Abstract
The rs3851179 which located at upstream of PICALM was reported to be associated with Alzheimer’s disease (AD); however, the relationship is still undefined. To gain a more precise understanding of the association, we conducted a meta-analysis: a comprehensive survey of 16 case-control studies that evaluated the role of rs3851179 gene variants in AD patients. The overall analysis revealed a significant association between the polymorphism and AD in the allelic, homozygote, heterozygote, dominant, and recessive models (p < 0.05). When stratified by ethnicity, a significant association was observed between AD development in Caucasian populations and the five-genetic models; Asian populations, however, featured a significant association in only the allelic, homozygote, and recessive models. We did not observe any influence of APOE ε4 carrier status on the incidence of AD and rs3851179 (p > 0.05). Our meta-analysis thus suggested that the PICALM rs3851179 polymorphism was associated with AD; the APOE ε4 status did not influence the relationship. Nevertheless, considering the limitations of our meta-analysis, further large-scale studies should be conducted to gain a more comprehensive understanding.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is a progressive neurodegenerative disease characterized by the decline of memory and other cognitive functions (Mawuenyega et al. 2010; Wang et al. 2017). As a prominent global health issue, AD has become a key epidemiological factor compromising the quality of life of the elderly: a total of 46.8 million people worldwide currently live with dementia and the number is projected to increase to 131.5 million by 2050 according to the “World Alzheimer Report 2015”. It was reported that nearly 5.3 million Americans have AD, of whom 5.1 million are over 65 years old (Alzheimer’s Association 2015). Possibly due to the complex roots of AD pathogenesis—genetic factors, lifestyle, and environmental conditions all contribute to its onset—available treatments slow disease progression only slightly (Tosto et al. 2016).
Researchers discovered that genetic factors had contributed to the development of AD (Bettens et al. 2013; Mengel-From et al. 2013). Recent genome-wide association studies (GWS) have also identified several putative candidate genes conferring risk for AD, such as phosphatidylinositol binding clatrin assembly protein (PICALM), clusterin (CLU), and ATP binding cassette subfamily A member 7 (ABCA7) (Zhu et al. 2017). Most of such risk genes are involved in neural apoptosis and the production, degradation, and clearance of Aβ. Among these genes, PICALM has been identified to play a crucial role in AD development (Thomas et al. 2016; Zhao et al. 2015).
PICALM is located on chromosome 11q14 and extends over 112 kb. It is involved in reversing the recruitment of clathrin and mediating endocytosis; it thus protects neurons against Aβ toxicity. A large-scale GWAS conducted by Harold et al. identified the rs3851179 (A > G) single nucleotide polymorphisms (SNPs) in PICALM; the study found that it was significantly associated with AD in Caucasian populations (Harold et al. 2009). A later study performed by Seshadri et al. reported similar results in Spanish populations (Seshadri et al. 2010). Liu et al. conducted two pooled meta-analyses and found the same significant association in Asian populations (Liu et al. 2013, 2017). These studies were, however, limited by their sample size and ethnic bias; the results could not define the relationship between rs3851179 and AD in either Caucasian or Asian populations.
Further confounding possible conclusions, results from recent investigations are inconsistent with those from the aforementioned studies: Shankarappa et al. and Liu et al. reported no correlation between rs3851179 susceptibility and AD in Indian and Chinese populations, respectively (Shankarappa et al. 2017; Li et al. 2011). The present study sought to further elucidate a possible correlation between rs3851179 polymorphism and AD; we performed a meta-analysis of case-control studies by pooling all eligible studies—including published theses—to explore the correlation.
Methods
Search strategy
This meta-analysis was performed according to the criteria for the Preferred Reporting Items for Systematic Reviews and Meta-analysis (PRISMA). Two investigators independently searched PubMed, Embase, Web of Knowledge, China National Knowledge Infrastructure (CNKI), and Wan Fang Data for studies published before 20 November 2017. MeSH and title/abstract were used to find eligible case-control studies according to the following format: (“Alzheimer’s disease”,“Alzheimer disease”, “AD” or “Dementia*”) AND (“phosphatidylinositol binding clatrin assembly protein”, “PICALM”, “rs3851179”) AND (“polymorphism”, “SNP”, “mutation”, “variant” or “genotype”).
