′; ΔMIOX5 5′-TTGCACAAGGCTGGAGCTTA-3′; 5′-AAGAGACATGTAGTACGGCTTAAC-3′).
Chemical analysis of the cell wall
The monosaccharide composition of the wall material, derived from leaves of 4-week-old Arabidopsis plants (greenhouse, 16-h light period, ~23°C), was determined by acid hydrolysis and derivatization of the resulting monosaccharides to alditol acetates followed by gas chromatographic analysis (York et al. 1985).
Ascorbic-acid measurements
Ascorbic-acid content was measured as previously described (Lorence et al. 2004). Leaves from 3- or 5-week-old Arabidopsis plants, grown in the greenhouse (16-h light period, ~23°C), were collected at noon on either a sunny or a cloudy day. Wild-type plants and MIOX knock-out lines were grown in the same tray to allow side-by-side comparison.
Results
Identification of the inositol oxygenase genes
In a previous study (Kanter et al. 2003), we purified the enzyme inositol oxygenase from the yeast Cryptococcus lactativorus and identified the gene by a PCR approach that used peptide sequence information for primer design. The amino acid sequence of the yeast MIOX is highly similar to a family of putative proteins in Arabidopsis. A BLAST search identified four MIOX-like proteins from Arabidopsis (P-value < 1 e−65)(see also Lorence et al. 2004). The hits also include a MIOX gene from pig, for which the enzyme was recently purified (Arner et al. 2001). It is important to note that the BLASTP search does not identify any other similar protein from Arabidopsis (using a cutoff of P<0.01), indicating that the MIOX sequences represent a unique class of proteins with no evolutionary close relatives. The amino acid sequences of the four identified MIOX proteins, which were named according to an earlier suggestion (Lorence et al. 2004), were aligned with the CLUSTALW program. The amino acid sequence varies between the four Arabidopsis isoforms, particularly within the N-terminal 50 amino acids, but is highly conserved for the large C-terminal part (~270 amino acids). A database search against the Arabidopsis ESTs and genomic sequences identified an additional hit on chromosome IV (between At4g26250 and At4g26260) with significant similarity to the MIOX sequences. Although this sequence is expressed (EST clone GenBank AY139989), it resembles a pseudogene due to a missing continuous open reading frame probably resulting from an ancient gene-duplication event (Fig. 2b). Using computer-based searches as well as manual inspection of the sequence, only fragments (motives) of the N-terminal 150 amino acid can be identified in the pseudogene. Thereafter, numerous stop codons in all three reading frames are present. The continuation of the sequence is the MIOX4 gene (At4g26260), which excludes a large intron or miss splicing as the reason for the missing intact ORF in the pseudogene.
Genomic structure of the MIOX gene family from Arabidopsis. a The exon/intron structures of the four expressed MIOX isoforms, obtained from RT-PCR experiments, are shown. The start codon (ATG) is marked by an arrow and the stop codon by an asterisk. The exons are positionally conserved but a few exons have merged into a single exon in MIOX1. b The Arabidopsis genome contains an expressed pseudogene, MIOX3, for which an EST clone is available in public databases. The gene is located 1.5 kbp upstream of the MIOX4 gene, and might be the highly mutated remain of an ancient gene-duplication event. c Neighbor-joining tree derived from the multiple alignment of MIOX protein sequences. The MIOX sequences cluster into three branches representing the kingdoms of animals, fungi, and plants. (At- MIOX1, At1g14520; At- MIOX2, At2g19800; At- MIOX4, At4g26260; At- MIOX5, At5g56640; Tomato1, TC116184; Tomato2, TC124407; Rice, TC186066; Wheat, TC148319; Barley, TC121151; Secale, TC2538; Vitis, TC33556; Medicago, TC92091; Soybean, TC190829)
The organization of the four Arabidopsis MIOX genes is shown in Fig. 2a. The transcripts are interrupted by several introns; some of them exhibit atypical splice donor and acceptor sites. The gene models in TAIR and MIPS differ in their prediction, due to the variable intron prediction. We therefore tested the models for each MIOX isoform by RT-PCR. A PCR product of the predicted size according to the TAIR model was obtained for all MIOX isoforms. Sequencing of the products confirmed the gene model predicted by TAIR.
