Abstract
Leaf rust, caused by Puccinia triticina Eriks, is one of the most common and persistent wheat diseases in the US Great Plains. We report that the Lr34 gene was mapped in the center of a QTL for leaf rust reaction and explained 18–35% of the total phenotypic variation in disease severity of adult plants in a Jagger × 2174 population of recombinant inbred lines (RILs) field-tested for 3 years. The sequence of the complete Lr34 gene was determined for the susceptible Jagger allele and for the resistant 2174 allele. The two alleles had exactly the same sequence as the resistant allele reported previously in Chinese Spring at three polymorphic sites in intron 4, exon 11, and exon 12. A G/T polymorphism was found in exon 22, where a premature stop codon was found in the susceptible Jagger allele (Lr34E22s), confirming a previous report, due to a point mutation compared with the resistant 2174 allele (Lr34E22r). We have experimentally demonstrated a tight association between the point mutation at exon 22 of Lr34 and leaf rust susceptibility in a segregating biparental population. A PCR marker was developed to distinguish between the Lr34E22r and Lr34E22s alleles. A survey of 33 local hard winter wheat cultivars indicated that 7 cultivars carry the Lr34E22s allele and 26 cultivars carry the Lr34E22r allele. This study significantly improves our genetic understanding of allelic variation in the Lr34 gene and provides a functional molecular tool to improve leaf rust resistance in a major US wheat gene pool.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Leaf rust is one of the most common and persistent diseases of winter wheat grown throughout the US Great Plains (Kolmer et al. 2009). Many contemporary cultivars in this region have either ineffective or no genes conferring stable and long-term leaf rust resistance (Bockus et al. 2009). Though they harbor resistance genes, popular cultivars such as Jagger and ‘Jagalene’ are susceptible to one or more races, and wheat breeders are challenged by a shrinking gene pool for new sources of resistance.
Two general types of resistance genes to leaf rust exist in wheat: race-specific and non-race specific. Some genes such as Lr1, Lr10, and Lr21 confer complete resistance to specific race(s) and remain effective throughout the life of the plant (Bennett 1984; Hsam and Zeller 2002). These genes, however, tend to be vulnerable to new races of pathogens; hence, their effects usually are of short duration (Oelke and Kolmer 2005). In contrast, some genes such as Lr34 and Lr46 confer partial and non-race specific resistance and remain mostly effective in adult plants (Bennett 1984; Hsam and Zeller 2002). The effect of the non-race specific genes is generally durable regardless of the development of new pathotypes due to mutation (Wolfe 1984). One example of durability is the Lr34 gene, which has remained effective worldwide since it was first described in 1966, and no P. triticina isolates with complete virulence to Lr34 have yet been found in North America (Kolmer et al. 2009).
The current strategy of breeding new cultivars resistant to leaf rust has shifted from selection for single race-specific genes to pyramiding of several genes (non-specific or race-specific), which may provide more durable resistance against multiple rusts of wheat (Krattinger et al. 2009; Lagudah et al. 2009). Efforts at CIMMYT have also shifted to accumulating multiple adult-plant resistance genes in genotypes that, with some gene combinations, can offer near-complete immunity (Singh et al. 2000).
The initial objective of this study was to identify genetic loci for resistance to multiple foliar diseases existing in locally adapted cultivars and then to pyramid them by molecular markers. The primary mapping was conducted in a population of recombinant inbred lines (RILs) generated from two local wheat cultivars, Jagger (Sears et al. 1997) and ‘2174’ (Carver et al. 2004). Jagger is highly susceptible to powdery mildew [caused by Blumeria graminis (DC) E.O. Speer f. sp. tritici] and to leaf rust, whereas 2174 has excellent resistance to powdery mildew and intermediate resistance to leaf rust. We have reported that the segregation of powdery mildew resistance in the Jagger × 2174 RIL population was controlled by Pm3a on chromosome 1A and modified by several other minor QTLs (Chen et al. 2009b). In the present study, we found that a QTL on the short arm of chromosome 7D co-segregated with leaf rust reaction in the Jagger × 2174 population. As the present study progressed, the Lr34 gene sequence was reported by Krattinger et al. (2009). We therefore extended the original study to identify allelic variation in Lr34 between the susceptible Jagger allele and the resistant 2174 allele.
The Lr34 gene encodes an adenosine triphosphate (ATP)-binding cassette (ABC) transporter, which is a member of the pleiotropic drug resistance transporter superfamily. Allelic variation in Lr34 between the resistant form in cv. Chinese Spring and the susceptible form in cv. Renan involves three polymorphisms (Krattinger et al. 2009). The first one is an A/T single-nucleotide polymorphism (SNP) in intron 4, the second one is a 3-bp ins/del in exon 11, and the third one is a C/T SNP in exon 12, where an amino acid is altered. No difference within 2 kb of the putative Lr34 promoter region was detected between the two alleles (Krattinger et al. 2009).
