Abstract
Aluminum toxicity in acidic soils is a major constraint to winter wheat productivity in the southern Great Plains, USA. In this study, a population of recombinant inbred lines generated from a cross between two winter wheat cultivars, ‘Jagger’ with high acidic soil tolerance and ‘2174’ with moderate acidic soil tolerance, was used to map genes for acidic soil tolerance in the field. As expected, a major QTL centered on TaALMT1 on chromosome 4DL was observed. Further sequencing, however, indicated that Jagger carried an allele having Type V of triplicated sequence repeats in TaALMT1-1, whereas 2174 carried an allele having Type IV of two (A–B) block sequences in TaALMT1-2. The Jagger TaALMT1-1-V allele was expressed at a significantly higher level than the 2174 TaALMT1-2-IV allele not only in roots as reported in previous studies but also in leaves found in this study. No sequence was found for genes to be homoeologous to TaALMT1 in any of the recently released genomic sequences of diploid, tetraploid, or hexaploid wheat, suggesting that neither TaALMT1-A nor TaALMT1-B exists in wheat. Therefore, TaALMT1 is a gene unique to genome D. In addition, Jagger had the favorable allele at a QTL mapped on chromosome 2DL, whereas 2174 had the favorable allele at a QTL mapped on chromosome 7BL. The combination of these three complementary QTLs/genes/alleles may help to generate winter wheat lines that confer greater tolerance to acidic soils.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Aluminum (Al) toxicity released from acidic soils is the major constraint to wheat productivity worldwide (Raman and Gustafson 2014), particularly in the southern Great Plains, USA, where soil acidity formation is frequent (Carver et al. 1988). Hard red winter wheat (HRW, Triticum aestivum, 2n = 6x = 42, AABBDD) is the primary crop in the southern Great Plains, but growth and development of HRW wheat are negatively affected by aluminum toxicity (Carver et al. 1993). The pH value of surface soils is 5.0–6.0 (moderately acidic) in most fields but can fall lower than 5.0 (strongly acidic) in some fields, a level which seriously threatens HRW wheat production and even results in failure of the wheat crop in central and western Oklahoma (Zhang and Raun 2006). Soil acidity is particularly critical for dual-purpose wheat, which is used as forage for cattle during the winter season and harvested for grain in the following spring, because soil acidity reduces both forage and grain yields (Carver et al. 1993; Kariuki et al. 2007). With utilization of ammonia N fertilizers and continued crop removal of soil basic cations, soil acidity will continue to develop in the HRW wheat region.
One way to ameliorate aluminum toxicity is to apply lime to neutralize soil acidity (Kochian et al. 2004; Ryan et al. 2010). Lime application is the most widely used long-term method of soil acidity amelioration, and its success has been well documented; however, liming may be impractical due to the high cost of transportation and the volume of lime needed across vast areas and also adverse due to subsequent encouragement of root rot diseases (Haynes 1982; Conyers et al. 1991; Scott et al. 2001). Another way to ameliorate the effects of aluminum toxicity is to improve the plant’s genetic constitution to enhance tolerance to acidic soil (Garvin and Carver 2003). The use of acidic soil-tolerant cultivars has been a successful but temporary alternative to liming in winter wheat production (Johnson et al. 1997a, b; Delhaize et al. 2004; Dai et al. 2013).
Sufficient genetic variation in acidic soil tolerance has been reported among wheat cultivars. On the basis of acidic soil tolerance on a field scale, rankings were established for winter wheat cultivars, including ‘Jagger’ and ‘2174,’ that have been extensively utilized in the southern Great Plains (Kariuki et al. 2007). Whereas Jagger was ranked as one of the most acidic soil-tolerant cultivars, 2174 was ranked as a moderately tolerant one, compared with cultivar ‘Custer’ that showed the lowest tolerance to acidic soils (Kariuki et al. 2007). In many instances, genetic variation in aluminum tolerance among wheat cultivars is due to a difference in a single gene (Kerridge and Kronstad 1968; Delhaize et al. 1993; Somers and Gustafson 1995; Riede and Anderson 1996; Sasaki et al. 2004; Raman et al. 2005), but evidence also suggests that more than one aluminum tolerance gene may exist in certain wheat cultivars (Berzonsky 1992; Carver and Ownby 1995; Zhou et al. 2007; Dai et al. 2013; Ryan et al. 2010). Genetic mechanisms underlying natural variation in HRW wheat cultivars produced in the southern Great Plains have not been revealed.
The first gene controlling natural variation in aluminum tolerance has been cloned from T. aestivum, and it is a gene that encodes an aluminum-activated malate transporter (TaALMT1) (Sasaki et al. 2004). Two alleles, TaALMT1-1 and TaALMT1-2, encode discernible amino acid sequences, but no association between the different alleles and aluminum tolerance was observed (Raman et al. 2005). Due to the insertion of duplicated or triplicated block sequences (Types I–VI), the two TaALMT1 alleles were found to have six different patterns in the upstream or promoter region (Sasaki et al. 2004; Raman et al. 2005; Sasaki et al. 2006). TaALMT1 expression levels were positively correlated with the copy number of the inserted block sequences and with aluminum tolerance among diverse and non-Japanese wheat lines but not in Japanese wheat lines (Sasaki et al. 2006). Wheat plants may possess different mechanisms in resistance to aluminum toxicity and acidic soils (Sasaki et al. 2004; Ryan et al. 2009).