The structure of the MIOX genes is compared in Fig. 2a. Though the number of the introns varies between MIOX1 and the three other isoforms, an evolutionary relationship between all family members can clearly be seen by the positionally conserved exon/intron boundaries. Three single exons of MIOX1 are represented by two neighboring exons, each in the other MIOX isoforms. This indicates a loss of an intron during the gene-duplication event for MIOX1. Alternatively, MIOX1 is the ancestral gene and the MIOX2, 4 and 5 isoforms acquired new introns during gene-duplication events. From the structure of the exon/intron organization, it is evident that an originally single gene for MIOX has duplicated three times during the evolution of Arabidopsis plants. The relationship of the Arabidopsis MIOX genes was further analyzed by comparing the gene loci with the segmental duplication events. The segmental duplication database (http://www.tigr.org/tdb/e2k1/ath1/ Arabidopsis_genome_dupication.shtml) showed that MIOX4 and MIOX5 lie in a region of segmental duplication between chromosomes IV and V. MIOX1 and MIOX2 were in the immediate vicinity of a segmental duplication between chromosomes I and II; however, they were not annotated as members of this duplication.
The MIOX genes are widely distributed in almost all eukaryotes. A multiple alignment of 27 MIOX sequences from animals, fungi and plants was used to generate a neighbor-joining tree (Fig. 2c). The NJ-tree distinguishes between the three kingdoms, and clusters all animal and fungal sequences into separate branches from the plant sequences. The non-plant MIOX sequences exhibit a variable N-terminal domain and a highly conserved long C-terminal domain similar to the sequence variability between the four Arabidopsis MIOX isoforms.
Expression pattern of the MIOX genes in Arabidopsis
The four MIOX genes in Arabidopsis are all expressed on the basis of conventional RT-PCR analysis (Fig. 3), real-time RT-PCR (Fig. 4), and data from microarray hybridization (data not shown). Real-time RT-PCR data indicate that the MIOX2 transcript is usually expressed at the highest level of all the MIOX genes (Fig. 4a). However, in flowers, MIOX4 is upregulated and represents the transcript expressed at the highest level. Similarly, for MIOX5, only small amounts of transcripts can be detected in roots, leaves or the stem of Arabidopsis WT plants, but again significant amounts of transcripts are detectable in flowers (Fig. 4a).
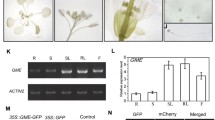
Semiquantitative RT-PCR for all four MIOX isoforms with samples from different developmental stages. The isoforms MIOX1 and 2, as well as the MIOX4 and 5 genes show a very similar pattern. RNA for RT-PCR was reverse transcribed from roots (1), leaves from 2-week-old plants (2), leaves from 5-week-old plants (3), stem (4), young flowers (preanthesis) (5), older flowers (postanthesis) (6), and siliques (7). The APT1 gene was amplified as a constitutively expressed gene to allow comparison of the template amount
Quantitative expression analysis of MIOX transcripts. a MIOX1 through MIOX5 transcript analysis in various plant organs by real-time RT-PCR. b MIOX2 transcript analysis in various plant organs of the different MIOX mutants and wild-type plants by real-time RT-PCR. Data are presented as relative expression values normalized to the average of Actin2 and adenine-phosphoribosyltransferase mRNA, which was set to 1. The values represent the mean ± SD of two measurements
Due to the possible family relations between the MIOX proteins and the rather flower-specific expression of MIOX4 and MIOX5 (Fig. 3), the most abundantly expressed MIOX2 and MIOX4 genes were chosen for a more detailed investigation. The staining of the GUS reporter indicates a tissue-specific but transient gene expression of MIOX2 (Fig. 5). The MIOX2 gene was found to be expressed in roots of seedlings. In general, the root tips from seedlings grown on MS-agar plates with sucrose exhibit a weak GUS activity (Fig. 5a). When the seedlings were grown on inositol MS plates the root tip stains intense blue (Fig. 5g). However, it should be noted that the roots remain very short (approximately one-half to one-fifth of the length compared to sucrose grown plants) when grown under this condition. The cortex cells of roots and hypocotyls show GUS activity often higher than that in the central cylinder (Fig. 5d). Etiolated seedlings usually exhibit only a small zone within the hypocotyl with MIOX2::GUS activity (Fig. 5c). Within the flowers, the MIOX2 gene is expressed in sepals and petals of very young flowers and later in the elongating filaments of the anthera (Fig. 5k). Activity remains visible in the connective tissue between the pollen sacs. The developing seeds in the pistil show some GUS expression.
