Abstract
Clusterin polymorphism (rs9331888) was reported to be associated with the susceptibility to alzheimer’s disease (AD). Nevertheless, the results were inconclusive. To derive a more precise estimation of this association, this meta-analysis was conducted. We’ve conducted a comprehensive search of PubMed, Embase, CNKI and AlzGene database for case-control studies published throughout October, 2016 that evaluated the role of rs9331888 gene variants in AD patients. Odds ratios (ORs) and their 95% confidence intervals (CIs) were calculated to assess the strength of associations between the rs9331888/C > G polymorphism and AD disease. A total of 9 studies were enrolled in the Meta Analysis. The overall analysis revealed a significant association between the rs9331888/C > G polymorphism and AD disease in the recessive model (GG vs. GC + CC: OR = 1.11, 95% CI: 1.05–1.18; P < 0.01). Sub-group analysis revealed that the Caucasian populations which with recessive model (GG vs. GC + CC: OR = 1.12, 95% CI: 1.06–1.2; P < 0.01) were dramatically related to AD, while no significant association was found in the Chinese populations among the five genetic models. Our meta-analysis demonstrated that the rs9331888/C > G polymorphism in the clusterin gene might contribute to AD susceptibility especially in Caucasian populations. Whereas the relationship of the polymorphism to the disease in Chinese populations was still in controversial. Additional well-designed studies, with larger sample sizes, are required to further elucidate this association.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Alzheimer’s disease’s (AD) is a chronic, multifactorial and personality changing of neurodegenerative disorder. As one of the most common types of neurodegenerative disease, people suffered from AD manifest as progressive and irreversible memory loss and cognitive decline (Scheltens et al. 2016). Approximately 1% of people who are over 65 years old are under threat of AD, while the number increases to 25%–35% among people over 85 years old (Lu et al. 2014). It is estimated that that there are now 46.8 million people living with dementia worldwide, with numbers projected to nearly double every 20 years, increasing to 74.7 million by 2030 and 131.5 million by 2050(World Alzheimer Report 2015:The Global Impact of Dementia). In American, an estimated 5.3 million Americans have AD, and of them 5.1 million are age 65 years (2015 Alzheimer's disease facts and figures 2015).
Although the potential cause of AD is still unclear, the involvement of heredity genetic risk factors in AD’s predisposition and progression is widely acknowledged. Recent advantages in multistage Genome Wide Association Studies (GWAS) have identified several loci conferring risk for AD (Bodily et al. 2016; Cuyvers and Sleegers 2016). Since 2009, a number of susceptibility genes, such as Bridging integrator 1 (BIN1), Clusterin (CLU), CD2 associated protein (CD2AP), and ATP binding cassette subfamily A member 7(ABCA7) were gradually reported to be correlated with AD disease (Zhang et al. 2016). Of all the reported loci, rs9331888 in the CLU gene was considered to be significantly associated with AD (Xing et al. 2012).
Clusterin which is also known as apolipoprotein J (Apo J), located at chromosome 8p21-p22 extending over 16 kb, is a multifunctional lipoprotein expressed in almost all mammalian tissues, especially in the brain. The protein binds to Aβ peptides and fibrils to prevent aggregation and also it can involve in Aβ clearance via binding to megalin receptors and increasing endocytosis by glial cells (Li et al. 2014; Zhou et al. 2014). Large-scale GWAS identified that rs9331888 (G > C) single nucleotide polymorphisms (SNPs) in CLU are significantly associated with AD in populations of Caucasian ancestry. And Merve Alaylıog˘ lu et al. also got the same results in Turkey populations (Alaylioglu et al. 2016). However, inconsistent results regarding this variant had been reported in Chinese populations. Lu et al. had reported that there was weak association or no association between the rs9331888 polymorphism and AD in southern Chinese populations (Lu et al. 2014). Meanwhile, Edina et al. found that there was no evidence of plasma clusterin lever correlated with Alzheimer”s disease (Silajdzic et al. 2012). Therefore, to confirm the association between rs933188 polymorphism and AD, we performed a meta-analysis of case-control studies by pooling all eligible studies to evaluate the overall risk and influence of ethnic factors to this disease.