The detectable difference in acidic soil tolerance between Jagger and 2174 and availability of a mapping population of recombinant inbred lines (RILs) generated from the two HRW cultivars provide an opportunity to identify genes that can be directly utilized in local HRW wheat breeding programs. Several hundred SSR markers were mapped in the RIL population for segregation in developmental traits (Chen et al. 2009a, b; 2010; Li et al. 2013), and in resistance to leaf rust (Cao et al. 2010), stripe rust (Fang et al. 2011), powdery mildew (Chen et al. 2009a, b), and Hessian fly (Tan et al. 2013), but acidic soil tolerance segregated in the population has not been reported. Single nucleotide polymorphism (SNP) markers have since been developed (Cavanagh et al. 2013). These SNP markers were utilized to genotype the Jagger × 2174 RIL population so that we can (1) better characterize the difference in acidic soil tolerance between two non-susceptible cultivars at the molecular level and (2) identify complimentary genes for acidic soil tolerance that may exist in HRW wheat.
Materials and methods
Jagger and 2174 were previously observed to differ in their reactions in Oklahoma acidic soils, with Jagger being more tolerant than 2174 (Kariuki et al. 2007). Therefore, the Jagger × 2174 population of 144 RILs was tested for acidic soil tolerance during the 2008–2009 crop season at two naturally low-pH locations—Stillwater and Enid, OK. The same RIL population was also evaluated at only Enid during the 2007–2008 crop season. Plots were arranged in the field in a completely randomized design with two replications for the RILs and six replications for parents. Each line was planted in two rows. The experimental sites were previously described by Johnson et al. (1997a, b) and Edwards et al. (2012) as having a soil–water pH of about 4.5 and aluminum saturation exceeding 10 %. Standard fertilization practices were followed at each location according to soil test recommendations, with the exception of zero lime application.
Visual ratings of acidic soil tolerance were collected from non-replicated trials with Jagger and 2174 interspersed as repeated checks. To further adjust for spatial variability inherent to field screening, a control plot of Jagger appeared once for every three plots containing the RILs. Plot size was one row 3.0 m in length, with a row spacing of 30 cm. The rating system focused on secondary effects of low-pH-restricted root growth in the aboveground foliage, such as highly prostrate growth and poor plant vigor during juvenile plant stages, leaf chlorosis and/or purpling, and poor tillering or spike production (Carver and Ownby 1995).
The rating scale ranged from 0 (highly tolerant, with no apparent symptoms of acidic soil stress) to 5 (highly susceptible, in which the majority of plants in a plot were non-recoverable or dead). Jagger was usually scored as ‘2,’ and 2174 was usually scored as ‘3’. In comparison, the acidic soil-tolerant cultivar ‘Endurance’ was consistently scored as ‘1’ and the acidic soil-susceptible cultivars TAM 110 and Custer were consistently scored as ‘5.’ Using this rating system, the RILs and the check plots of Jagger and 2174 were scored across multiple developmental stages. Readings were taken at Enid on November 2, 2007 (Feekes 3.0), November 3, 2008 (Feekes 3.0), February 6, 2009 (Feekes 4.0), and March 17, 2009 (Feekes 6.0–7.0), and Stillwater on May, 18 2009 (Feekes 11.0).
Approximately 1500 pairs of simple sequence repeat (SSR) primers for genome-wide markers were selected to screen the parents, and 400 SSR markers were eventually mapped in the Jagger × 2174 RIL population; however, no SSR marker was mapped with linkage to a gene on chromosome 4DL for tolerance to acidic soils. A high-throughput array to interrogate 9000 gene-associated single nucleotide polymorphisms (Wheat 9K iSelect SNP assay) in wheat was developed to detect key genomic regions (Cavanagh et al. 2013). The population of 144 Jagger × 2174 recombinant inbred lines (RILs) was genotyped using the 9K SNP markers to saturate the previous SSR genetic maps. Those SNP markers that had less than 20 % missing data were selected for final genetic maps, and the 8-digit SNP codes served as the reference number for each SNP. The SNP mapping method was described in a recent study (Li et al. 2013). Briefly, the SNP markers were used to make linkage groups using JoinMap 4.0 (Van Ooijen 2006), and the interval mapping program was run to locate QTLs for the acidic soil tolerance using MapQTL 6.0 (Van Oojjen 2009). The SNP markers in the targeted region were integrated with SSR markers mapped in previous studies, and the Kosambi mapping function was used to estimate the distance among the markers within the targeted QTLs using MapMaker 3.0. The interval mapping program was run to locate QTLs for the acidic soil tolerance using WinQTL Cart 2.5. Logarithm of the odds (LOD) threshold for significance was 2.5 for the presence of a putative QTL. Maximum LOD values were used to estimate QTL peak positions.
