Abstract
The association between PICALM rs3851179 variant and Alzheimer’s disease (AD) has been well established by previous genome-wide association studies (GWAS) and candidate gene studies in European population. Recent studies investigated the association between PICALM rs3851179 and AD susceptibility in Chinese population. However, these studies reported consistent and inconsistent results. Here, we selected 9435 samples including 3704 AD cases and 5731 controls from previous studies and evaluated this association using a meta-analysis method for additive model. We did not observe significant genetic heterogeneity in Chinese population. Our results indicate significant association between PICALM rs3851179 and AD in Chinese population. The sensitivity analysis indicates that the association between rs3851179 and AD did not vary substantially. The regression analysis suggests no significant publication bias. In summary, this updated meta-analysis highlights the involvement of PICALM rs3851179 variant in Alzheimer’s disease susceptibility in Chinese population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease (AD) is the most common neurodegenerative disease in the elderly population [1, 2]. It is reported that genome-wide association studies (GWAS) have been widely used to investigate the pathogenesis of AD and have yielded important new insights into the genetic mechanisms of AD [1, 3–5]. In 2009, the PICALM rs3851179 variant was originally identified to be significantly associated with AD in European population (P = 1.30E-09) [6]. Meanwhile, the potential association of rs3851179 with AD was also widely investigated in Chinese populations.
In 2011–2012, Yu, Li and Chen et al. analyzed 609, 1065, and 812 Chinese samples and reported no significant association between rs3851179 and AD [7–9]. In 2013, we conducted a pooled analysis of these three studies and still did not identify significant association using both genotype and allele tests [10].
In 2014, Wang et al. evaluated this association using 1509 individuals comprising two independent Chinese case-control cohorts including the cohort from Southwest China (333 AD patients and 334 cognitively healthy controls) and the cohort from East China (416 AD patients and 426 cognitively healthy individuals) [11]. In the Southwest China population, they identified that rs3851179 was significantly associated with AD [11]. They further identified significant association between rs3851179 and AD by a meta-analysis of Asian populations (Chinese and Japanese populations) [11].
In 2015, Xiao et al. selected 459 Chinese sporadic AD patients and 751 Chinese cognitively normal controls [12]. They did not report any significant association of rs3851179 with AD (P = 0.194 and 0.177) [12]. In 2015, Jiao et al. screened 58 SNPs using 229 AD cases and 318 controls from mainland China [13]. Their results show no association between rs3851179 and AD with P = 0.413 [13].
In summary, previous and recent studies reported consistent and inconsistent results about the association between rs3851179 and AD in Chinese populations. It is still unclear whether rs3851179 contributes to AD susceptibility in Chinese population. Here, we performed an updated meta-analysis to evaluate the association between rs3851179 and AD using 9435 samples from previous studies.
Materials and Methods
Comprehensive Literature Search
We conducted a comprehensive literature search in four databases including the PubMed (http://www.ncbi.nlm.nih.gov/pubmed), AlzGene (http://www.alzgene.org/), China National Knowledge Infrastructure (CNKI, http://www.cnki.net/), and Google Scholar (https://scholar.google.com/) databases. We selected all possible studies with key words “Alzheimer’s disease,” “PICALM,” and “China or Chinese.” The literature search was updated on January 2, 2016. More detailed information about the inclusion and exclusion criteria is described in the following inclusion and exclusion criteria section.
Inclusion and Exclusion Criteria
The inclusion criteria include the following: (1) the selected studies must use a case-control design, (2) the selected studies must evaluate the association between rs3851179 and AD, (3) the selected studies must provide the number of rs3851179 genotypes or sufficient data to calculate the number of rs3851179 genotypes, or (4) the selected studies must provide an odds ratio (OR) with a 95 % confidence interval (CI) or sufficient data to calculate the OR and a 95 % CI. Any study that does not meet the inclusion criteria above is excluded.
Data Extraction
We extracted (1) the name of the first author, (2) the year of publication, (3) the population, (4) the numbers of AD cases and controls, (5) the genotype numbers of rs3851179 in cases and controls, and (6) the OR with 95 % CI or, if not provided, calculated the OR and 95 % CI.