Inclusion and exclusion criteria
Eligible reports met the following criteria: (1) the study evaluated the association between PICALM rs3851179 polymorphism and risk of AD, (2) the report focused on detailed genotype frequencies among human beings of late onset sporadic AD, and (3) the investigation was a case-control study. Accordingly, the exclusion criteria were as follows: (1) comment, review, and editorial articles; (2) studies without detailed genotype data; and (3) reports with overlapping data.
Data extraction
Two investigators independently extracted relevant information from all eligible articles by using a standardized form. If available, the following data were collected from each study: primary author, year of publication, country of origin, ethnicity of subjects, source of controls, frequency of genotypes among AD patients and controls, and evidence of Hardy-Weinberg equilibrium among controls. If multiple publications conducted a study on the same population, the study with the most thorough analysis was selected. Any discrepancy was resolved through discussion among the two investigators until a consensus was reached. If dissent remained, a third investigator resolved the dispute.
Quality assessment
The quality of the literature was evaluated independently by two investigators using quality scoring criteria modified from a previous study (Zhang et al. 2017). Quality scores ranged from 0 (worst) to 10 (best). Studies scoring higher than 5 were classified as having adequate quality.
Statistics analysis
All statistical analyses were performed by using the STATA version 12.0 (STATA Corporation, College Station, TX, USA). Hardy-Weinberg equilibrium (HWE) was assessed among controls using a χ2 test. A P value <0.05 was considered significant. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated to assess the strength of associations between PICALM polymorphism and AD susceptibility. Pooled ORs were obtained from a combination of single studies by homozygote comparison (AA vs.GG), heterozygote comparison (AG vs. GG), dominant model (AG + GG vs. AA), recessive model (GG vs. AG + AA), and allelic model (A vs. G). Heterogeneity was evaluated by Q statistic and I2 statistic. The fixed effect model (Mantel–Haenszel method) was used to calculate the pooled ORs (Q-test >0.10 or I2 < 50%); for all other analyses, the random-effect model (DerSimonian–Laird method) was used. The significance of the pooled ORs was assessed by a Z-test, where P < 0.05 indicated statistical significance. Subgroup analyses were conducted based on ethnicity and source of control. Sensitivity analyses were performed to display possible variability. Begg’s and Egger’s linear regression tests were applied to assess potential publication bias. We further evaluated the number of missing studies in the meta-analysis by applying the trim and fill method; we recalculated the pooled risks with the addition of the missing studies.
Results
Characteristics of the studies
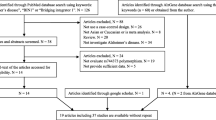
A total of 189 relevant studies were identified from an initial search through databases. Thirteen duplicated publications were removed in the preliminary screening. After screening titles and abstracts, 139 irrelevant articles were further excluded. The remaining articles were subjected to full-text review by two independent investigators. Finally, 16 eligible articles were included in this meta-analysis. The flow diagram of the search process is illustrated in Fig. 1. The studies were published between 2009 and 2017. They encompassed a total of 6972 AD patients and 10,199 controls; the former was composed of 3227 Asians and 3745 Caucasians. The characteristics of the enrolled studies are summarized in Table 1. The genotype and allele distribution among AD cases and controls are summarized in Table 2. The distributions of the genotype frequencies of the controls were all consistent with the Hardy-Weinberg equilibrium (HWE) (all P > 0.05)).
Meta-analysis results
Heterogeneity was identified by Q-test and I-squared statistic in five genetic models. As is shown in Table 3, serious heterogeneity was found only in the heterozygote and recessive models (I2 = 59.8%, I2 = 57.1%); the random-effect model was therefore employed in the analysis. The fixed model was used in the homozygote, dominant and allelic models (I2 = 22.3%, I2 = 18.6%, I2 = 43.3%). The results revealed that there was a significant association between the PICALM rs3851179 polymorphism and AD in all five genetic models. The pooled ORs revealed that the allelic, homozygote, and heterozygote models showed a decreased risk of AD (OR = 0.894, 95% CI: 0.865–0.923; OR = 0.773, 95% CI: 0.720–0.829; OR = 0.878, 95% CI: 0.838–0.920; OR = 1.213, 95% CI: 1.135–1.296; and OR = 1.162, 95% CI: 1.074–1.258, respectively). The dominant (OR = 1.213, 95% CI: 1.135–1.296) and recessive models (OR = 1.162, 95% CI: 1.074–1.258) showed an increased risk of AD. Subgroup analysis based on ethnic descent showed that rs3851179 polymorphism was strongly associated with AD among Caucasians in the five genetic models, while the association in Asians populations was only significant in the allelic (OR = 0.918,95% CI: 0.860–0.981), homozygote (OR = 0.822, 95% CI: 0.714–0.947), and dominant models (OR = 1.172, 95% CI: 1.030–1.333) (Table 3 and Fig. 2 ). We subsequently performed a comparison of the risk of PICALM rs3851179 polymorphism for AD development between the APOE є4+ group and the APOE є4- group to explore the potential effect of APOE є4 status on AD development. However, we did not observe any correlation between the polymorphism and AD in either the APOE є4+ group or the APOE є4- group in any of the five genetic models (Table 4).