Allelic variation in TaALMT-1
The published cDNA sequences of TaALMT1-1 from ‘ET8’ (GenBank accession AB081803) and TaALMT1-2 from ‘ES8’ (GenBank accession AB081804) were used as references to design primers and obtain the genomic sequences of TaALMT1 from the Jagger and 2174 alleles. The complete TaALMT1 gene for each allele was isolated using the primers ALMT1-F5 5′-CGCGGCCAGGAATTCGATCAC-3′ and ALMT1-R1 5′-CTTCCTCCGTCACATCGTACA-3′. The PCRs were performed using LongAmp Taq DNA polymerase (New England BioLabs) and 35 thermal cycles after denature at 95 °C for 5 min, each cycle consisting of 94 °C for 30 s, 56 °C for 30 s, and 65 °C for 6 min. The resulting PCR products were cloned into a PCR-XL-TOPO vector for sequencing. Four clones for each allele were sequenced. Primer ALMT-F5 was paired with another primer ALMT-In2R1 CGGAGGAAGTAATTAAAAAGAGAGTCTCAAAGGTAC to map the promoter region. The PCRs were performed using the protocol as described above except annealing temperature that was changed to 60 °C and extension time that was changed to 1 min 40 s. A new PCR marker was developed to map a SNP in exon 6 using ALMT-MF1 5′-GATCGGAGGGAGTAGCTTTCATTTATTC-3′ and ALMT-MR2 5′-AGCTAGAGTTATACCTGGGTTTTTGAGG-3′. The PCR products were digested with restriction enzyme Bcl I. The PCRs were performed using the protocol as described above except annealing temperature that was changed to 57 °C and extension time that was changed to 30 s.
TaALMT1 gene expression
RNA samples were collected from the entire roots and shoots from five plants of five- and six-leaf seedlings grown in commercial soil. RNAs were extracted from leaves using the TRIzol method (Invitrogen) and reverse-transcribed to cDNA using a poly(dT) primer. RT-PCRs were performed to determine TaALMT1 transcriptional levels using SYBR Green I kit (Bio-Rad). The primers ALMT-RT-F1 5′-AAGAGCGTCCTTAATTCG-3′ and ALMT-RT-R2 5′-TAAGCACCTTGGAGGAATGC-3′ amplified a 274-bp fragment of the gene. The forward primer was the same as reported in the previous study (Sasaki et al. 2004), but the reverse primer was modified to avoid a SNP in the primer region between the Jagger and 2174 alleles. The same cDNA sample was used for TaALMT1 and two endogenous control genes: β-tubulin gene (Sasaki et al. 2004) and actin gene (Li et al. 2013). The RT-PCRs were performed using 40 thermal cycling after denature at 95 °C for 2 min, each cycle consisting of 95 °C for 15 s, 55 °C for 20 s and 72 °C for 31 s.
Results
Complementary effects of three QTLs on aluminum tolerance
Wide phenotypic segregation in acidic soil tolerance was observed among the Jagger × 2174 RILs, and the segregated phenotypes were used to map QTLs for acidic soil tolerance. Of the approximately 400 SSR markers used to genotype the population in previous studies, two SSR markers Xbarc105 and Xgpw94042 were mapped to chromosome 4DL, but neither of them showed an association with the phenotypes. After SNP markers were mapped in the population, three QTLs for acidic soil tolerance were detected.
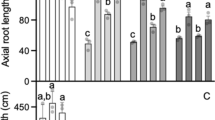
The first one was located on chromosome 4DL (QAlmt.osu-4D). Nineteen SNP markers were assembled into a group that included the two mapped SSR markers Xbarc105 and Xgpw94042. This linkage group spanned 50.9 cM and produced a major QTL accounting for up to 38.5 % of the total phenotypic variation (Fig. 1a). The effect of this QTL was most noticeable following the onset of stem elongation—hence in the adult plant stages (Table 1; Fig. 1a). Both Xbarc105 and Xgpw94042 were on the border of this QTL, explaining why no QTL was formed using SSR markers. QAlmt.osu-4D showed consistent effects on acidic soil tolerance across environments.
Genetic effects of three QTLs for acidic soil tolerance in the Jagger × 2174 RIL population. a QAlmt.osu-4D on chromosome 4DL. TaALMT1 was mapped under the peak of the QTL. b QAlmt.osu-2D on chromosome 2DL. c QAlmt.osu-7B on chromosome 7BL. EN3.0-2007 represents the phenotype collected at Feekes stage 3.0 at Enid in 2007. EN3.0-2008, EN4.0-2009, and EN6.0-2009 represent phenotypes collected at Feekes stages 3.0, 4.0, and 6.0 at Enid in the 2008–2009 crop season. ST11.0-2009 represents the phenotype collected at Feekes stages 11.0 at Stillwater. The 8-digit SNP codes served as the reference number for each SNP (Cavanagh et al. 2013). Logarithm of the odds (LOD) threshold for significance was 2.5 for the presence of the three QTLs
The second QTL was located on chromosome 2DL (QAlmt.osu-2D), where a group of seven SSR markers and 15 SNP markers spanned 11.7 cM across the QTL region (Fig. 1b). QAlmt.osu-2D showed variable effects on acidic soil tolerance across developmental stages, years, and locations. Its effect was most pronounced in the vegetative stages, where it accounted for 29 and 32 % of the total phenotypic variation at Feekes stage 3.0 at Enid in 2007 and 2008, respectively. It showed much less or no significant effects on phenotypes characterized in the adult stages.