Genetic Model
We used the additive genetic model to evaluate the association between rs3851179 and AD: the A allele versus the G allele.
Hardy-Weinberg Equilibrium Test
We evaluate the Hardy-Weinberg equilibrium (HWE) of rs3851179 in controls for each study using a chi-square test, which was conducted using R (http://www.r-project.org/). The significance threshold is 1.00E-03. A HWE test P value less than 1.00E-03 indicates significant deviation from the HWE.
Heterogeneity Test
The heterogeneity test was conducted using both Cochran’s Q statistic and \( {I}^2=\raisebox{1ex}{$\left(Q-\left(k-1\right)\right)$}\!\left/ \!\raisebox{-1ex}{$Q$}\right.\times 100\% \) [14]. Q statistic approximately follows a χ 2 distribution with k-1 degrees of freedom (k stands for the number of studies for analysis). The P value from Cochran’s Q statistic <0.1 and I 2 > 50 % indicate significant heterogeneity [14, 15].
Meta-analysis
If there was no significant heterogeneity, the pooled OR was calculated by the fixed effect model (Mantel-Haenszel). Otherwise, it is calculated by random-effects (DerSimonian-Laird). The significance of OR was determined using the Z test. All statistical methods in meta-analysis were performed using the RevMan (v.5.1) and R (http://www.r-project.org/).
Sensitivity and Publication Bias Analyses
The relative influence of each study on the pooled OR and significance were evaluated by omitting each study at a time. We used the funnel plot to evaluate the potential publication bias [16]. A regression based approach proposed by Egger is used to test for publication bias to provide statistical evidence [17]. The significance level was 0.01. All statistical tests were computed using R (http://www.r-project.org/).
Results
Comprehensive Literature Search
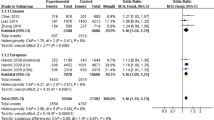
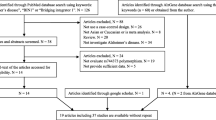
We got 14, 2, 2, and 3 articles from PubMed, AlzGene, CNKI, and Google Scholar databases, respectively. In the end, we excluded the overlapping studies and finally selected ten articles including 11 studies for the following meta-analysis. More detailed information is described in Fig. 1. The main characteristics of the included studies are described in Table 1.
HWE, Heterogeneity, and Meta-analysis
With the significance level P < 1.00E-03, we do not identify significant deviation from the HWE (a HWE P value less than 1.00E-03). The heterogeneity test showed no significant genetic heterogeneity of rs3851179 polymorphism in Chinese population with P = 0.55 and I 2 = 0 %. We calculated the overall OR by the fixed effect model. Our results showed significant association between rs3851179 and AD with P = 5.20E-03, OR = 0.92, and 95 % CI 0.86–0.97. Detailed results are described in Fig. 2.
Sensitivity Analysis and Publication Bias Analysis
Using sensitivity analysis, we identified that the association between rs3851179 and AD did not vary substantially by excluding any one study (Table 2).
The funnel plot is symmetrical inverted funnel (Fig. 3). The regression analysis provides statistical evidence, suggesting no significant publication bias for additive model (regression P = 0.94).
The funnel plots for publication bias analysis of the selected studies investigating the association between rs3851179 and AD using the additive genetic model. The x-axis stands for the ORs and the y-axis is the standard error for each of the 11 studies. A linear regression based approach proposed by Egger et al. is used to evaluate the asymmetry of the funnel plot
Discussion
The association between PICALM rs3851179 and AD has been well established by previous GWAS and candidate gene studies in European population. Meanwhile, the potential role of PICALM in aging and AD was widely investigated. It is reported that PICALM is associated with an earlier age at midpoint of cognitive decline [18]. It is well known that the β-amyloid (Aβ) peptides are involved in the pathogenesis of AD [19]. PICALM could regulate amyloid precursor protein internalization and subsequent Aβ generation [20]. In the hippocampus, it is reported that PICALM knockdown can decrease soluble and insoluble Aβ levels and amyloid plaque load and PICALM overexpression can increase Aβ levels and amyloid plaque load [20].