Publication bias
No significant publication bias was found in the Begg’s test or Egger’s test (P > 0.05)(Fig. 3 ). The trim and fill method was also employed to further determine a possible publication bias. Negligible changes in OR and 95% CI were observed between the different (Table 5).
Sensitivity analysis
We performed a sensitivity analysis to assess the influence of each individual study on the pooled OR by sequentially removing each eligible study. The results indicated that only the removal of the study by Lambert et al. led to the loss of a significant association between PICALM rs3851179 polymorphism and the risk of AD in the pooled population. No other single study influenced the quality of the pooled ORs in the sensitivity analyses (Fig. 4).
Disscussion
Ubiquitously expressed in the central nervous system, especially at presynaptic and postsynaptic structures, PICALM has been shown to be associated with the morbidity of AD. Bushlin et al. observed that the reduction in PICALM levels in cultured embryonic hippocampal neurons resulted in dendritic dystrophy, reduced endocytosis, and disrupted secretory transport (Bushlin et al. 2008), and Kanatsu et al. reported that the reduction in PICALM levels decreased Aβ deposition, as well as brain levels of insoluble Aβ1–42 in vivo (Kanatsu et al. 2016).
Since Harold et al. first reported on the possible association of the rs3851179 polymorphism with PICALM, several investigations have published contradictory results. We found that rs3851179 polymorphism was associated with AD in the domain and recessive models in the overall meta-analysis; in the allelic, homozygote, and heterozygote models, however, the polymorphism reflected a reduced risk of AD. The subgroup analysis of the Caucasian population showed a trend similar to the overall analysis in all models, while no association was found in the heterozygote and recessive models in the Asian population; the former finding agreed with those of Harold et al., while the latter was in accordance with those of Wang et al. (2016). Our results diverged from those of Liu et al., however, which showed that the polymorphism was associated with AD in the recessive model (Liu et al. 2013); the difference may be caused by the numbers of enrolled studies.
The present study sought to further contribute to the literature by examining the difference in the APOE є4 status between AD patients and healthy controls. As one of the risk factors for AD, APOE є4 can form deposits in neuritic plaques and neurofibrillary tangles; it thus augments the effect of other factors promoting disease progression (Michaelson 2014). It was reported that APOE є4 may account for approximately 50% or more of the late onset Alzheimer’s disease cases in the USA. Nevertheless, we did not find any significant differences between APOE є4 carriers and non-carriers. Our results were in accordance with those reported by Tomoyuki Ohara et al., which showed no significant association between PICALM and APOE ε4 carrier status (p = 0.68) (Ohara et al. 2012).
Compared to prior studies, our meta-analysis made use of a more comprehensive collection of references; we enrolled investigations from not only the Alzgene database, but also recently published studies and theses. The present study thus performed a thorough analysis of the relationship between the rs3851179 polymorphism and AD in the Caucasian and Asian population; our findings provide new support for the GWAS results of Harold et al. We also add to the literature by having used the obtained data to evaluate of the association of APOE ε4 with AD and the rs3851179 polymorphism. Upon selecting eligible studies, a methodological quality assessment was conducted; all studies were of acceptable quality.
Due to potential limitations of the present meta-analysis, however, results from this study should be interpreted with caution. Specifically, there were no unified detection methods: serious heterogeneity was observed even in our subgroup analyses, possibly accounting for the negative results. Further, publication bias – though none was detected – or other confounding factors may have further distorted the meta-analysis. Our investigation into the association between PICALM rs3851179 polymorphisms and APOE є4 status featured another limitation: insufficient data precluded a comprehensive meta-analysis. Small sample sizes in each study may underlie the failure to achieve statistical significance.