The third QTL was located on chromosome 7BL (QAlmt.osu-7B), where 4 SSR markers and 35 SNP markers were grouped and spanned 15.7 cM covering the QTL region (Fig. 1c). Similar to QAlmt.osu-2D, QAlmt.osu-7B also showed variable effect on acidic soil tolerance across developmental stages and environments. While this QTL explained up to 25.1 % of the total phenotypic variation (Feekes stage 4.0 at Enid in 2009), the effect was most evident prior to full stem extension.
The genetic effects of the three QTLs on acidic soil tolerance are summarized in Table 1. No significant interaction was observed among the three QTLs. Whereas Jagger showed a tolerant allele at TaALMT1 and QAlmt.osu-7B, 2174 showed a tolerant allele at QAlmt.osu-2D. A full complement of tolerant alleles at all three QTLs made some RILs more tolerant than others and the parental lines. Among 141 RILs that were phenotyped at Feekes stage 11.0 at Stillwater in 2009, 39 RILs were highly tolerant and scored as ‘1,’ and 13 RILs were highly susceptible and scored as ‘5’ for acidic soil tolerance.
Allelic variation in TaALMT-1
To test whether TaALMT1 is linked with the gene causing QAlmt.osu-4D, the complete TaALMT1 gene consisting of six exons and five introns was isolated and sequenced from each of the two alleles. Sequencing results showed that the Jagger TaALMT1-1 allele is 5345 bp in length, including 1286 bp from the translational start codon, 3968 bp from the start codon to the stop codon, and 91 bp after the stop codon. The 2174 TaALMT1-2 allele is 5048 bp in length, including 1006 bp from the translational start codon, 3951 bp from the start codon to the stop codon, and 91 bp after the stop codon. TaALMT1-1 was an allele that had the same sequence from the translational start codon to the stop codon as haplotype ALMT1-1 reported in ET8 and Atlas66, and TaALMT1-2 was an allele that had the same sequence from the translational start codon to the stop codon as haplotype ALMT1-2 reported in ES8, CS, and Scout 66. TaALMT1-1 and TaALMT1-2 were distinguished in the gene region by 44 SNPs or small insertions/deletions (indels) including six single nucleotide polymorphisms (SNPs) in the coding region resulting in different TaALMT1 proteins.
TaALMT1 in T. tauschii has the promoter Type I (DQ072271), which has the simplest structure, while other accessions tested in the previous studies have block A, B, C, and/or D sequences that are duplicated or triplicated in different arrangements (Sasaki et al. 2006). Further sequence analysis indicated that the Jagger allele had triplicates of blocks A–B producing the pattern A–B, A–B, A–B, C, and D in the promoter region (Type 5) (Fig. 2a), but 2174 had duplicates of blocks A–B producing the pattern A–B, A–B, C, and D in the promoter (Type IV) (Fig. 2b). Hence, the Jagger TaALMT1-1-V allele and the 2174 TaALMT1-2-IV allele were distinguished from one another by one more or less copy of block A (172 bp) and block B (108 bp). A total difference of 280 bp of blocks A–B in the TaALMT1 promoter size was detectable in an agarose gel as shown in Fig. 2c. This marker was used to map TaALMT1 in the population (Fig. 1a). Another PCR marker for a SNP in exon 6 was also developed (Fig. 2d), and it mapped TaALMT1 to chromosome 4DL.
Gene structure and PCR markers of TaALMT1. a Diagram of TaALMT1-1-V gene structure. The Jagger TaALMT1-1-V allele consists of the Type V promoter and the TaALMT1-1 gene region that has six exons and five introns. b Diagram of TaALMT1-2-IV gene structure. The 2174 TaALMT1-2-IV allele consists of the Type IV promoter and the TaALMT1-1 gene region that has six exons and five introns. Promoter Type V has triplicates of blocks A (shaded box in black) and B (shaded box in dark gray), whereas promoter Type IV has duplicates of blocks A and B. c A PCR marker for promoters differing in length between TaALMT1-1 and TaALMT1-2-IV. D. A PCR marker for one SNP (X) in exon 6. M denotes a DNA marker. Lanes 1 and 2 represent the Jagger TaALMT1-1-V allele, whereas lanes 3 and 4 represent the 2174 TaALMT1-2-IV allele
Expression of TaALMT1 not only in roots but also in leaves
TaALMT1 gene expression was tested for five promoter types, except for Type IV that was not found in any of 34 lines tested by Sasaki et al. (2006). In order to test whether there was differential expression between TaALMT1-1-V and TaALMT1-2-IV, a regular PCR assay was used to qualitatively characterize gene expression. Surprisingly, both of the two alleles were found to be expressed not only in roots (Fig. 3a, lanes 5–8) but also in leaves (lanes 1–4) of seedling plants. Moreover, TaALMT1 was hardly observed in roots of adult plants (lanes 13–16) but were clearly detectable in leaves (lanes 9–12) as well as in spikes (lanes 17–20).