Methods
Literature search and inclusion criteria
To identify all relevant publications focus on the risk for AD and rs9331888 polymorphism, we conducted a comprehensive literature search of electronic databases, including the Pubmed, Embase and China National Knowledge Infrastructure (CNKI). Eligible case-control studies were extracted with the last search update on October 1, 2016. The following terms were used: “Alzheimer’s disease”, “demential”, “Clusterin,”, “CLU”, “APO J”, OR “rs933188” and “polymorphism” OR “Variant” without any limitation applied. The reference lists of retrieved studies and recent reviews were also manually searched for further relevant studies. AlzGene database (www.alzgene.org, updated April 18, 2011) was also utilized in our searching process.
Inclusion and exclusion criteria
Studies in this meta-analysis must meet the following inclusion criteria: (1) evaluation of the association between rs933188 polymorphism and the AD; (2) case-control study; (3) studies focusing on human being; (4) providing detail genotype frequencies; Exclusion criteria: (1) duplication of previous publications; (2) comment, review and editorial; (3) study without detailed genotype data.
Data extraction
The data of the eligible studies were extracted in duplicate by two investigators (Zhu and Zhang) independently in duplicate with a standard data-collection form. The following data was collected from each study if available: (1)first author’s name; (2) years of publication; (3) country of origin; (4) participants ethnicity; (5) Hardy-Winberg equilibrium; (6) AD diagnosis criteria; (7) genotyping method; (8) numbers of cases and controls; (9) counts of cases and controls for each genotype. When there were multiple publications from the same population, only the one with largest study was included. Any discrepancy was resolved through discussion until a consensus was reached. If the dissent still existed, the third investigators would be involved to resolve the dispute.
Quality assessment
The literature quality was evaluated by using the quality scoring criteria modified from previous study (Zhang et al. 2017) by two authors independently. Quality scores ranged from 0 point (worst) to 10 points (best). Studies scoring higher than 5 points were classified with adequate quality. Disagreement was settled through discussion among four of the investigators.
Statistics analysis
Hardy-Weinberg equilibrium was evaluated using Chi-square test in control groups and a P value <0.05 was considered significant disequilibrium. The odds ratio (OR) and 95% confidence interval (95% CI) were calculated to evaluate the strength of AD susceptibility and CLU polymorphism. Pooled ORs were obtained from combination of single study by heterozygote comparison (GC vs. CC), homozygote comparison (GG vs. CC), dominant model (GG + GC vs. CC), recessive model (GG vs. GC + CC), and allelic model (G vs. C) respectively. Heterogeneity was evaluated by Q statistic and I2 statistic. Once, Q-test >0.10 or I2 < 50%, the fixed-effect model (Mantel–Haenszelmethod) was used to calculate the pooled ORs, otherwise, the random-effect model (DerSimonian–Laird method) was used. The significance of the pooled ORs was assessed by Z-test, where P < 0.05 indicated statistically significant. Publication bias was assessed by Begg’s funnel plots and Begg’s test quantitatively. If publication bias was indicated, we further evaluated the number of missing studies in a meta-analysis by applying the trim and fill method and recalculated the pooled risks estimate with the addition of those missing studies. All statistical analyses were performed using the STATA software version 12.0 (STATA Corporation, College Station, TX, USA).
Results
Characteristics of studies
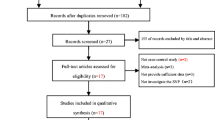
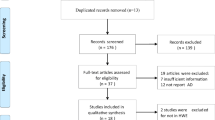
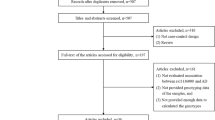
The initial search identified a total of 179 citations, 44 of which were duplicated. 19 citations were included for further review by title and abstract screening of the remaining 135 citations. After examination of full text articles, 9 case-control studies were identified as being eligible for analysis. The PRISMA flow diagram is illustrated in Fig. 1. Among the studies, 8 were Caucasians and 5 were Chinese population (including five sub studies). Multiple genotyping methods were performed in the studies, including PCR-RFLP, TaqMan and DNA sequencing. All studies were complied with Hardy–Weinberg equilibrium (HWE) in the controls and the genotyping distribution was in agreement with HWE. The characteristics of involved articles were summarized in Tables 1 and 2.