Evidence shows that PICALM rs3851179 polymorphism plays a significantly protected role against rapid AD progression [21]. Parikh et al. evaluated the association between rs3851179 and PICALM expression in three cell lines including microvessels, neurons, and astrocytes [22]. The results showed that PICALM was expressed robustly in microvessels and moderately in other cell types [22]. The rs3851179 protective allele (A) is associated with modestly increased PICALM expression [22].
Recently, Zhao et al. identified reduced expression of PICALM in AD and found that PICALM deficiency diminished Aβ clearance across the murine blood-brain barrier (BBB) and accelerated Aβ pathology [23]. By PICALM re-expression, this situation will be further reversed [23]. Using inducible pluripotent stem cell-derived human endothelial cells, Zhao et al. further showed that rs3851179 protective allele (A) regulated increased PICALM expression and enhanced Aβ clearance [23]. They conclude that PICALM regulates Aβ BBB transcytosis and clearance, which has implications for Aβ brain homeostasis and clearance therapy [23]. Moreau et al. show that PICALM modulates autophagy activity and tau accumulation [24].
Evidence also shows that in case-control association studies, a deviation from HWE in cases may indicate a genetic association. Here, HWE test was only used in controls but not AD cases [25, 26]. Until recently, there are several studies evaluating the potential association between rs3851179 and AD in Asian or Chinese populations, such as three studies from Yu, Li, and Chen et al. [7–9], our previous studies using the pooled samples from these three studies above [10, 14], Wang et al. [11], Xiao et al. [12], and Jiao et al. [13]. There is a major difference between our study and previous studies. This difference is that all above studies evaluate the association between rs3851179 and AD. However, all these studies reported consistent and inconsistent association results.
Here, based on the inconsistent results reported by recent studies in Chinese population, we performed an updated meta-analysis using 7678 individuals from Chinese population. Our results showed no significant genetic heterogeneity of rs3851179 polymorphism in Chinese population. Meta-analysis using the fixed effect model further showed significant association between rs3851179 and AD in Chinese population. The sensitivity analysis indicates that the association between rs3851179 and AD did not vary substantially. The regression analysis suggests no significant publication bias using the additive model. In summary, this updated meta-analysis highlights the involvement of PICALM rs3851179 variant in Alzheimer’s disease susceptibility in Chinese population.
References
Bao X, Liu G, Jiang Y, Jiang Q, Liao M, Feng R, Zhang L, Ma G, Zhang S, Chen Z, Zhao B, Wang R, Li K (2015) Cell adhesion molecule pathway genes are regulated by cis-regulatory SNPs and show significantly altered expression in Alzheimer’s disease brains. Neurobiol Aging 36:2904 e2901–2907
Liu G, Bao X, Jiang Y, Liao M, Jiang Q, Feng R, Zhang L, Ma G, Chen Z, Wang G, Wang R, Zhao B, Li K (2015) Identifying the association between Alzheimer’s disease and Parkinson’s disease using genome-wide association studies and protein-protein interaction network. Mol Neurobiol 52:1629–1636
Liu G, Yao L, Liu J, Jiang Y, Ma G, Chen Z, Zhao B, Li K (2014) Cardiovascular disease contributes to Alzheimer’s disease: evidence from large-scale genome-wide association studies. Neurobiol Aging 35:786–792