MIOX2::GUS reporter gene expression in transgenic Arabidopsis seedlings. a–c Etiolated seedlings grown on MS medium with sucrose (suc; a) or MS medium with inositol (ino; b, c); d, e roots of 5-day- / 2-week-old seedlings (suc); f, g light-grown seedlings, grown on suc (f) or ino (g); h rosette leafs of a 3-week-old Arabidopsis plant; i flower manually opened; j young flower with sepals and petals removed; k older flower
The expression pattern in the MIOX4::GUS plants differs from that of MIOX2::GUS plants (Figs. 5, 6). MIOX4 reporter gene activity can be detected only in floral structures. Staining of a series of developmental stages is shown in Fig. 6. Activity is detected in young sepals and petals and, in particular, in a short phase during ripening of the pollen in the pollen sacs. Within the pistil, the integuments of the developing seeds show GUS activity as does the wall of the silique.
MIOX4::GUS reporter gene expression in transgenic Arabidopsis seedlings. The MIOX4 gene is expressed only in flower structures. a Young bud; b young flower manually opened; c–f progressively older flowers, showing transient expression of the MIOX4 gene, for example in pollen sacs. Note that the younger stamen in e does not yet show MIOX expression; g, h flowers after fertilization show MIOX activity in the integuments; i young siliques
For MIOX2, MIOX4 and MIOX5, affymetrix microarray elements could be identified in the TAIR Microarray Element Search Tool (http://www.arabidopsis.org). These were used to search the AtGenExpress developmental dataset. Here, the obtained data were in good agreement with the RT-PCR and GUS experiments (data not shown).
MIOX ESTs from plants
A database search for EST sequence coding for MIOX leads to significant hits in the EST list of a wide array of plant species. These include the moss Physcomitrella, the fern Ceratopteris, gymnosperm trees, and many angiosperm plants from monocots and dicots. A comparison of the frequency of MIOX ESTs with ESTs coding for UGD of the alternative pathway is an indicator of the relative expression level of the genes coding for the enzymes of the irreversible entry reactions of each pathway. Three patterns can be observed:
-
1.
The number of UGD ESTs largely exceeds the number of MIOX ESTs (>12-fold), found for instance in sugar cane and Medicago.
-
2.
The number of UGD ESTs is moderately higher (approximately twofold) than the number of MIOX ESTs. This pattern is widely observed and typical for plants like barley, wheat, soybean, poplar and Arabidopsis.
-
3.
The number of MIOX ESTs exceeds the number of UDG ESTs (twofold to tenfold). This pattern is, so far, observed only for different tomato species.
Protein properties of MIOX
MIOX is a soluble enzyme as indicated by previous biochemical studies (Reddy et al. 1981; Arner et al. 2001; Kanter et al. 2003). An analysis of the amino acid sequences of the MIOX enzymes using computer software from the EXPASY website gave no hint for targeting of the protein to organelles (PSORT: http://www.psort.org/; TargetP: http://www.cbs.dtu.dk/services/TargetP/). The MIOX protein has no predictable signal peptide and also no transmembrane helix (TMHMM: http://www.cbs.dtu.dk/services/TMHMM-2.0/; TMpred: http://www.ch.embnet.org/software/TMPRED_form.html). MIOX is therefore considered to be a soluble cytoplasmic enzyme.