Conclusion
In conclusion, the results of this meta-analysis suggest that the PICALM rs3851179 polymorphism is associated with the susceptibility to AD among Asians and Caucasians. However, as confounding factors may exist, our results were not consistent with several prior case-control studies. Future research should analyze larger populations with different ethnicities and prioritize data including APOE є4 status in order to explore the broader role that polymorphisms play in the pathogenesis of AD.
References
Alzheimer’s Association (2015) 2015 Alzheimer's disease facts and figures. Alzheimers Dement 11:332–384
Bettens K, Sleegers K, Van Broeckhoven C (2013) Genetic insights in Alzheimer's disease. Lancet Neurol 12:92–104. https://doi.org/10.1016/S1474-4422(12)70259-4
Bushlin I, Petralia RS, Wu F, Harel A, Mughal MR, Mattson MP et al (2008) Clathrin assembly protein AP180 and CALM differentially control axogenesis and dendrite outgrowth in embryonic hippocampal neurons. J Neurosci 28:10257–10271. https://doi.org/10.1523/JNEUROSCI.2471-08.2008
Carrasquillo MM, Belbin O, Hunter TA, Ma L, Bisceglio GD, Zou F et al (2010) Replication of CLU, CR1, and PICALM associations with alzheimer disease. Arch Neurol 67:961–964. https://doi.org/10.1001/archneurol.2010.147
Chen LH, Kao PY, Fan YH, Ho DT, Chan CS, Yik PY et al (2012) Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer's disease in a southern Chinese population. Neurobiol Aging 33:210–211. https://doi.org/10.1016/j.neurobiolaging.2011.09.016
Ding D (2012) Population-based prevalence survey and genetic epidemiology of cognitive impairment among elderly. Fudan University
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML et al (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 41:1088–1093. https://doi.org/10.1038/ng.440
Hui J (2014) Association analysis of eight gen variations with alzheimer's disease susceptibility in Northen Chinese population. Ningxia Medical University
Kanatsu K, Hori Y, Takatori S, Watanabe T, Iwatsubo T, Tomita T (2016) Partial loss of CALM function reduces Abeta42 production and amyloid deposition in vivo. Hum Mol Genet 25:3988–3997. https://doi.org/10.1093/hmg/ddw239
Klimkowicz-Mrowiec A, Sado M, Dziubek A, Dziedzic T, Pera J, Szczudlik A et al (2013) Lack of association of CR1, PICALM and CLU gene polymorphisms with Alzheimer disease in a Polish population. Neurol Neurochir Pol 47:157–160
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M et al (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 41:1094–1099. https://doi.org/10.1038/ng.439
Li HL, Shi SS, Guo QH, Ni W, Dong Y, Liu Y et al (2011) PICALM and CR1 variants are not associated with sporadic Alzheimer's disease in Chinese patients. J Alzheimers Dis 25:111–117. https://doi.org/10.3233/JAD-2011-101917
Liu XY (2014) The Association analysis of late-onset Alzheimer's disease and susceptibility genes in Chinese Han population. Central South University
Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y et al (2013) PICALM gene rs3851179 polymorphism contributes to Alzheimer's disease in an Asian population. NeuroMolecular Med 15:384–388. https://doi.org/10.1007/s12017-013-8225-2
Liu G, Xu Y, Jiang Y, Zhang L, Feng R, Jiang Q (2017) PICALM rs3851179 variant confers susceptibility to Alzheimer's disease in Chinese population. Mol Neurobiol 54:3131–3136. https://doi.org/10.1007/s12035-016-9886-2
Mawuenyega KG, Sigurdson W, Ovod V, Munsell L, Kasten T, Morris JC et al (2010) Decreased clearance of CNS beta-amyloid in Alzheimer's disease. Science 330:1774. https://doi.org/10.1126/science.1197623
Mengel-From J, Thinggaard M, Lindahl-Jacobsen R, McGue M, Christensen K, Christiansen L (2013) CLU genetic variants and cognitive decline among elderly and oldest old. PLoS One 8:e79105. https://doi.org/10.1371/journal.pone.0079105
Michaelson DM (2014) APOE epsilon4: the most prevalent yet understudied risk factor for Alzheimer's disease. Alzheimers Dement 10:861–868. https://doi.org/10.1016/j.jalz.2014.06.015