Meta-analysis results
Heterogeneity was firstly identified by Q-test and I-squared statistic in five genetic models. As is showed in the Fig. 2, serious heterogeneity were found in Allele model (I2 = 70.8%), Homozygote model (I2 = 70.4%) and Dominant model (I2 = 65.1%), thus random effect model was applied. The results revealed that no significant associations between the rs9331888/C > G polymorphism and AD disease in above three genotype distributions (G vs. C: OR = 0.968,95% CI: 0.887–1.056, P = 0.464; GG vs. CC: OR = 0.959, 95% CI: 0.790–1.163, P = 0.669; GG + GC vs. CC:0.987, 95% CI: 0.839–1.160, P = 0.872). Fixed effect model were used in the Heterozygote model (I2 = 46.7%) and Recessive model (I2 = 47.7%), and results showed that the recessive model (GG vs. GC + CC: OR = 1.11, 95% CI: 1.05–1.18; P < 0.01) was significantly associated with AD whereas no effect was found in Heterozygote model (CG vs. CC: OR = 0.962, 95% CI: 0.876–1.056, P = 0.416) with AD development.
Subgroups based on ethnicity were utilized to further analyze the relationship of polymorphism with AD. In Caucasian populations, AD was proved to be correlated with rs9331888 polymorphism under the recessive model (GG vs. GC + CC: OR = 1.12, 95% CI: 1.06–1.2; P < 0.01). Nevertheless, no significant differences were observed in any other genetic models (G vs. C: OR = 0.939, 95% CI: 0.863–1.023, P = 0.149; CG vs. CC: OR = 0.942, 95% CI: 0.797–1.112, P = 0.479; GG vs. CC: OR = 0.905, 95% CI: 0.732–1.118, P = 0.353; GG + GC vs. CC:OR = 0.923, 95% CI: 0.764–1.114, P = 0.404); In Chinese populations, we did not observed any correlations of the clusterin polymorphism under five genetic models (G vs. C: OR = 0.991, 95% CI: 0.805–1.220, P = 0.931; GG vs. CC: OR = 1.008, 95% CI: 0.683–1.490,P = 0.966; CG vs. CC: OR = 1.098, 95% CI: 0.888–1.358, P = 0.390; GG + GC vs. CC:OR = 1.066, 95% CI: 0.812–1.399, P = 0.644; GG vs. GC + CC: OR = 0.931, 95% CI: 0.850–1.019, P = 0.711) (Fig. 3).
Publication bias
Publication bias was evaluated by using the Begg and Egger tests. Allele model and Homozygote model were found with significant evidence of publication bias (Allele model: Begg P = 0.044,Egger P = 0.028; Homozygote model: Begg P = 0.059, Egger P = 0.018) (Fig. 4). Nevertheless, the application of the trim and fill method did not change the risk estimate (Allele model: P = 0.859, 95% confidence interval:-0.168 to 0.201;Homozygote model:P = 0.669, 95% confidence interval:-0.235 to 0.151). No missing studies were imputed in the contour enhanced funnel plots.
Sensitivity analysis
We performed sensitivity analysis to assess the influence of each individual study on the pooled OR by sequentially removing each eligible study. The results indicated that the removal of the study by Lambert et al. led to the loss of a significant association with the risk of AD in the overall pooled population under a recessive model. However, this effect was attributed to loss of power of the meta-analysis due to the overall high weight of this study (30.25%). We did not find that any single study influence the quality of the pooled ORs in the sensitivity analyses(Fig. 5).
Discussion
Alzheimer’s disease is the most common form of dementia in elder, accounting for 50% of all dementia. It had been proved to be one of common diseases with strong genetic component. Study on SNP provided a compelling evidence for a role of genetic variation in AD risk assessment, diagnosis and development of new therapies (Jiao et al. 2015). Clusterin gene located in chromosome 8p21-p12 which is a chromosomal region of interest in AD and it may explain around 9% of the late-onset AD attributable risk (Tan et al. 2016). The study by Lacour et al. found the rs11136000and rs9331888 polymorphism of CLU gene showed strong association with AD (Lacour et al. 2017). Haplotype analyses found that rs9331888 in combination with rs11136000 is likely to play a key role in the progress of AD. Mengel et al. had found that patients who carriers of the common rs11136000 and rs9331888 TTC haplotype in the CLU gene performed cognitively better than non-carriers and carriers of a rare TCC haplotype performed worse on the cognitive composite score(Mengel-From et al. 2013). Considering of the importance of rs9331888 in AD, we believed that it is critically important to explore the relationship of haplotype and the genotype in the morbidity of AD.