Xiang Z, Xu M, Liao M, Jiang Y, Jiang Q, Feng R, Zhang L, Ma G, Wang G, Chen Z, Zhao B, Sun T, Li K, Liu G (2015) Integrating genome-wide association study and brain expression data highlights cell adhesion molecules and purine metabolism in Alzheimer’s disease. Mol Neurobiol 52:514–521
Liu G, Jiang Y, Wang P, Feng R, Jiang N, Chen X, Song H, Chen Z (2012) Cell adhesion molecules contribute to Alzheimer’s disease: multiple pathway analyses of two genome-wide association studies. J Neurochem 120:190–198
Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A, Jones N, Thomas C, Stretton A, Morgan AR, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Morgan K, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Love S, Kehoe PG, Hardy J, Mead S, Fox N, Rossor M, Collinge J, Maier W, Jessen F, Schurmann B, Heun R, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Hull M, Rujescu D, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Carrasquillo MM, Pankratz VS, Younkin SG, Holmans PA, O’Donovan M, Owen MJ, Williams J (2009) Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet 41:1088–1093
Yu JT, Song JH, Ma T, Zhang W, Yu NN, Xuan SY, Tan L (2011) Genetic association of PICALM polymorphisms with Alzheimer’s disease in Han Chinese. J Neurol Sci 300:78–80
Li HL, Shi SS, Guo QH, Ni W, Dong Y, Liu Y, Sun YM, Bei W, Lu SJ, Hong Z, Wu ZY (2011) PICALM and cr1 variants are not associated with sporadic Alzheimer’s disease in chinese patients. J Alzheimers Dis 25:111–117
Chen LH, Kao PY, Fan YH, Ho DT, Chan CS, Yik PY, Ha JC, Chu LW, Song YQ (2012) Polymorphisms of cr1, CLU and PICALM confer susceptibility of Alzheimer’s disease in a southern chinese population. Neurobiol Aging 33:210 e211–217
Liu G, Zhang L, Feng R, Liao M, Jiang Y, Chen Z, Zhao B, Li K (2013) Lack of association between PICALM rs3851179 polymorphism and Alzheimer’s disease in chinese population and apoeepsilon4-negative subgroup. Neurobiol Aging 34:1310 e1319–1310 e1310
Wang HZ, Bi R, Hu QX, Xiang Q, Zhang C, Zhang DF, Zhang W, Ma X, Guo W, Deng W, Zhao L, Ni P, Li M, Fang Y, Li T, Yao YG (2014) Validating gwas-identified risk loci for Alzheimer’s disease in han chinese populations. Mol Neurobiol
Xiao Q, Liu ZJ, Tao S, Sun YM, Jiang D, Li HL, Chen H, Liu X, Lapin B, Wang CH, Zheng SL, Xu J, Wu ZY (2015) Risk prediction for sporadic Alzheimer’s disease using genetic risk score in the han chinese population. Oncotarget 6:36955–36964
Jiao B, Liu X, Zhou L, Wang MH, Zhou Y, Xiao T, Zhang W, Sun R, Waye MM, Tang B, Shen L (2015) Polygenic analysis of late-onset Alzheimer’s disease from mainland china. PLoS One 10:e0144898
Liu G, Zhang S, Cai Z, Ma G, Zhang L, Jiang Y, Feng R, Liao M, Chen Z, Zhao B, Li K (2013) PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromolecular Med 15:384–388
Higgins JP, Thompson SG (2002) Quantifying heterogeneity in a meta-analysis. Stat Med 21:1539–1558
Jiang Y, Zhang R, Zheng J, Liu P, Tang G, Lv H, Zhang L, Shang Z, Zhan Y, Lv W, Shi M (2012) Meta-analysis of 125 rheumatoid arthritis-related single nucleotide polymorphisms studied in the past two decades. PLoS One 7:e51571
Song F, Khan KS, Dinnes J, Sutton AJ (2002) Asymmetric funnel plots and publication bias in meta-analyses of diagnostic accuracy. Int J Epidemiol 31:88–95
Sweet RA, Seltman H, Emanuel JE, Lopez OL, Becker JT, Bis JC, Weamer EA, DeMichele-Sweet MA, Kuller LH (2012) Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the cardiovascular health study. Am J Psychiatry 169:954–962