Expression profiles of TaALMT1. a Tissue-specific expression profiles of TaALMT1 in different tissues in seedling plants. Odd numbers indicate the Jagger TaALMT1-1-V allele, whereas even numbers indicate the 2174 TaALMT1-2-IV allele. Lanes 21 and 22 represent gDNA used as a control. b Differential transcriptional levels in roots and leaves, with standard error bars, between TaALMT1-1-V and TaALMT1-2-IV. Tubulin was used as an endogenous gene control. c Differential transcriptional levels in roots, with standard error bars, between TaALMT1-1-V and TaALMT1-2-IV. d Differential transcriptional levels in leaves, with standard error bars, between TaALMT1-1-V and TaALMT1-2-IV. c, d Actin was used as an endogenous gene control. Sixteen PCRs for each allele were performed
Real-time quantitative PCR was used to quantitatively determine the difference in transcriptional level between the Jagger TaALMT1-1-V allele and the 2174 TaALMT1-2-IV allele, and tubulin was used as an endogenous control for each sample (Fig. 3b). Compared with tubulin, TaALMT1-1-V was expressed at a very high level in roots, whereas TaALMT1-2-IV was minimally expressed in roots. In leaves, TaALMT1-1-V was clearly expressed, but TaALMT1-2-IV was expressed at a very low level.
To further test the possibility that low expression of the 2174 TaALMT1-2-IV allele was caused by variation in tubulin primer sequences, actin was used as an endogenous control to quantitatively determine the difference in transcriptional level between the Jagger TaALMT1-1-V allele and the 2174 TaALMT1-2-IV allele. As shown in Fig. 3c, in roots, TaALMT1-1-V was expressed twice as much as TaALMT1-2-IV. Although TaALMT1-1 was detected at a much lower transcriptional level in leaves than in roots, the difference in gene expression between the two alleles was significant (Fig. 3d).
No homoeologous TaALMT1 sequences in wheat
Hexaploid wheat has three homoeologous genomes, and each gene is expected to have three homoeologous genes. In order to determine whether there is homoeologous TaALMT-A1 or homoeologous TaALMT-B1 in wheat, the complete TaALMT1 gene sequence was used to search the IWGSC databases (http://www.wheatgenome.org), and two sequence contigs were hit. One is IWGSC_chr4DL_V3_ab_k71_contigs_longerthan_200_14215826 (3301 bp), and the other is IWGSC_chr4DL_V3_ab_k71_contigs_longerthan_200_14459193 (3090 bp). The two sequences were from two parts of the same TaALMT-D1 gene. However, no homoeologous TaALMT-A1 or TaALMT-B1 gene was found in any contig on genome-wide sequences.
The complete TaALMT1 gene sequence was used to search for TaALMT-A1 and TaALMT-B1 in transcriptome sequences of T. urartu containing genome A and T. turgidum containing genomes A and B in GrainGenes databases, genome sequences of T. urartu, and all other databases in GenBank, but neither TaALMT-A1 nor TaALMT-B1 was found in these databases.
Discussion
In this study, two locally adapted cultivars known to differ in tolerance to acidic soils were used to generate a biparental population and to map acidic soil tolerance genes. As a result, the aluminum tolerance gene TaALMT1 was found to be one of the candidate genes for segregation of acidic soil tolerance in the population. The tolerant cultivar, Jagger, carries TaALMT1-1-V, the same allele present in ET8 and Atlas66. 2174, with moderate or intermediate tolerance, carries TaALMT1-2-IV, which may carry the same TaALMT1-2 as TaALMT1-2-IV that was described but not sequenced in a previous study (Raman et al. 2008). TaALMT1-2 was reported to have promoter Types I, II, III, V, and VI, and TaALMT1-1 was reported to have promoter Types I and V only (Sasaki et al. 2006; Raman et al. 2008). Promoter Type IV was reported to exist in 4 of 73 lines screened in a previous study, including Bariacora M92, CD87, Currawong, and Genaro-1 (Sasaki et al. 2006), but the sequence of the gene region of TaALMT1 in these four lines was not reported. 2174 has the same sequences in the gene region of TaALMT1-2 as ES8, CS, and Scout 66, but different sequences in the promoter. Gene expression was increased when a TaALMT-1 allele contained tandem repeated elements in its promoter (Sasaki et al. 2006). The effect of tandem repeated block sequences in enhancing gene expression has been demonstrated in transformation experiments (Ryan et al. 2010). The Jagger TaALMT1-1-V allele has triplicated sequence repeats (Type V), whereas the 2174 TaALMT1-2-IV allele has duplicated sequence repeats (Type IV). This study has experimentally demonstrated that TaALMT1-1-V had the higher transcriptional level and conferred greater tolerance to acidic soils compared to TaALMT1-2-IV probably due to greater aluminum tolerance. The positive association of the number of block sequence repeats in the ALMT1 promoters with gene transcriptional level and acidic soil tolerance was observed in non-Japanese lines but not in Japanese lines (Sasaki et al. 2006).