The rs9331888 polymorphism is located in the first exon of transcript NM_203339 and is one of the functional DNA variants underlying this association between CLU and AD. It regulates Aβ fibril formation and toxicity and facilitates amyloid-β (Aβ) transport across the blood-brain barrier (Bettens et al. 2015; Lidstrom et al. 1998). However, inconsistent results reported in Chinese and Caucasian populations. Thambisetty et al. showed that CLU influenced longitudinal changes in brain function and faster cognitive decline and GWAS studies have also identified that rs9331888 in CLU was substantially associated with AD risk in individuals of Caucasian ancestry and other populations (Thambisetty et al. 2013). Tan et al. showed that AD risk rs9331888 allele was associated with a decrease in CLU plasma levels in Chinese populations (Tan et al. 2016). Nevertheless, other researches such as Lu et al. showed that the rs9331888 variants may not be an AD susceptibility factor in southern Chinese Han population. A meta-analysis by Zhang et al. also reported that there was no significant association with East Asian population (Zhang et al. 2016). To solve the problem, we conduct a meta-analysis to investigate the pooled effect size of association between CLU SNP rs9331888 and AD in different ethnic backgrounds.
Our results implied a conspicuously significant relationship between the CLU polymorphism and AD risk under the recessive genetic model but not in other model. To further analysis the influence of ethnic factor to AD risk, a subgroup was employed in our research. The results revealed that in recessive model Caucasian populations are even more easily to progress to AD (P < 0.05), which is in agreement with previous observations (Jiao et al. 2015). Nevertheless, we did not found any association of polymorphism of rs933188 with AD risk in Chinese population, which was in accordance with the results drawn by Shuai et al., and these suggesting that the SNP may be an ethnicity-dependent factor in AD progress. Additional, subgroup analysis results of Caucasian populations were with difference, they had just showed that there were significant association of SNP rs9331888 with AD among allelic model, homozygote model, recessive model, and dominant model, whereas in our research we had showed that only the recessive model were significant associated with AD risk.
There were fewer researches to study the relationship of rs9331888 polymorphism with AD. Prior to this study, two groups performed meta-analyses to detect the correlation of rs9331888 polymorphism in patients with AD, and our results are in accordance with the two studies which suggest that rs9331888 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian but not in Chinese populations (Shuai et al. 2015). However, compared with the prior study, our study was an update of the former researches. In this meta-analysis, we had enrolled more researches not only including in the Alzgene database but also recently published studies especially in Chinese populations. Totally, 9292 AD patients and 11,958 controls were included in the study, which can provide enough statistical power and strengthened the reliability of our results. Upon including eligible studies, a methodological quality assessment was conducted and all studies had acceptable quality.
Due to several limitations of the present meta-analysis, the results of our study should be interpreted with caution. To be specific, publication biases exist in the 9 studies in the five genetic models, which indicated that there might be problems due to insufficient articles. In addition, small sample size in each study might be the cause of the failure to achieve statistical significance. Additional, as there were not unified detection methods, serious heterogeneity were observed in our study even in subgroup analysis which may be one of the main risk factor explain for the negative results.
In summary, our data from an independent and large case-control sample revealed that SNP rs9331888 displayed significant association with AD in Caucasian populations under the Recessive model. However, limitations still exist, such as that we did not observed the influence of rs11136000 and rs9331888 TTC haplotype in the progress of AD development. Additional, as with limited sample size, the relationship of rs9331888 polymorphism with AD susceptibility in Chinese populations was worth further exploration. In future, more studies with genotype and haplotype data are required to further verify the results.