Corbett GT, Gonzalez FJ, Pahan K (2015) Activation of peroxisome proliferator-activated receptor alpha stimulates adam10-mediated proteolysis of APP. Proc Natl Acad Sci USA
Xiao Q, Gil SC, Yan P, Wang Y, Han S, Gonzales E, Perez R, Cirrito JR, Lee JM (2012) Role of phosphatidylinositol clathrin assembly lymphoid-myeloid leukemia (PICALM) in intracellular amyloid precursor protein (APP) processing and amyloid plaque pathogenesis. J Biol Chem 287:21279–21289
Ruiz A, Hernandez I, Ronsende-Roca M, Gonzalez-Perez A, Rodriguez-Noriega E, Ramirez-Lorca R, Mauleon A, Moreno-Rey C, Boswell L, Tune L, Valero S, Alegret M, Gayan J, Becker JT, Real LM, Tarraga L, Ballard C, Terrin M, Sherman S, Payami H, Lopez OL, Mintzer JE, Boada M (2013) Exploratory analysis of seven Alzheimer’s disease genes: disease progression. Neurobiol Aging 34:1310 e1311–1317
Parikh I, Fardo DW, Estus S (2014) Genetics of PICALM expression and Alzheimer’s disease. PLoS One 9:e91242
Zhao Z, Sagare AP, Ma Q, Halliday MR, Kong P, Kisler K, Winkler EA, Ramanathan A, Kanekiyo T, Bu G, Owens NC, Rege SV, Si G, Ahuja A, Zhu D, Miller CA, Schneider JA, Maeda M, Maeda T, Sugawara T, Ichida JK, Zlokovic BV (2015) Central role for PICALM in amyloid-beta blood-brain barrier transcytosis and clearance. Nat Neurosci 18:978–987
Moreau K, Fleming A, Imarisio S, Lopez Ramirez A, Mercer JL, Jimenez-Sanchez M, Bento CF, Puri C, Zavodszky E, Siddiqi F, Lavau CP, Betton M, O’Kane CJ, Wechsler DS, Rubinsztein DC (2014) PICALM modulates autophagy activity and tau accumulation. Nat Commun 5:4998
Wang MJ, Liu RP, Kong ZJ (2016) Re: PICALM gene rs3851179 polymorphism contributes to Alzheimer’s disease in an Asian population. Neuromol Med
Xu Y, Jiang Q, Liu G (2016) PICALM rs3851179 variant and Alzheimer’s disease in Asian population. Neuromol Med
Ding D (2012) Population-based prevalcence survey and genetic epidemiology of cognitive impairment among elderly. China National Knowledge Infrastructure http://cdmd.cnki.com.cn/Article/CDMD-10246-1013102653.htm
Hui J (2014) Association analysis of eight gene variations with Alzheimer’s disease susceptibility in Northen Chinese populations. China National Knowledge Infrastructure http://cdmd.cnki.com.cn/Article/CDMD-10752-1015087724.htm
Liao YC, Lee WJ, Hwang JP, Wang YF, Tsai CF, Wang PN, Wang SJ, Fuh JL (2014) ABCA7 gene and the risk of Alzheimer’s disease in Han Chinese in Taiwan. Neurobiol Aging 35:2423 e2427–2423 e2413
Liu X (2014) Association analysis of late-onset Alzheimer’s disease and susceptibility genes in Chinese Han populations. China National Knowledge Infrastructure http://cdmd.cnki.com.cn/Article/CDMD-10533-1014407146.htm
Acknowledgments
This work was supported by funding from the National Nature Science Foundation of China (Grant No. 81300945 and 61571152). This work was partially supported by the National High-Tech Research and Development Program (863) of China (Grant No. 2012AA02A601, 2012AA02A602, 2012AA020404, 2012AA020409, 2012AA02A604, 2014AA021505, 2015AA020101, and 2015AA020108) and the National Science and Technology Major Project (Grant No. 2013ZX03005012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Additional information
Guiyou Liu and Yining Xu contributed equally to this work.
Rights and permissions
About this article
Cite this article
Liu, G., Xu, Y., Jiang, Y. et al. PICALM rs3851179 Variant Confers Susceptibility to Alzheimer’s Disease in Chinese Population. Mol Neurobiol 54, 3131–3136 (2017). https://doi.org/10.1007/s12035-016-9886-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-016-9886-2