Malate is the predominant organic acid that is released in aluminum-tolerant but not aluminum-sensitive genotypes of ALMT1 (Raman et al. 2005; Sasaki et al. 2006). ALMT1 in crops encodes a membrane transporter that mediates malate efflux underlying wheat aluminum tolerance in roots (Delhaize et al. 1993, 2012). TaALMT1 was reported to be constitutively expressed in root apices in a previous study, in which northern blots were used to investigate the transcriptional profiles (Sasaki et al. 2004). However, extraction of RNAs from root tissue makes it inconvenient to study TaALMT1 function. This study used the more sensitive quantitative PCRs to test the TaALMT1 expression and showed that TaALMT1 was expressed not only in roots but also in seedling leaves, as well as in spikes of adult plants. It is likely that TaALMT1 was predominantly in roots and partly expressed in shoots due to the specificity of the TaALMT1 promoter, but the possibility that the RNA transcripts detected in leaves were transported from roots cannot be excluded. When grown in acidic soils, plants can accumulate aluminum in leaves in wheat. The presence of more TaALMT1 transcripts in tolerant wheat cultivars may effectively detoxify internal aluminum by forming aluminum-organic acid complexes (Ma et al. 2001). The difference in the TaALMT1 transcriptional level in leaves between the TaALMT1-1-V and TaALMT1-2-IV alleles was significant and consistent. Therefore, RNAs from leaves can be used to determine TaALMT1 transcriptional levels for allele variation or experimental effects in future studies.
The genetic effects of TaALMT1 were observed in several populations (Ma et al. 2005; Raman et al. 2005; Sasaki et al. 2004; Zhou et al. 2007; Dai et al. 2013). This locus explained various proportions of the total phenotypic variation in several biparental populations, including 84 % for HSS × NRG (Dai et al. 2013), approximately 50 % for Atlas 66 × Century (Ma et al. 2005), 45 % for Atlas 66 × Chisholm (Zhou et al. 2007), and 51 % for FSW × ND35 (Cai et al. 2008). The maximum effect was 38 % as observed in the Jagger × 2174 RIL population. Across studies, the large variation in genetic effect of TaALMT1 is related not only to experimental soil conditions (field) and phenotypic methods (aboveground traits or root staining pattern) but also to the parental difference relative to aluminum tolerance and genetic backgrounds of tested materials. TaALMT1 in wheat behaved as a monogene controlling aluminum tolerance under some genetic backgrounds (Raman et al. 2005; Riede and Anderson 1996) but as part of multiple gene effects under other genetic backgrounds (Berzonsky 1992; Cai et al. 2008; Zhou et al. 2007). Other genes/loci related to aluminum tolerance included those on 1A, 1B, 2A, 2B, 2D, 3A, 3B, 4A, 4B, 4D, 5B, 6A, 6B, 7A, and 7B that were identified by genome-wide association analysis and those on 2A, 3B, and 4B by QTL mapping (Dai et al. 2013). The observation of genetic effects of genes/QTLs on 4A or 4B for aluminum tolerance raised the hypothesis whether there is homoeologous TaALMT-A1 on 4A or homoeologous TaALMT-B1 that plays a similar role as TaALMT1 on 4DL. Interestingly, no sequence for such a gene was found in any databases of wheat genome sequences currently available. A gene on chromosome 4B for aluminum tolerance was cloned but it encodes a MATE transport protein and belongs to a different family from the ALMT proteins (Tovkach et al. 2013), supporting the hypothesis that a close homolog of TaALMT1 is not present on chromosome 4B or the entire genome of hexaploid wheat. An extensive screen of aluminum resistance in Triticum germplasm showed no aluminum tolerance from either A or B genomes in tetraploid species T. turgidum and T. dicoccoides or from only the A genome or its relative in diploid T. urartu and T monococcum, tetraploid T. timopheevii, and hexaploid T. zhukovskyi showed on resistance to aluminum, but significant or moderate aluminum tolerance from T. tauschii that is the donor of the D genome in bread wheat (Ryan et al. 2010). The frequent observation of aluminum tolerance in diverse populations irrespective of their geographic origins (Luo and Dvorák 1996; Riede and Anderson 1996; Ma et al. 2005; Raman et al. 2005, 2006, 2008; Ryan et al. 2009) suggested that TaALMT1 is a gene unique to genome D in wheat.
Many previously reported QTLs/genomic regions were identified for aluminum tolerance that were phenotyped based on staining of roots tested in hydroponic solutions containing aluminum. This study collected the phenotypic data from field and identified the same major QTL as reported on chromosome 4DL, indicating the phenotyping method was feasible. The two new QTL for acidic soil tolerance identified in the same system can be utilized in winter wheat breeding programs. The variability of acidic soil tolerance may be caused by different seasons, in addition to developmental stages of plants and years and locations of experiments. For example, plant shoots may be less dependent on roots in taking up water from deep soils in a wet season than a dry season. The peak of the QAlmt.osu-2D was in a 5.7-cM region between CFD16 and SNP51738501, and the peak of the QAlmt.osu-7B was in a 4.0-cM region between SNP55699328 and GWM577. These flanking markers and their linked markers can be used in molecular breeding for improvement of acidic soil tolerance in wheat.