The functional Lr34 allele has been reported to be present in the hard red winter wheat cultivar Sturdy (Dyck et al. 1966) and other contemporary North American cultivars, by identification of the marker csLv34 associated with Lr34 (Kolmer et al. 2008). However, the csLv34 marker has limited utility in Jagger deritatives. Here we report that segregation of resistance to leaf rust in the Jagger × 2174 population was associated with the Lr34 gene. Comparison of the complete Lr34 gene sequence between the susceptible Jagger allele and the resistant 2174 allele showed one SNP in exon 22, where a point mutation resulted in a premature stop codon in Jagger. The point mutation in Lr34 in Jagger was described in a recent study (Lagudah et al. 2009), who reported a pair of primers that was used to detect this mutation in Jagger. This study not only validated the function of Lr34 in US hard winter wheat but also developed a new PCR marker with greater efficiency and less cost to track the presence, or absence, of the mutated Lr34 allele in wheat.
Materials and methods
Plant materials
A population of RILs was generated from a single cross of two winter wheat cultivars, Jagger and 2174. This RIL population was previously used to map the onset of stem elongation stage (Chen et al. 2009a) and reaction to powdery mildew (Chen et al. 2009b). Jagger was resistant to leaf rust when it was released (Sears et al. 1997), but it has shown an increasingly susceptible reaction in the field in Oklahoma since 2003. 2174 has consistently shown an intermediate resistance to reaction to leaf rust since it was released in 1997 (PI 602595; USDA-ARS 2009). Therefore, the Jagger × 2174 population was used to map genes for resistance to leaf rust.
Phenotyping resistance to leaf rust
A population of 152 RILs and the two parental lines were evaluated for 3 years in the field at the North Central Research Station near Lahoma, OK, in 2007 and at the Stillwater Agronomy Research Station in 2008 and 2009. The lines were arranged in a replicates-in-sets design, with two replicates in four sets. Each set, which contained 38 RILs and the two parents, was arranged in the field as a randomized complete-block design. Planting dates were 6 October 2006, 30 October 2007, and 19 November 2008. Under natural infection and when sporulation appeared most pronounced, reaction to leaf rust was recorded for 96 of the 152 lines on the flag and penultimate leaves of adult plants. Ratings were collected on 10 May 2007 and 20 May 2009 using a 1 (resistant)-to-9 (highly susceptible) scale. In 2008, ratings were collected independently by two individuals (9 May and 14 May) to confirm consistency of ratings.
Mapping genetic loci resistant to leaf rust
A total of 246 SSR markers were mapped for the stem elongation trait using the same 96 RILs by Chen et al. (2009a). The marker number was increased to 310 for mapping powdery mildew resistance (Chen et al. 2009b). Forty markers were added in this study to increase marker density in the previous linkage maps and to better enable detection of QTLs for leaf rust reaction. The 350 SSR markers were assembled in linkage groups using the MapMaker 3.0 program. QTLs were claimed when the logarithm of the odds (LOD) score exceeded the threshold value of 2.5 using interval mapping (IM) and composite interval mapping (CIM) (via WinQTLCart 2.5, North Carolina State University, Raleigh, NC). An analysis of variance (ANOVA) was used to compare means for leaf rust rating separated by a single marker.
Development of a PCR marker for Lr34
As we discovered a major QTL for leaf rust reaction associated with the Lr34 locus on the short arm of chromosome 7D, nine markers that were reported to be linked with or flanking the Lr34 gene (Lagudah et al. 2006; Krattinger et al. 2009) were tested for PCR product polymorphisms between Jagger and 2174. Three SNP markers (SWSNP1, SWSNP2, and SWSNP3) showed no difference in sequence between the Jagger and 2174 alleles. Three indel markers (SWDEL1, SWDEL2, and SWDEL3), two SSR markers (SWM10 and csLVMS), and one RFLP-converted PCR marker (csLV34) did not show a detectable difference between the two parental lines. Therefore, a gene marker for the Lr34 gene was developed by sequencing the Jagger allele and the 2174 allele.
The promoter region and introns of the complete Lr34 gene were initially prioritized for sequencing to find polymorphisms between the Jagger and 2174 alleles, since they should have a higher probability of harboring polymorphisms than exons or coding regions. Primers were designed according to the sequence of the Chinese Spring allele. PCR products of different fragments of the Lr34 gene were directly sequenced, or they were cloned for sequencing if sequence reactions for PCR products did not succeed due to the presence of multiple Lr34 genes from genome A, B and D. Initially, no attempt was made to sequence the complete Lr34 gene, since the Chinese Spring allele (GenBank accession # FJ436983) contains 11,805 bp between the translational start and stop codons. However, when one SNP was found in exon 22, the complete gene from each parent was sequenced to explain any difference between the functional and non-functional Lr34 alleles. The complete gene was assembled using fragments from different regions of the Lr34 gene.