Mutant lines for MIOX
For some of the MIOX genes, T-DNA insertion lines (ΔMIOX1, ΔMIOX2 and ΔMIOX5) were isolated from the SALK collection (Alonso et al. 2003) or the GABI-KAT collection (Rosso et al. 2003, http://www.gabi-kat.de). Homozygous knock-outs in single isoforms were analyzed for phenotypic changes compared to the wild type. The growth of all knock-out mutants was observed to be normal in size and height, and all mutant lines produced viable seeds. Cell walls derived from leaves of the wild type and mutant lines were analyzed for neutral monosaccharide composition. Wall material derived from wild-type plants consisted of glucose (7.3%), galactose (34.8%), mannose (5.8%), xylose (18.8%), arabinose (16.1%), fucose (3%) and rhamnose (14.3%). However, no significant differences were observed in the neutral sugar composition of the MIOX knock-out plants. Moreover, when the transcripts of the four MIOX genes were analyzed by real-time RT-PCR in the mutants, not many compensatory changes were detected (data not shown). However, in ΔMIOX2 mutants, barely any MIOX2 transcript was detectable by real-time RT-PCR (Fig. 4b). Similarly, in the ΔMIOX1 and ΔMIOX5 lines no transcripts for MIOX1 or MIOX5 were detectable by conventional RT-PCR, suggesting a functional knock-out of the respective genes.
We therefore tested the incorporation of radio-labeled inositol into cell-wall polymers of 7-day-old seedlings. Inositol is readily taken up by germinating Arabidopsis seedlings and incorporated into polymeric (70% ethanol insoluble) structures. Acid hydrolysates of such a wall material were separated by thin layer chromatography, and products were compared to authentic standards (Seitz et al. 2000). Labeling occurred in the sugars arabinose, xylose and the uronic acids (galacturonic and glucuronic acid), whereas no significant labeling of glucose and galactose was detected (data not shown). These data are consistent with the conversion of inositol into glucuronic acid via the MIOX enzyme activity. The total incorporation of the inositol pathway-derived monosaccharides in the walls of ΔMIOX1 and ΔMIOX2 seedlings is strongly reduced, whereas no reduction was observed for the ΔMIOX5 plants (Fig. 7). These data are in agreement with expression data from the RT-PCR (Figs. 3, 4) and published microarray data which suggest no (or very low) expression of the MIOX4/5 genes in seedlings.
Functional assay for the incorporation of 3H-inositol into cell-wall polymers of 7-day-old Arabidopsis seedlings. The seedlings (ten seedlings each) were extracted repeatedly with aqueous ethanol to remove inositol containing low-molecular-weight compounds like membrane lipids. The remaining radioactivity was determined by liquid scintillation counting. The values represent the mean ± SD of six experiments. The ΔMIOX1 and the ΔMIOX2 plants show a strong reduction of inositol incorporation into cell walls
Ascorbate content in MIOX mutants
The level of the antioxidant ascorbic acid was measured as glucuronic acid is a potential precursor of ascorbate synthesis. In addition, the overexpression of a single MIOX gene has been reported to result in elevated levels of ascorbic acid in plants (Lorence et al. 2004). The amounts of ascorbate in wild-type and ΔMIOX2 or ΔMIOX5 knock-out plants were between 2 and 3.5 μmol g−1 fresh weight in individual experiments, depending on the environmental conditions, and differed less than 5% in several independent measurements. No differences were also observed for the amounts of total ascorbate (ascorbic acid + dehydroascorbate), excluding the possibility of changes in the redox potential of the cells in MIOX knock-out plants.
Discussion
In Arabidopsis, the major central intermediate for precursors of cell-wall polymers is UDP-GlcA based on polymer analysis of cell walls (Zablackis et al. 1995). This nucleotide sugar is at a transition point between sugars of a storage pathway to sucrose and a nucleotide sugar interconversion pathway nearly exclusively used to synthesize matrix polysaccharides of cell walls. The control of the transition is realized by an entry enzyme reaction, catalyzing an irreversible oxidation step.