This study indicates that the tolerance of winter wheat cultivars grown in the southern Great Plains to acidic soils is mainly controlled by three genes: two in Jagger (TaALMT1 and QAlmt.osu-7B) and one in 2174 (QAlmt.osu-2D). Sufficient levels of tolerance have been observed in hard red winter wheat cultivars (Kariuki et al. 2007), which may be inadvertently retained or selected by local breeders. Instead, breeders would be better served with selection tools that enable favorable alleles to be combined across multiple loci into a single genotype. Development of molecular markers for the presence of the resistant allele at all of the three genes/loci will accelerate their utilization in future breeding of novel winter wheat cultivars adapted to acid soil conditions throughout wheat plant development.
References
Berzonsky WA (1992) The genomic inheritance of aluminum tolerance in ‘Atlas 66’ wheat. Genome 35:689–693
Cai S, Bai GH, Zhang D (2008) Quantitative trait loci for aluminum resistance in Chinese wheat landrace FSW. Theor Appl Genet 117:49–56
Cao S, Carver BF, Zhu X, Fang T, Chen Y, Hunger RM, Yan L (2010) A single-nucleotide polymorphism that accounts for allelic variation in the Lr34 gene and leaf rust reaction in hard winter wheat. Theor Appl Genet 121:385–392
Carver BF, Ownby JD (1995) Acid soil tolerance in wheat. In: Sparks DL (ed) Advances in agronomy, vol 54. Academic Press, San Diego, pp 117–173
Carver BF, Inskeep WP, Wilson NP, Westerman RL (1988) Seedling tolerance to aluminum toxicity in hard red winter wheat germplasm. Crop Sci 28:463–467
Carver BF, Whitmore WE, Smith EL, Bona L (1993) Registration of four aluminum-tolerant winter wheat germplasm and two susceptible near-isolines. Crop Sci 33:1113–1114
Cavanagh C, Chao S, Wang S, Huang BE, Stephen S et al (2013) Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci 110:8057–8062
Chen Y, Carver BF, Wang S, Zhang F, Yan L (2009a) Genetic loci associated with stem elongation and dormancy release in winter wheat. Theor Appl Genet 118:881–889
Chen Y, Hunger RM, Carver BF, Zhang H, Yan L (2009b) Genetic characterization of powdery mildew resistance in U.S. hard winter wheat. Mol Breed 24:141–152
Chen Y, Carver BF, Wang S, Cao S, Yan L (2010) Genetic regulation of developmental phases in winter wheat. Mol Breed 26:573–582
Conyers MK, Poile GJ, Cullis BR (1991) Lime responses by barley as related to available soil aluminum and manganese. Aust J Agric Res 42:379–390
Dai GB, Zhang D, Hong DL (2013) Validation of quantitative trait loci for aluminum tolerance in Chinese wheat landrace FSW. Euphytica 192:172–179
Delhaize E, Craig S, Beaton CD et al (1993) Aluminum tolerance in wheat (Triticum aestivum L.). I. Uptake and distribution of aluminum in root apices. Plant Physiol 103:685–693
Delhaize E, Ryan PR, Hebb DM, Yamamoto Y, Sasaki T, Matsumoto H (2004) Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc Natl Acad Sci 101:15249–15254
Delhaize E, James RA, Ryan PR (2012) Aluminum tolerance of root hairs underlies genotypic differences in rhizosheath size of wheat (Triticum aestivum) grown on acid soil. New Phytol 195:609–619
Edwards JT, Hunger RM, Smith EL, Horn GW, Chen M-S, Yan L, Bai G, Bowden RL, Klatt AR, Rayas-Duarte P, Osburn RA, Kolmer JA, Jin Y, Porter DR, Giles KL, Seabourn BW, Bayles MB, Carver BF (2012) ‘Duster’ wheat: a durable, dual-purpose cultivar adapted to the southern Great Plains of the USA. J Plant Regul 6:37–48
Fang T, Campbell KG, Liu Z, Chen X, Wan A, Li S, Liu S, Cao S, Chen Y, Robert LB, Carver BF, Yan L (2011) Stripe rust resistance in the wheat cultivar Jagger is due to Yr17 and a novel resistance gene. Crop Sci 51:2455–2465
Garvin DF, Carver BF (2003) Role of genotypes tolerant of acidity and aluminum toxicity. In: Rengel Z (ed) Handbook of soil acidity. Marcel Dekker, New York, pp 387–406
Haynes RJ (1982) Effects of liming on phosphate availability in acid soils. Plant Soil 68:289–308
Johnson JP, Carver BF, Baligar VC (1997a) Productivity in Great Plains acid soils of wheat genotypes selected for aluminum tolerance. Plant Soil 188:101–106
Johnson JP, Carver BF, Baligar VC (1997b) Expression of aluminum tolerance transferred from Atlas 66 to hard winter wheat. Crop Sci 37:103–108
Kariuki H, Zhang H, Schroder JL, Edwards J, Payton M, Carver BF, Raun WR, Krenzer EG (2007) Hard red winter wheat cultivar responses to a pH and aluminum concentration gradient. Agron J 99:88–98
Kerridge PC, Kronstad WE (1968) Evidence of genetic resistance to aluminum toxicity in wheat. Agron J 60:710–711