The SNP in exon 22 found between Jagger and 2174 was attributed to a mutation in Jagger, which produces a premature stop codon TGA (Fig. 1). This mutated site was the same as that reported by Lagudah et al. (2009), but they reported that the purified PCR products of the Jagger Lr34 allele by primers (cssfr7_f and cssfr7_r) could be digested with NlaIII (5′-CATG-3′, the digested site between G and the first nucleotide downstream) (Fig. 1).
Positions and sequences of forward primers used to detect a SNP in exon 22 of Lr34. The SNP in exon 22 was found between Jagger and 2174, highlighted and underlined ‘G’ in 2174 and ‘T’ in Jagger. cssfr7_f is a forward primer used to detect the mutation in Jagger by digestion of purified PCR products with NlaIII [(5′-CATG-3′, from the fifth nucleotide downstream of cssfr7_f) (Lagudah et al. 2009)]. In this study, a new forward primer Lr34F was developed to detect the SNP in exon 22 of Lr34. The nucleotide “C” at position 21 of the primer was artificially mutated from the original nucleotide “T”. PstI (5′-CTGCAG-3′) can be used to digest the PCR products from the 2174 Lr34E22r allele but not from the Jagger Lr34E22s allele
A new PCR marker was developed to detect the only SNP in Lr34 between the Jagger and 2174 alleles using an approach called derived or degenerated cleaved amplified polymorphic site (dCAPS) (Konieczny and Ausubel 1993). A new forward primer Lr34F (5′-TTGTAATGTATCGTGAGAGATCTGCA-3′) was designed, in which the nucleotide “C” at position 21 was artificially mutated from the original nucleotide “T” (Fig. 1). The reverse primer was Lr34R was the same as cssfr7_r (5′-CATAGGAATTTGTGTGCTGTCC-3′). The mismatched PCR products from the 2174 allele would include the sequence 5′-TTGTAATGTATCGTGAGAGATCTGCAGNNNNN-3′, in which N is the sequence up to the reverse primer, and the raw PCR products can be digested with PstI (5′-CTGCAG-3′, the digested site between A and G). The mismatched PCR products from the Jagger allele would include the sequence 5′-TTGTAATGTATCGTGAGAGATCTGCATNNNNN-3′, in which N is sequence up to the reverse primer, and the raw PCR product cannot be digested with PstI due to an incompatible sequence at the restriction site, 5′-CTGCAT-3′.
The same set of 31 contemporary cultivars, including Jagger and 2174, that were genotyped for Pm3a (Chen et al. 2009b) were again genotyped here using the Lr34 PCR marker described above. Two additional Great Plains cultivars ‘Protection CL’ and ‘Ripper’ were included in this study.
Results
Leaf rust reaction in parental lines and their RILs
Jagger rapidly developed symptoms of leaf rust in the field each year, producing ratings of 5–7 on a 1 (resistant)-to-9 (susceptible) scale. In contrast, 2174 produced ratings of 2–4, with delayed development of symptoms typical of slow-rusting. Mean ratings for the RILs in 2007 varied from 1.0 to 8.5, with a population mean of 3.7 (data not shown). Weather conditions were more conducive in 2008 to disease spread and greater severity of symptoms, as the RILs varied in rating from 1.5 to 9.0 with a population mean of 5.2. In 2009, ratings varied from 1.0 to 9.0, with a population mean of 4.3. Hence, disease pressure was of sufficient magnitude each year to allow phenotypic assessment of leaf rust response across multiple environments. Moreover, reactions were consistent across environments based on between-year correlation coefficients of 0.63 (2007 vs. 2008), 0.71 (2007 vs. 2009), and 0.58 (2008 vs. 2009) (P < 0.001).
QTL mapping of leaf rust resistance
A total of 350 markers was mapped and assembled into linkage groups to identify QTLs for leaf rust reaction. Several QTLs were found, but when the threshold was set at 2.5 for LOD value, all QTLs appeared in one year but not another, except for the QTL associated with Lr34. Those loci showing inconsistent effects were not further characterized in this study.
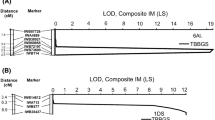
A QTL tightly linked with Lr34 on the short arm of chromosome 7D was consistently detected across 3 years (Fig. 2). This QTL was initially mapped to a linkage group including two SSR markers Xbarc352 and Xgwm130 that were reported by others to be tightly linked to the Lr34 locus (Schnurbusch et al. 2004; Suenaga et al. 2003). Eleven SSR markers were mapped in this linkage group; mapping of the vernalization gene VRN-D3 in the same group validated chromosomal location of this QTL. Final mapping in this group of a PCR marker developed directly from the Lr34 gene sequence suggested that Lr34 was responsible for the QTL on chromosome 7D.