Heterologous expression of MIOX4 in bacteria resulted in an enzyme with MIOX activity (Lorence et al. 2004). Hence, MIOX is the key enzyme of the inositol oxygenase pathway and is remarkably conserved between all eukaryotes (Fig. 2). The MIOX from Cryptococcus lactativorus (Kanter et al. 2003) and the corresponding Arabidopsis sequences still show ~57% homology including conserved substitutions despite the long evolutionary separation of plants and fungi. MIOX is a unique group of proteins with no other relatives in the database and thus the genes for these proteins could be clearly identified.
Whether the anabolic metabolism from GlcA to UDP-GlcA occurs outside the plant kingdom is unclear. In the yeast Cryptococcus lactativorus, from which we have recently purified the MIOX enzyme, GlcA is catabolized to CO2. This yeast can grow on minimal mineral medium with inositol as the only carbon source. In principle, the anabolic route to UDP-GlcA might also be active in fungi because GlcA is found in the cell-wall polymers of several fungi (Ahrazem et al. 2000). In animals, inositol oxygenation is clearly part of a degradation pathway for inositol and the MIOX enzyme activity is only found in kidneys. In addition, GlcA is an intermediate in the biosynthesis of ascorbic acid in animals (Smirnoff et al. 2001). In contrast to animals, plants seem to synthesize ascorbate via an L-galactose dependent pathway (Smirnoff et al. 2001). The recent overexpression of an ArabidopsisMIOX gene in Arabidopsis, however, revealed that glucuronic acid might also be an intermediate in another route to ascorbic acid (Lorence et al. 2004). In contrast, our ΔMIOX2 and ΔMIOX5 knock-out plants show no reduced level of ascorbate under low (~100 μE m−2 s−1) or moderate (~250 μE m−2 s−1) light conditions. This either reflects the genetic redundancy in the expression of MIOX genes (compare Figs. 3 and 4) or more likely the dominance of the L-galactose pathway to ascorbate biosynthesis.
MIOX is functional for nucleotide sugar biosynthesis
Sterile-grown Arabidopsis seedlings take up inositol from the medium and incorporate it into cell-wall polymers (Seitz et al. 2000). This confirms earlier studies in which the conversion of inositol into GlcA-derived sugar monomers was clearly demonstrated by labeling experiments (Wakabayashi et al. 1989; Kroh and Loewus 1968). In both studies, no significant amount of radioactivity from inositol was found in the glucose fraction after cell-wall hydrolysis. We have repeated these experiments and also found a good incorporation of 3H-inositol into cell-wall polymers. Separation of products after hydrolysis of the cell wall revealed radioactivity only in the uronic acids, arabinose, and xylose, which are predicted to be labeled (see Fig. 1). A proof for the role of MIOX in providing nucleotide sugars for cell-wall polymers came from the analysis of ΔMIOX1/2 mutant lines. Both MIOX knock-out plants showed a drastic reduction of the 3H-inositol incorporation into cell walls, suggesting that MIOX1 and 2 are functional in this pathway. The MIOX5 gene is expressed in seedlings only at a very low level, and therefore it is not surprising that incorporation of inositol in the MIOX5 knock-out line was not reduced. In summary, the data provide strong evidence for a functional inositol oxygenation pathway in plants involved in the biosynthesis of cell-wall polysaccharides.