Kochian LV, Hoekenga OA, Piñeros MA (2004) How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu Rev Plant Biol 55:459–493
Li G, Yu M, Fang T, Cao S, Carver BF, Yan L (2013) Vernalization requirement duration in winter wheat is controlled by TaVRN-A1 at the protein level. Plant J 76:742–753
Luo MC, Dvořák J (1996) Molecular mapping of an aluminum tolerance locus on chromosome 4D of Chinese Spring wheat. Euphytica 91:31–35
Ma JF, Ryan PR, Delhaize E (2001) Aluminium tolerance in plants and the complexing role of organic acids. Trends Plant Sci 6:273–278
Ma HX, Bai GH, Carver BF, Zhou LL (2005) Molecular mapping of a quantitative trait locus for aluminum tolerance in wheat cultivar Atlas 66. Theor Appl Genet 112:51–57
Raman H, Gustafson P (2014) Genetic dissection of aluminum tolerance in the Triticeae. In: Tuberosa R, Graner A, Frison E (eds) Genomics of plant genetic resources. Springer, Netherlands, pp 211–231
Raman H, Zhang KR, Cakir M, Appels R, Garvin DF, Maron LG, Kochian LV, Moroni JS, Raman R, Imtiaz M, Drake-Brockman F, Waters I, Martin P, Sasaki T, Yamamoto Y, Matsumoto H, Hebb DM, Delhaize E, Ryan (2005) Molecular characterization and mapping of ALMT1, the aluminum-tolerance gene of bread wheat (Triticum aestivum L.). Genome 48:781–791
Raman H, Raman R, Wood R, Martin P (2006) Repetitive indel markers within the ALMT1 gene conditioning aluminum tolerance in wheat (Triticum aestivum L.). Mol Breed 18:171–183
Raman H, Ryan PR, Raman R, Stodart BJ, Zhang K, Martin P, Wood R, Sasaki T, Yamamoto Y, Mackay M, Hebb DM, Delhaize E (2008) Analysis of TaALMT1 traces the transmission of aluminum resistance in cultivated common wheat (Triticum aestivum L.). Theor Appl Genet 116:343–354
Riede CR, Anderson JA (1996) Linkage of RFLP markers to an aluminum tolerance gene in wheat. Crop Sci 36:905–909
Ryan PR, Raman H, Gupta S, Horst Walter J, Delhaize E (2009) A second mechanism for aluminum resistance in wheat relies on the constitutive efflux of citrate from roots. Plant Physiol 149:340–351
Ryan PR, Raman H, Gupta S, Sasaki T, Yamamoto Y, Delhaize E (2010) The multiple origins of aluminium resistance in hexaploid wheat include Aegilops tauschii and more recent cis mutations to TaALMT1. Plant J 64:446–455
Sasaki T, Yamamoto Y, Ezaki B, Katsuhara M, Ahn SJ, Ryan PR, Matsumoto H (2004) A wheat gene encoding an aluminum activated malate transporter. Plant J 37:645–653
Sasaki T, Ryan PR, Delhaize E, Hebb DM, Ogihara Y, Kawaura K, Yamamoto Y (2006) Sequence upstream of the wheat (Triticum aestivum L.) ALMT1 gene and its relationship to aluminum resistance. Plant Cell Physiol 47:1343–1354
Scott BJ, Fisher JA, Cullins BR (2001) Aluminum tolerance and lime increase wheat yield on the acidic soils of central and southern New South Wales. Aust J Exp Agric 41:523–532
Somers DJ, Gustafson JP (1995) The expression of aluminum stress induced polypeptides in a population segregating for aluminum tolerance in wheat (Triticum aestivum L.). Genome 38:1213–1220
Tan CT, Carver BF, Chen M, Gu Y, Yan L (2013) Genetic association of OPR and LOX genes with resistance to Hessian fly in hexaploid wheat. BMC Genom 14:369
Tovkach A, Ryan PR, Richardson AE, Lewis DC, Rathjen TM, Ramesh S, Tyerman SD, Delhaize E (2013) Transposon-mediated alteration of TaMATE1B expression in wheat confers constitutive citrate efflux from root apices. Plant Physiol 161:880–892
Van Ooijen JW (2006) JoinMap 4, software for the calculation of genetic linkage maps in experimental populations. Kyazma B.V, Wageningen
Van Oojjen JW (2009) MapQTL 6, software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyazma B.W, Wageningen
Zhang H, Raun WR (2006) Oklahoma soil fertility handbook. Dept of Plant and Soil Sciences, Oklahoma State University, Stillwater
Zhou LL, Bai GH, Ma HX, Carver BF (2007) Quantitative trait loci for aluminum resistance in wheat. Mol Breed 19:153–161
Acknowledgments
This work was supported by USDA-NIFA T-CAP Grant No. 2011-68002-30029, the Oklahoma Wheat Research Foundation, and the Oklahoma Agricultural Experiment Station.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no conflict of interest.
Additional information
Meiyan Liu and Ming Yu have contributed equally to the research work.
Rights and permissions
About this article
Cite this article
Liu, M., Yu, M., Li, G. et al. Genetic characterization of aluminum tolerance in winter wheat. Mol Breeding 35, 205 (2015). https://doi.org/10.1007/s11032-015-0398-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-015-0398-y