Chromosomal location of a QTL for leaf rust disease reaction in the recombinant inbred lines (RILs) of the Jagger × 2174 population. The QTL was mapped on the short arm of chromosome 7D. The solid line indicates a QTL based on reactions determined at the North Central Research Station at Lahoma, OK, in 2007; the dashed line indicates a QTL based on reactions determined at the Stillwater Research Station in 2008 (YC and BC were two individuals who independently made ratings); the dotted line indicates a QTL based on reactions determined at the Stillwater Research Station in 2009. Molecular markers along the chromosome are placed as centimorgans on the vertical axis. The horizontal solid line represents a common threshold of 2.5 for the LOD value
A G/T polymorphism was found in exon 22 (see below), which facilitated development of PCR markers for Lr34 by creating a mutation in upstream position 3 from the polymorphic site (Fig. 1). After digestion with PstI, the polymorphic PCR products could be visibly detected on an agarose gel (Fig. 3). The PCR marker was used to detect the difference at the polymorphic site among the RILs, allowing direct mapping of Lr34 in the Jagger and 2174 population.
Difference in the digested PCR products with PstI between the Jagger Lr34E22s allele and the 2174 Lr34E22r allele. 1 M represents DNA marker Quick-Load® 100 bp DNA Ladder (NEB); 2 Jagger; 3 2174. PCR was performed using primers (Lr34F and Lr34R) for 40 cycles (94°C for 15 s, 65°C for 20 s, and 72°C for 15 s per cycle) followed by a 10-min final extension at 72°C. 10-μl raw PCR products were digested directly in the PCR buffer with 0.5-μl PstI (#R0140S, New England BioLabs)
Lr34 was mapped at the center of the peaks of the QTLs for leaf rust ratings in each of 3 years (Fig. 2). All LOD values of the QTLs exceeded the common threshold of 2.5 LOD value, indicating the effect of this genetic locus was stable and consistent. The LOD value for Lr34 was 8.1 for ratings taken in Lahoma in 2007, which explained 35.2% of the total phenotypic variation. The LOD values for Lr34 were 3.8 and 4.1 for independent ratings taken by two individuals in Stillwater in 2008, which on average accounted for 18.1% of the total phenotypic variation. Corresponding values in 2009 at Stillwater were 3.9 (LOD value) and 18.2%. Across all 3 years, Lr34 explained 23.8% of the total phenotypic variation, which would be expected of a single gene within the mapped QTLs conferring adult-plant resistance to leaf rust.
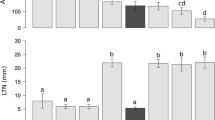
Furthermore, RILs carrying the Jagger Lr34 allele showed a significantly higher leaf rust rating than RILs carrying the 2174 Lr34 allele (Fig. 4). Averaged across years, respective mean ratings for the two RILs groups were 5.7 and 3.8, indicating that the Jagger allele was associated with greater susceptibility to leaf rust than the 2174 allele.
Allelic variation in Lr34
A total length of 15,977 bp from Lr34 was retrieved from each of the Jagger and 2174 alleles. Both sequences included 11,805 bp between the translational start and stop codons, and 3,339 bp upstream of the start codon and 833 bp downstream of the stop codon. By comparison with sequences of a resistant allele of Chinese Spring (GenBank accession # FJ436983) and a susceptible allele of Renan (GenBank accession # FJ436985) (Wicker et al. 2009), several salient features were observed in the Jagger and 2174 sequences (Fig. 5).
Diagram for mutated sites at Lr34. Star symbol indicates positions of three reported polymorphisms in intron 4, exon 11, and exon 12 in the susceptible allele (Renan) compared with the resistant allele of Chinese Spring (CS) (Krattinger et al. 2009; Wicker et al. 2009). Plus symbol indicates positions of four mutations present in the promoter of Lr34 in Renan only. Dot symbol indicates positions of four polymorphic sites in the 2174 allele only. Heart symbol indicates the position of the polymorphism in exon 22 of Lr34 in the susceptible allele (Jagger) compared with the resistant allele (2174). The position of the premature stop codon TGA resulting in a lack of 185 amino acids in Jagger is indicated. ‘R’ represents ‘resistant’ and ‘S’ represents ‘susceptible’
First, both Jagger and 2174 alleles had the nucleotide ‘A’ in position 675 of intron 4 of Lr34, as did the resistant allele in Chinese Spring. This nucleotide was replaced by ‘T’ in the susceptible allele of Renan. Second, both Jagger and 2174 alleles had the same sequence as the Chinese Spring resistant allele in exon 11; the susceptible allele of Renan had ‘TTC’ insertion at positions 106–108 in this exon. Third, both Jagger and 2174 alleles had the nucleotide ‘C’ at position 56 of exon 12 of Lr34, as did the resistant allele in Chinese Spring; this nucleotide was replaced by ‘T’ in the susceptible allele of Renan.