Gene-expression pattern of MIOX isoforms
MIOX1 and 2 are expressed in the roots of seedlings, in some tissues of etiolated hypocotyls, developing cotyledons, leaves, and some reproductive tissues. Though most of the inositol, converted into UDP-sugars for matrix polymers, is likely to be synthesized from Glc-6-P, the cotyledons contain additional inositol as a breakdown product from the phosphate storage phytate (Raboy 2003). This might explain the MIOX2::GUS staining in cotyledons (Fig. 5g), which otherwise show little UGD-gene expression (data not shown). In contrast, MIOX4 and 5 show an exclusive gene expression in generative structures, beginning with petal development and continuing to embryo development in seeds. During flower development, several MIOX isoforms are expressed in the same tissues, for example MIOX2 and MIOX4 are both expressed in growing sepals and petals (Figs. 5, 6). The most prominent expression of a MIOX4::GUS reporter is found in pollen (Fig. 6). In fact, labeling of germinating Lilium pollen with 14C-inositol showed the highest incorporation in pectins of the pollen tube (Kroh and Loewus 1968).
MIOX versus UGD pathway
The fraction of ESTs coding for MIOX is extremely high in tomato plants/pollen. Clustering of ESTs from the Lycopersicon pennellii cDNA library lists MIOX among the ten most highly expressed genes from this pollen-specific library with a frequency of ~1.3% of all sequences (data not shown). The abundant expression of the MIOX genes in plant pollen suggests a function of the MIOX pathway to synthesize UDP GlcA with an advantage over the UGD pathway. Nevertheless, the UGD pathway is expressed in generative tissues including pollen, but at a lower level. This is indicated by UGD::GUS stains in Arabidopsis (M. Klinghammer and R. Tenhaken, unpublished data) and also from the results of the clustering of the tomato pollen-specific EST library.
One difference between both routes for UDP-GlcA biosynthesis is the formation of NADH in the UGD pathway, whereas the MIOX enzyme transfers the electrons directly to molecular oxygen. As a consequence, the preference of the MIOX pathway will contribute to a more oxidized redox potential compared to an alternative scenario, in which UDP-GlcA is made through the UGD pathway. This concept needs to be tested in Arabidopsis knock-out plants for MIOX in the future but will likely require the multiple knock-out of several MIOX genes.
The study by Lorence et al. (2004) suggests the participation of GlcA for the biosynthesis of ascorbate in plants. However, single knock-outs in MIOX2 or 5 genes do not lower the ascorbate concentration in plants. This can be explained by the redundancy in gene expression of the MIOX genes, which showed a very similar tissue-specific expression for the pair MIOX1/2 and MIOX4/5. It is also well conceivable that GlcA is involved in the ascorbate bioynthesis only under certain conditions, e.g. when a surplus of free inositol is available.
The monomer composition of neutral polysaccharides from cell walls is very similar in wild-type plants and in single knock-out plants for MIOX genes. Again, this might be explained in part by the gene redundancy within the MIOX-gene family but also by the fact that the UGD pathway provides the same product, UDP-GlcA. Although the UGD genes are controlled at the transcriptional level (Seitz et al. 2000), biochemical feedback inhibition of the UGD enzymes by UDP-Xyl is superimposed. Under physiological conditions, most of the UGD enzyme activity is likely inhibited by UDP-Xyl (K i <10 μM; Hinterberg et al. 2002), thus providing a buffering capacity to refill the pool for UDP-GlcA-derived nucleotide sugars.
The flux of carbohydrates into a pool of nucleotide sugars via UDP-GlcA is strictly controlled in plants. The most obvious reason is simply the fact that the metabolite UDP-GlcA cannot be reconverted into carbohydrates used to synthesize storage compounds like sucrose. Thus, the pool for storage compounds and the pool for sugars used to synthesize cell walls are strictly separated.