References:
Alaylioglu M, Gezen-Ak D, Dursun E, Bilgic B, Hanagasi H, Ertan T, Gurvit H, Emre M, Eker E, Uysal O, Yilmazer S (2016) The association between clusterin and APOE polymorphisms and late-onset Alzheimer disease in a Turkish cohort. J Geriatr Psychiatry Neurol 29(4):221–226
Alzheimer's disease facts and figures (2015) 2015 Alzheimer's disease facts and figures. Alzheimers Dement 11(3):332–384
Bettens K, Vermeulen S, Van Cauwenberghe C, Heeman B, Asselbergh B, Robberecht C, Engelborghs S, Vandenbulcke M, Vandenberghe R, De Deyn PP, Cruts M, Van Broeckhoven C, Sleegers K (2015) Reduced secreted clusterin as a mechanism for Alzheimer-associated CLU mutations. Mol Neurodegener 10:30
Bodily PM, Fujimoto MS, Page JT, Clement MJ, Ebbert MT, Ridge PG (2016) A novel approach for multi-SNP GWAS and its application in Alzheimer's disease. BMC Bioinf 17(Suppl 7):268
Cuyvers E, Sleegers K (2016) Genetic variations underlying Alzheimer's disease: evidence from genome-wide association studies and beyond. Lancet Neurol 15(8):857–868
Gu H, Wei X, Chen S, Kurz A, Muller U, Gasser T, Dodel RC, Farlow MR, Du Y (2011) Association of clusterin gene polymorphisms with late-onset Alzheimer's disease. Dement Geriatr Cogn Disord 32(3):198–201
Jiao B, Liu X, Zhou L, Wang MH, Zhou Y, Xiao T, Zhang W, Sun R, Waye MM, Tang B, Shen L (2015) Polygenic analysis of late-onset Alzheimer's disease from mainland China. PLoS One 10(12):e144898
Kamboh MI, Minster RL, Demirci FY, Ganguli M, DeKosky ST, Lopez OL, Barmada MM (2012) Association of CLU and PICALM variants with Alzheimer's disease. Neurobiol Aging 33(3):518–521
Lacour A, Espinosa A, Louwersheimer E, Heilmann S, Hernandez I, Wolfsgruber S, Fernandez V, Wagner H, Rosende-Roca M, Mauleon A, Moreno-Grau S, Vargas L, Pijnenburg YA, Koene T, Rodriguez-Gomez O, Ortega G, Ruiz S, Holstege H, Sotolongo-Grau O, Kornhuber J, Peters O, Frolich L, Hull M, Ruther E, Wiltfang J, Scherer M, Riedel-Heller S, Alegret M, Nothen MM, Scheltens P, Wagner M, Tarraga L, Jessen F, Boada M, Maier W, van der Flier WM, Becker T, Ramirez A, Ruiz A (2017) Genome-wide significant risk factors for Alzheimer's disease: role in progression to dementia due to Alzheimer's disease among subjects with mild cognitive impairment. Mol Psychiatry 22(1):153–160
Lambert JC, Heath S, Even G, Campion D, Sleegers K, Hiltunen M, Combarros O, Zelenika D, Bullido MJ, Tavernier B, Letenneur L, Bettens K, Berr C, Pasquier F, Fievet N, Barberger-Gateau P, Engelborghs S, De Deyn P, Mateo I, Franck A, Helisalmi S, Porcellini E, Hanon O, de Pancorbo MM, Lendon C, Dufouil C, Jaillard C, Leveillard T, Alvarez V, Bosco P, Mancuso M, Panza F, Nacmias B, Bossu P, Piccardi P, Annoni G, Seripa D, Galimberti D, Hannequin D, Licastro F, Soininen H, Ritchie K, Blanche H, Dartigues JF, Tzourio C, Gut I, Van Broeckhoven C, Alperovitch A, Lathrop M, Amouyel P (2009) Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 41(10):1094–1099
Li X, Ma Y, Wei X, Li Y, Wu H, Zhuang J, Zhao Z (2014) Clusterin in Alzheimer's disease: a player in the biological behavior of amyloid-beta. Neurosci Bull 30(1):162–168
Lidstrom AM, Bogdanovic N, Hesse C, Volkman I, Davidsson P, Blennow K (1998) Clusterin (apolipoprotein J) protein levels are increased in hippocampus and in frontal cortex in Alzheimer's disease. Exp Neurol 154(2):511–521