In addition, both Jagger and 2174 alleles had exactly the same sequences as Chinese Spring within the region 3,339 bp upstream from the translation start codon, but there were four polymorphic sites compared with Renan. A motif of 11 ‘C’ occurred at positions 2,266–2,276 in Jagger, 2174, and Chinese Spring, but Renan contained a motif of 14 ‘C’. Finally, a 1-bp deletion at positions 3,181, 3,209, and 3,237 occurred in Renan but not in the other three cultivars.
Three SNPs and one indel polymorphism were found in non-coding regions between the Jagger and 2174 Lr34 alleles. In intron 4, the Jagger allele had the nucleotide ‘T’ at each of position 868 and position 1,228, whereas 2174 had ‘C’ at both of the two positions. In intron 6, the Jagger allele had the nucleotide ‘T’ at position 48 and 11 ‘A’ at positions 58–67, whereas 2174 had ‘C’ and 10 ‘A’ at these two positions. The susceptible Jagger allele had exactly the same sequences as the resistant Chinese Spring allele at these three SNPs and one indel polymorphic site. Hence, any of these polymorphic sites cannot be accountable for loss of the Jagger Lr34 function.
However, one polymorphism was found between Jagger and 2174 in the coding region. This occurred at position 136 of exon 22, where the nucleotide ‘G’ in the 2174 Lr34 allele was mutated into the nucleotide ‘T’ in the Jagger Lr34 allele, resulting in the formation of a premature stop codon (TGA) at positions 137–139 of exon 22 in the Jagger Lr34 allele (Figs. 1, 5). Consequently, the predicted Jagger Lr34 protein lacks 185 amino acids of the C-terminus (Fig. 5). In order to distinguish between this allelic variation and other polymorphisms, the susceptible Jagger allele was designated Lr34E22s and the resistant 2174 allele was designated Lr34E22r.
Genotypes of local winter wheat cultivars
To screen cultivars in the southern Great Plains for their putative Lr34 allele, we used the PCR marker for the Lr34E22 alleles. Because no sequences were determined for these genotypes, those that had the same genotype as Jagger were believed to carry the Lr34E22s allele, whereas those that had the same genotype as 2174 were believed to carry the Lr34E22r allele.
Among 33 local hard winter wheat cultivars analyzed, 7 cultivars carried the Jagger Lr34E22s allele (Fuller, Jagalene, Jagger, Ok Bullet, Protection CL, Santa Fe, and Shocker), and 26 cultivars carried the Lr34E22r allele (2174, Above, Centerfield, Cimarron, Custer, Cutter, Danby, Deliver, Doans, Duster, Endurance, Fannin, Guymon, Hatcher, Intrada, JEI 110, Lakin, Neosho, Ok102, Okfield, Overley, Ripper, TAM 110, TAM 111, TAM 112, and Trego).
Discussion
So far approximately 60 leaf rust-resistance genes have been identified in wheat and its wild relatives (Sun et al. 2009). These genetic loci for leaf rust were mapped using QTL analyses in various populations (Messmer et al. 2000; Navabi et al. 2005; Rosewarne et al. 2008; Schnurbusch et al. 2004; Suenaga et al. 2003; William et al. 1997). Other than Lr1, Lr10, and Lr21 which confer resistance to specific races, the only non-race specific resistance gene to be cloned has been Lr34 (Krattinger et al. 2009).
The Lr34 gene has provided partial, but effective, durable, adult-plant resistance to leaf rust worldwide for more than 50 years (Dyck et al. 1966; Singh 1992; Krattinger et al. 2009). The gene was mapped on the short arm of chromosome 7D by different markers, such as SSR markers Xbarc310, Xbarc352, Xgwm130, Xgwm295, Xgwm1220, SWM10, and csLV34 (Bossolini et al. 2006; Schnurbusch et al. 2004; Suenaga et al. 2003). Once one QTL for leaf rust reaction was associated with any of these markers on chromosome 7DS, Lr34 was naturally and reasonably considered the gene responsible for the QTL. Obviously, the QTL mapped between SSR markers Xbarc352 and Xgwm130 for leaf rust reaction in the Jagger × 2174 population was caused by the Lr34 gene. The recent availability of the Lr34 sequence allowed us to pursue perfect markers to test the association of the QTL and Lr34 gene.