Abbreviations
- GlcA:
-
Glucuronic acid
- GUS:
-
β-Glucuronidase (uidA-gene from E. coli)
- MIOX:
-
Myo-inositol oxygenase
- UDP-Glc:
-
Uridine-diphospho-glucose
- UDP-GlcA:
-
Uridine-diphospho-glucuronic acid
- UGD:
-
UDP-glucose dehydrogenase
References
Ahrazem O, Gómez-Miranda B, Prieto A, Bernabé M, Leal JA (2000) Heterogeneity of the genus Myrothecium as revealed by cell wall polysaccharides. Arch Microbiol 173:296–302
Alonso JM et al (2003) Genome-insertional mutagenesis of Arabidopsis thaliana. Science 301:653–657
Arner RJ, Prabhu KS, Thompson JT, Hildenbrandt GR, Liken AD, Reddy CC (2001) Myo-inositol oxygenase: molecular cloning and expression of a unique enzyme that oxidizes myo-inositol and d-chiro-inositol. Biochem J 360:313–320
Burget EG, Verma R, Mølhøj M, Reiter WD (2003) The Biosynthesis of l-arabinose in plants: molecular cloning and characterization of a Golgi-localized UDP-d-xylose 4-epimerase encoded by the MUR4-gene of Arabidopsis. Plant Cell 15:523–531
Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162:156–159
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Czechowski T, Bari RP, Stitt M, Scheible W-R, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38:366–379
Gibeaut DM (2000) Nucleotide sugars and glycosyltransferases for synthesis of cell wall matrix polysaccharides. Plant Physiol Biochem 38:69–80
Harper AD, Bar-Peled M (2002) Biosynthesis of UDP-xylose. Cloning and characterization of a novel Arabidopsis gene familiy, UXS, encoding soluble and putative membrane-bound UDP-glucuronic acid decarboxylase isoforms. Plant Physiol 130:2188–2198
Hinterberg B, Klos C, Tenhaken R (2002) Functional characterization of recombinant UDP-glucose dehydrogenase from soybean. Plant Physiol Biochem 40:1011–1017
Jefferson RA (1987) Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep 5:387–405
Kanter U, Becker M, Friauf E, Tenhaken R (2003) Purification, characterization and functional cloning of inositol oxygenase from Cryptococcus. Yeast 20:1317–1329
Kroh M, Loewus F (1968) Biosynthesis of pectic substance in germinating pollen: labeling with myoinositol-2–14C. Science 160:1352–1354
Leibowitz MD, Bickinson DB, Loewus FA, Moewus M (1977) Partial purification and study of pollen glucuronokinase. Arch Biochem Biophys 179:559–564
Loewus FA, Murthy PPN (2000) myo-inositol metabolism in plants. Plant Sci 150:1–19
Loewus FA, Kelly S, Neufeld EF (1962) Metabolism of myo-inositol in plants: conversion to pectin, hemicellulose, D-xylose and sugar acids. Proc Natl Acad Sci U S A 48:421–425
Logeman J, Schell J, Willmitzer L (1987) Improved method for the isolation of RNA from plant tissues. Anal Biochem 163:21–26
Lorence A, Chevone BI, Mendes P, Nessler CL (2004) myo-inositol oxygenase offers a possible entry point into plant ascorbate biosynthesis. Plant Physiol 134:1200–1205
Mølhøj M, Verma R, Reiter WD (2003) The biosynthesis of the branched-chain sugar D-apiose in plants: functional cloning and characterization of a UDP-d-apiose/UDP-d-xylose synthase from Arabidopsis. Plant J 35:693–703
Mølhøj M, Verma R, Reiter WD (2004) The biosynthesis of d-galacturonate in plants. Functional cloning and characterization of a membrane-anchored UDP-d-glucuronate 4-epimerase from Arabidopsis. Plant Physiol 135:1221–1230
Majerus PW (1992) Inositol phosphate biochemistry. Annu Rev Biochem 61:225–250
Moffat BA, McWhinne EA, Agarwhal SK, Schaff DA (1994) The adenine phosphoribosyltransferase-encoding gene of Arabidopsis thaliana. Gene 143:211–216
O’Neill MA, Eberhard S, Albersheim P, Darvill AG (2001) Requirement of borate cross-linking of cell wall rhamnogalacturonan II for Arabidopsis growth. Science 294:846–849
Raboy V (2003) myo-inositol-1,2,3,4,5,6-hexa kisphosphate. Phytochemistry 64:1033–1043