Liu, XY (2014). Association analysis of late onset Alzheimer’s disease and Susceptibility genes in Chinese Han population, Central South University
Lu SJ, Li HL, Sun YM, Liu ZJ, Yang P, Wu ZY (2014) Clusterin variants are not associated with southern Chinese patients with Alzheimer's disease. Neurobiol Aging 35(11):2656–2659
Mengel-From J, Thinggaard M, Lindahl-Jacobsen R, McGue M, Christensen K, Christiansen L (2013) CLU genetic variants and cognitive decline among elderly and oldest old. PLoS One 8(11):e79105
Scheltens P, Blennow K, Breteler MM, de Strooper B, Frisoni GB, Salloway S, Van der Flier WM (2016) Alzheimer's disease. Lancet 388(10043):505–517
Shuai P, Liu Y, Lu W, Liu Q, Li T, Gong B (2015) Genetic associations of CLU rs9331888 polymorphism with Alzheimer's disease: a meta-analysis. Neurosci Lett 591:160–165
Silajdzic E, Minthon L, Bjorkqvist M, Hansson O (2012) No diagnostic value of plasma clusterin in Alzheimer's disease. PLoS One 7(11):e50237
Tan L, Wang HF, Tan MS, Tan CC, Zhu XC, Miao D, Yu WJ, Jiang T, Tan L, Yu JT (2016) Effect of CLU genetic variants on cerebrospinal fluid and neuroimaging markers in healthy, mild cognitive impairment and Alzheimer's disease cohorts. Sci Rep 6:26027
Thambisetty M, Beason-Held LL, An Y, Kraut M, Nalls M, Hernandez DG, Singleton AB, Zonderman AB, Ferrucci L, Lovestone S, Resnick SM (2013) Alzheimer risk variant CLU and brain function during aging. Biol Psychiatry 73(5):399–405
Wang, B. (2014). Association of polymorphisms in Alzheimer’S disease and CLU and clusterin in people with mild cognitive impairment, Fudan University
World Alzheimer Report (2015) The global impact of dementia. World Alzheimer Report 2015:The Global Impact of Dementia. http://www.alz.co.uk/research/WorldAlzheimerReport2015.pdf
Xing YY, Yu JT, Cui WZ, Zhong XL, Wu ZC, Zhang Q, Tan L (2012) Blood clusterin levels, rs9331888 polymorphism, and the risk of Alzheimer's disease. J Alzheimers Dis 29(3):515–519
Yu JT, Li L, Zhu QX, Zhang Q, Zhang W, Wu ZC, Guan J, Tan L (2010) Implication of CLU gene polymorphisms in Chinese patients with Alzheimer's disease. Clin Chim Acta 411(19–20):1516–1519
Zhang S, Li X, Ma G, Jiang Y, Liao M, Feng R, Zhang L, Liu J, Wang G, Zhao B, Jiang Q, Li K, Liu G (2016) CLU rs9331888 polymorphism contributes to Alzheimer's disease susceptibility in Caucasian but not east Asian populations. Mol Neurobiol 53(3):1446–1451
Zhang S, Wang XB, Han YD, Wang C, Zhou Y, Zheng F (2017) Certain polymorphisms in SP110 Gene confer susceptibility to tuberculosis: a comprehensive review and updated meta-analysis. Yonsei Med J 58(1):165–173
Zhou Y, Hayashi I, Wong J, Tugusheva K, Renger JJ, Zerbinatti C (2014) Intracellular clusterin interacts with brain isoforms of the bridging integrator 1 and with the microtubule-associated protein tau in Alzheimer's disease. PLoS One 9(7):e103187
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81402814).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Bin Zhu and Rui Min Wang contributed equally to this work.
Rights and permissions
About this article
Cite this article
Zhu, B., Wang, R.M., Wang, J.T. et al. Correlation of rs9331888 polymorphism with Alzheimer’s disease among Caucasian and Chinese populations: a meta-analysis and systematic review. Metab Brain Dis 32, 981–989 (2017). https://doi.org/10.1007/s11011-017-9957-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-017-9957-8