The Lr34 gene was cloned using a map-based cloning approach, and the final candidate region was the 0.15-cM interval flanked by XSWSNP3/XcsLVA1 and XcsLVE17, or the 363-kb physical interval containing eight candidate genes (Krattinger et al. 2009). However, no difference in either these markers or the previously reported maker csLv34 (Kolmer et al. 2008; Krattinger et al. 2009) was detected between Jagger and 2174, preventing the direct mapping of the Lr34 gene in the RIL population. The complete sequences of the Lr34 gene from two alleles suggested that the polymorphic site in exon 22 uniquely resulted in the loss of the Jagger Lr34 allele function. This point mutation in Lr34 in Jagger was described in a very recent study, and a total of 185 amino acids of the C-terminus were missing in the predicted Jagger Lr34 protein (Lagudah et al. 2009). The complete sequence we obtained from the Jagger susceptible allele confirms the point mutation in Jagger is the only polymorphism in the coding region of Lr34 compared with the resistant allele in Chinese Spring and 2174. The point mutation in the Jagger Lr34 allele could result in a non-functional protein, since the missing part includes the majority of the second hydrophobic transmembrane domain of the Lr34 protein (Krattinger et al. 2009). This study also is the first to demonstrate a tight association between the point mutation at Lr34 and adult-plant leaf rust resistance in a segregating biparental population.
Jagger has shown an increasingly susceptible reaction, since it was released (Sears et al. 1997). Genotypes of the Lr34E22 marker for Jagger’s parents would provide important information for when the point mutation in exon 22 of the Lr34 gene in Jagger occurred. Jagger was created by a cross of ‘Stephens’ with KS82W418 carried out by the Kansas Agricultural Experiment Station. Stephens has a resistant Lr34E22r allele, but unfortunately, seed of KS82W418 is no longer available. Therefore, no conclusion can be made whether the Lr34E22s allele in Jagger was mutated from Stephens or if the Lr34E22s allele was derived from the original KS82W418 line.
Based on variation at three polymorphic sites in intron 4 and exons 11 and 12, two haplotypes were previously found in spring wheat from North and South America and European winter wheat (Krattinger et al. 2009), and a third haplotype was found in European winter wheat and spelt wheat (Lagudah et al. 2009). A larger number of accessions was investigated in those studies, and all of the tested accessions were subjected to classification of the three haplotypes. However, both Jagger and 2174 have the same sequences at the three polymorphic sites previously described. Jagger carried a susceptible Lr34E22s allele whereas 2174 carried a resistant Lr34E22r allele, which was caused by the G/T polymorphism at a new polymorphic site in exon 22. Hence according to this new allelic variation in Lr34 exon 22, Jagger is a new haplotype (Lagudah et al. 2009). 2174 had similar sequences as Chinese Spring with exception of three SNPs and one indel polymorphism in intron 4 and intron 6 found in this study. Hence, 2174 is also a new haplotype.
An artificial mutation to degenerate a dCAPS is an easy way to develop a PCR marker with less labor and expense for simple locus assays (Konieczny and Ausubel 1993). In the previous study by Lagudah et al. (2009), 10 units of NlaIII (New England BioLabs) were required to digest 40 μl purified PCR products of the Lr34 gene. We modified this protocol for two reasons. First, the requirement for purified PCR product prior to digestion will hinder use of this marker in breeding programs. Second, NlaIII has low activity and therefore low sample load, i.e., 500 units per 100 μl at a cost of $61 (#R01256S, New England BioLabs, 2007–2008 Catalog), which will accommodate only 50 samples. With the modified protocol, however, PCR products were directly digested with PstI without requirement of purification. In addition, PstI has high activity and high sample load potential, i.e., 10,000 units per 100 μl at a cost of $58 (#R0140S, New England BioLabs, 2007–2008 Catalog), which will accommodate 200 samples. This modified protocol will facilitate screening for or against a specific allele of Lr34 in breeding programs.
A cultivar carrying the Lr34E22s allele carries a form of the non-functional allele and may be susceptible to leaf rust, depending on the presence of alternative resistance genes. Most of the cultivars classified with the susceptible Lr34E22s allele are believed not to carry an effective allele at the Lr34 locus according to their field reactions, such as OK Bullet, Jagalene, and Protection CL (Edwards et al. 2009). The occurrence of the Lr34E22s allele in these cultivars may indicate that the point mutation was a single event in US winter wheat, because they shared Jagger as a parent. This mutation event was not found in any cultivar tested in previous studies except for Jagger (Krattinger et al. 2009; Lagudah et al. 2009). To achieve at least a base level of adult-plant resistance, breeders should select against the Lr34E22s allele in breeding new cultivars.