Ohara T, Ninomiya T, Hirakawa Y, Ashikawa K, Monji A, Kiyohara Y et al (2012) Association study of susceptibility genes for late-onset Alzheimer's disease in the Japanese population. Psychiatr Genet 22:290–293. https://doi.org/10.1097/YPG.0b013e3283586215
Piaceri I, Bagnoli S, Lucenteforte E, Mancuso M, Tedde A, Siciliano G et al (2011) Implication of a genetic variant at PICALM in Alzheimer's disease patients and centenarians. J Alzheimers Dis 24:409–413. https://doi.org/10.3233/JAD-2011-101791
Santos-Reboucas CB, Goncalves AP, Dos SJ, Abdala BB, Motta LB, Laks J et al (2017) rs3851179 polymorphism at 5′ to the PICALM gene is associated with Alzheimer and Parkinson diseases in Brazilian population. NeuroMolecular Med. https://doi.org/10.1007/s12017-017-8444-z
Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M et al (2010) Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA 303:1832–1840. https://doi.org/10.1001/jama.2010.574
Shankarappa BM, Kota LN, Purushottam M, Nagpal K, Mukherjee O, Viswanath B et al (2017) Effect of CLU and PICALM polymorphisms on AD risk: a study from South India. Asian J Psychiatr 27:7–11. https://doi.org/10.1016/j.ajp.2016.12.017
Thomas RS, Henson A, Gerrish A, Jones L, Williams J, Kidd EJ (2016) Decreasing the expression of PICALM reduces endocytosis and the activity of beta-secretase: implications for Alzheimer's disease. BMC Neurosci 17:50. https://doi.org/10.1186/s12868-016-0288-1
Tosto G, Bird TD, Bennett DA, Boeve BF, Brickman AM, Cruchaga C et al (2016) The role of cardiovascular risk factors and stroke in familial Alzheimer disease. JAMA Neurol 73:1231–1237. https://doi.org/10.1001/jamaneurol.2016.2539
Wang HZ, Bi R, Hu QX, Xiang Q, Zhang C, Zhang DF et al (2016) Validating GWAS-identified risk loci for Alzheimer's disease in Han Chinese populations. Mol Neurobiol 53:379–390. https://doi.org/10.1007/s12035-014-9015-z
Wang Y, Liu S, Wang J, Zhang J, Hua Y, Li H et al (2017) Association between LRP1 C766T polymorphism and Alzheimer's disease susceptibility: a meta-analysis. Sci Rep 7:8435. https://doi.org/10.1038/s41598-017-08335-w
Yu JT, Song JH, Ma T, Zhang W, Yu NN, Xuan SY et al (2011) Genetic association of PICALM polymorphisms with Alzheimer's disease in Han Chinese. J Neurol Sci 300:78–80. https://doi.org/10.1016/j.jns.2010.09.027
Zhang S, Wang XB, Han YD, Wang C, Zhou Y, Zheng F (2017) Certain polymorphisms in SP110 gene confer susceptibility to tuberculosis: a comprehensive review and updated meta-analysis. Yonsei Med J 58:165–173. https://doi.org/10.3349/ymj.2017.58.1.165
Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K et al (2015) Central role for PICALM in amyloid-beta blood-brain barrier transcytosis and clearance. Nat Neurosci 18:978–987. https://doi.org/10.1038/nn.4025
Zhu B, Wang RM, Wang JT, Chen RL, Zheng YF, Zhang L et al (2017) Correlation of rs9331888 polymorphism with Alzheimer's disease among Caucasian and Chinese populations: a meta-analysis and systematic review. Metab Brain Dis 32:981–989. https://doi.org/10.1007/s11011-017-9957-8
Acknowledgments
This work was supported by Beijing Municipal Administration of Hospitals’ Youth Programme (grant number: QML20170703), China Postdoctoral Science Foundation (No.2017M620700), Beijing Natural Science Foundation (grant number:7164256), The National Key Research and Development Program of China (grant number: 2016YFC1306300) and The Key Project of Natural Science Foundation of Beijing, China (grant number:4161004).
Author information
Authors and Affiliations
Contributions
BZ. and ZGZ designed this study and had full access to all of the data in the study; LXL and SYA acquisition of data, LZ, YT and SSG analysis and interpretation of data. WZ Critical revision of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Rights and permissions
About this article
Cite this article
Zhu, B., Li, LX., Zhang, L. et al. Correlation of PICALM polymorphism rs3851179 with Alzheimer’s disease among Caucasian and Chinese populations: a meta-analysis and systematic review. Metab Brain Dis 33, 1849–1857 (2018). https://doi.org/10.1007/s11011-018-0291-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-018-0291-6