Reddy CC, Swan JS, Hamilton GA (1981) myo-inositol oxygenase from hog kidney I. J Biol Chem 256:8510–8518
Reiter W, Vanzin GF (2001) Molecular genetics of nucleotide sugar interconversion pathways in plants. Plant Mol Biol 47:95–113
Roberts RM (1971) The formation of uridine diphosphate-glucuronic acid in plants. Uridine diphosphate-glucuronic acid pyrophosphorylase from barley seedlings. J Biol Chem 246:4995–5002
Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53:247–259
Seifert GJ (2004) Nuceleotide sugar interconversions and cell wall biosynthesis: how to bring the inside to the outside. Curr Opin Plant Biol 7:277–284
Seifert GJ, Barber C, Wells B, Dolan L, Roberts K (2002) Galactose biosynthesis in Arabidopsis. Genetic evidence for substrate channeling from UDP-D-galactose into cell wall polymers. Curr Biol 12:1840–1845
Seitz B, Klos C, Wurm M, Tenhaken R (2000) Matrix polysaccharide precursors in Arabidopsis cell walls are synthesised by alternate pathways with organ specific expression patterns. Plant J 21:537–546
Smart CC, Fleming AJ (1993) A plant gene with homology to D-myo-inositol-3-phosphate synthase is rapidly and spatially up-regulated during an abscisic-acid-induced morphogenic response in Spirodela polyrrhiza. Plant J 4:279–293
Smirnoff N, Conklin PL, Loewus FA (2001) Biosynthesis of ascorbic acid in plants: a Renaissance. Annu Rev Plant Physiol Plant Mol Biol 52:437–467
Tenhaken R, Thulke O (1996) Cloning of an enzyme that synthesizes a key nucleotide sugar precursor of hemicellulose biosynthesis from soybean: UDP-glucose dehydrogenase. Plant Physiol 112:1127–1134
Usadel B, Schluter U, Mølhøj M, Gipmans M, Verma R, Kossmann J, Reiter W-D, Pauly M (2004a) Identification and characterization of a UDP-D-glucuronate 4-epimerase in Arabidopsis. FEBS Lett 569:327–331
Usadel B, Kuschinsky AM, Rosso MG, Eckermann N, Pauly M (2004b) RHM2 is involved in mucilage pectin synthesis and is required for the development of the seed coat in Arabidopsis. Plant Physiol 134:286–295
Vernon D, Bohnert HJ (1992) A novel methyl transferase induced by osmotic stress in the facultative halophyte Mesembryanthemum crystallinum. EMBO J 11:2077–2085
Wakabayashi K, Sakurai N, Kuraishi S (1989) Effects of ABA on synthesis of cell-wall polsaccharides in segments of etiolated squash hypocotyl. I Changes in incorporation of glucose and myo-inositol onto cell-wall components. Plant Cell Physiol 30:99–105
Western TL, Young DS, Dean GH, Tan WL, Samuels AL, Haughn GW (2004) Mucilage-modified4 encodes a putative pectin biosynthetic enzyme developmentally regulated by APETALA2, TRANSPARENT TESTA GLABRA1, and GLABRA2 in the Arabidopsis seed coat. Plant Physiol 134:296–306
York WS, Darvill AG, McNeil T, Stevenson TT, Albersheim P (1985) Isolation and characterization of plant cell walls and cell wall components. Methods Enzymol 118:3–40
Zablackis E, Huang J, Muller B, Darvill AG, Albersheim P (1995) Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol 107:1129–1138
Acknowledgements
U. Kanter and B. Usadel contributed equally to this paper. We would like to thank the anonymous reviewers for helpful suggestions and Mark-Daniel Ziemann for the reading the manuscript. This work was financially supported by the German Science Foundation (DFG) and the Max-Planck Society.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kanter, U., Usadel, B., Guerineau, F. et al. The inositol oxygenase gene family of Arabidopsis is involved in the biosynthesis of nucleotide sugar precursors for cell-wall matrix polysaccharides. Planta 221, 243–254 (2005). https://doi.org/10.1007/s00425-004-1441-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00425-004-1441-0