Any genotype carrying the Lr34E22r allele could be resistant to leaf rust, but this recommendation can only be made in the context of the G/T SNP in exon 22. Some cultivars such as Above, Cutter, and TAM 110, had the Lr34E22r allele, but they are susceptible to leaf rust as indicated by their field performance (Edwards et al. 2009); nor are these varieties expected to carry an effective Lr34 allele based on their parentage. Three other polymorphisms in intron 4, exon 11, and exon 12 have been associated with resistance/susceptibility to leaf rust races (Krattinger et al. 2009; Lagudah et al. 2009). Further work is underway in investigating if cultivars that have the Lr34E22r genotype but a susceptible phenotype have any mutations at the three known polymorphic sites or other unknown sites in the Lr34 gene.
Wheat cultivars containing Lr34 occupy more than 26 million ha in various developing countries alone and contribute substantially to yield savings in epidemic years (Marasas et al. 2004). In some cases, Lr34 alone does not render plants resistant to the pathogen, but it can enhance the resistance found in some cultivars (Vanegas et al. 2008). High levels of resistance can be achieved by combining multiple adult-plant resistance genes in the same cultivar (Singh et al. 2000). Near-immune resistance in adult plants can be achieved by combination of Lr34 with other non-specific genes such as Lr46 on chromosome 1B (Suenaga et al. 2003; Lillemo et al. 2008). The application of molecular markers for the Lr34E22 alleles will provide a more reliable tool for breeders to eliminate the susceptible Lr34E22s allele in breeding populations and allow more concentrated selection for other traits in the enriched materials.
References
Bennett AGA (1984) Resistance to powdery mildew in wheat: a review of its use in culture and breeding programs. Plant Pathol 33:279–300
Bockus WW, Bowden RL, Hunger RM, Morrill WL, Murray TD, Smiley RM (eds) (2009) Compendium of wheat diseases and insects, 3rd edn. APS Press, St. Paul, MN
Bossolini E, Krattinger SG, Keller B (2006) Development of simple sequence repeat markers specific for the Lr34 resistance region of wheat using sequence information from rice and Aegilops tauschii. Theor Appl Genet 113:1049–1062
Carver BF, Krenzer EG, Hunger RM, Porter DR, Smith EL, Klatt AR, Verchot-Lubicz J, Rayas-Duarte P, Guenzi AC, Bai G, Martin BC (2004) Registration of ‘Ok102’. Wheat Crop Sci 44:1468–1469
Chen Y, Carver BF, Wang S, Zhang F, Yan L (2009a) Genetic loci associated with stem elongation and winter dormancy release in wheat. Theor Appl Genet 118:881–889
Chen Y, Hunger RM, Carver BF, Zhang H, Yan L (2009b) Genetic characterization of powdery mildew resistance in US hard winter wheat. Mol Breed 24:141–152
Dyck PL, Samborski DJ, Anderson RG (1966) Inheritance of adult-plant leaf rust resistance derived from the common wheat varieties Exchange and Frontana. Can J Genet Cytol 8:665–671
Edwards J, Hunger RM, Carver B, Royer T. (2009). Wheat variety comparison. Okla Coop Ext Rep PSS-2142, Stillwater, OK. Available at http://osufacts.okstate.edu
Hsam SLK, Zeller FJ (2002) Breeding for powdery mildew resistance in common wheat (Triticum aestivum L.). In: Belanger RR, Bushnell WR, Dik AJ, Carver TLW (eds) The powdery mildews. A comprehensive treatise. St. Paul, MN, USA, pp 219–238
Kolmer J, Singh RP, Garvin DF, Viccars L, William HM, Huerta-Espino JH, Obonnaya FC, Raman H, Orford S, Bariana HS, Lagudah ES (2008) Analysis of the Lr34/Yr18 rust resistance region in wheat germplasm. Crop Sci 48:1841–1852
Kolmer J, Chen X, Jin Y (2009) Diseases which challenge global wheat production—the wheat rusts. In: Carver BF (ed) Wheat: science and trade. Wiley–Blackwell, IA, pp 89–124
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410
Krattinger SG, Lagudah ES, Spielmeyer W, Singh RP, Huerta-Espino J, McFadden H, Bossolini E, Selter LL, Keller B (2009) A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science 323:1360–1363
Lagudah ES, McFadden H, Singh RP, Huerta-Espino J, Bariana HS, Spielmeyer W (2006) Molecular genetic characterization of the Lr34/Yr18 slow rusting resistance gene region in wheat. Theor Appl Genet 114:21–30
Lagudah ES, Krattinger SG, Herrera-Foessel S, Singh RP, Huerta-Espino J, Spielmeyer W, Brown-Guedira G, Selter LL, Keller B (2009) Gene-specific markers for the wheat gene Lr34/Yr18/Pm38 which confers resistance to multiple fungal pathogens. Theor Appl Genet 118:889–898
Lillemo M, Asalf B, Singh RP, Huerta-Espino J, Chen XM, He ZH, Bjørnstad Å (2008) The adult plant rust resistance loci Lr34/Yr18 and Lr46/Yr29 are important determinants of partial resistance to powdery mildew in bread wheat line Saar. Theor Appl Genet 116:1155–1166
Marasas CN, Smale M, Singh RP (2004) The economic impact in developing countries of leaf rust in CIMMYT-related spring wheat. Econ. Program Rap. 04-01. CIMMYT, DF, Mexico
Messmer MM, Seyfarth S, Keller M, Schachermayr G, Winzeler M, Zanetti S, Feuillet C, Keller B (2000) Genetic analysis of durable leaf rust resistance in winter wheat. Theor Appl Genet 100:419–431
Navabi A, Tewari JP, Singh RP, McCallum B, Laroche A, Briggs KG (2005) Inheritance and QTL analysis of durable resistance to stripe and leaf rusts in an Australian cultivar, Triticum aestivum ‘Cook’. Genome 48:97–106
Oelke LM, Kolmer JA (2005) Genetics of leaf rust resistance in spring wheat cultivars Olsen and Norm. Phytopathology 95:773–778
Rosewarne GM, Singh RP, Huerta-Espino J, Robetzke GJ (2008) Quantitative trait loci for slow-rusting resistance in wheat to leaf rust and stripe rust identified with multi-environment analysis. Theor Appl Genet 116:1027–1034
Schnurbusch T, Paillard S, Schori A, Messmer M, Schachermayr G, Winzeler M, Keller B (2004) Dissection of quantitative and durable leaf rust resistance in Swiss winter wheat reveals a major resistance QTL in the Lr34 chromosomal region. Theor Appl Genet 108:477–484
Sears RG, Moffatt JM, Martin TJ, Cox TS, Bequette RK, Curran SP, Chung OK, Heer WF, Long JH, Witt MD (1997) Registration of ‘Jagger’ wheat. Crop Sci 37:1010
Singh RP (1992) Association between gene Lr34 for leaf rust resistance and leaf tip necrosis in wheat. Crop Sci 32:874–878
Singh RP, Huerta-Espino J, Rajaram S (2000) Achieving near-immunity to leaf rust and stripe rust in wheat by combining slow rusting resistance genes. Acta Phytopathol Entomol 35:133–139
Suenaga K, Singh RP, Huerta-Espino J, William HM (2003) Microsatellite markers for genes Lr34/Yr18 and other quantitative trait loci for leaf rust and stripe rust resistance in bread wheat. Phytopathology 93:881–890
Sun X, Bai G, Carver BF (2009) Molecular markers for wheat leaf rust resistance gene Lr41. Mol Breed 23:311–321
USDA-ARS (2009) National Genetic Resources Program, Germplasm Resources Information Network (GRIN) National Germplasm Resources Laboratory, Beltsville, M. Available at http://www.ars-grin.gov/cgi-bin/npgs/acc/display.pl?1553866 (verified 8 April 2009)
Vanegas CD, Garvin GDF, Kolmer JA (2008) Genetics of stem rust resistance in the spring wheat cultivar Thatcher and the enhancement of stem rust resistance by Lr34. Euphytica 159:391–401
Wicker T, Krattinger SG, Lagudah ES, Komatsuda T, Pourkheirandish M, Matsumoto T, Cloutier S, Reiser L, Kanamori H, Sato K, Perovic D, Stein N, Keller B (2009) Analysis of intraspecies diversity in wheat and barley genomes identifies breakpoints of ancient haplotypes and provides insight into the structure of diploid and hexaploid Triticeae gene pools. Plant Physiol 149:258–270
William HM, Hoisington D, Singh RP, Gonzalez-de-Leon D (1997) Detection of quantitative trait loci associated with leaf rust resistance in bread wheat. Genome 40:253–260
Wolfe MS (1984) Trying to understand and control powdery mildew. Plant Pathol 33:451–466
Acknowledgments
This study was supported by the National Research Initiative of the USDA-Cooperative State Research, Education, and Extension Service (CAP grant 2006-55606-16629), by the Oklahoma Center of Advanced Science and Technology (OCAST). This research project was partially funded by the Oklahoma Agricultural Experiment Station and the Oklahoma Wheat Research Foundation.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by J. Snape.
Rights and permissions
About this article
Cite this article
Cao, S., Carver, B.F., Zhu, X. et al. A single-nucleotide polymorphism that accounts for allelic variation in the Lr34 gene and leaf rust reaction in hard winter wheat. Theor Appl Genet 121, 385–392 (2010). https://doi.org/10.1007/s00122-010-1317-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00122-010-1317-6