Abstract
Breast cancer is one of the most common malignancies worldwide. Circular RNAs (CircRNAs) were revealed to be implicated in the development of breast cancer. In this research, we aimed to investigate the role and underlying mechanism of circ_0008500 in the development and radiosensitivity of breast cancer. Using real-time quantitative PCR (RT-qPCR) and western blot, we found that hsa_circ_0008500 (circ_0008500) and profilin 2 (PFN2) were increased, while microRNA-758-3p (miR-758-3p) was decreased in breast cancer tissues and cells. Cell viability, the number of colonies, proliferation and apoptosis were detected using CCK-8, colony formation, EdU assays and flow cytometry, respectively. Dual-luciferase reporter and RNA immunoprecipitation (RIP) assays were devoted to test the interaction between miR-758-3p and circ_0008500 or PFN2. The results showed that circ_0008500 knockdown inhibited cell growth, and facilitated cell apoptosis and radiosensitivity in breast cancer cells in vitro. Moreover, circ_0008500 regulated PFN2 expression by sponging miR-758-3p. Functionally, circ_0008500 knockdown regulated cell behaviors and radiosensitivity by targeting miR-758-3p to downregulate PFN2 expression in vitro. Additionally, in vivo tumor formation assay and immunohistochemistry (IHC) assay demonstrated that circ_0008500 knockdown enhanced the radiosensitivity and repressed tumor growth in vivo. In conclusion, circ_0008500 inhibition promoted the radiosensitivity and restrained the development of breast cancer by downregulating PFN2 expression via targeting miR-758-3p.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
With over 2 million newly diagnosed cases and more than 626,000 deaths annually, breast cancer is a common type of human cancers among women globally [1,2,3]. Although early diagnosis methods and comprehensive treatment, including molecular targeted drug applications, radiotherapy, chemotherapy and radical surgery, have gained enormous improvements, the recurrence and mortality rates are still disappointing in women with breast cancer [4, 5]. It was well known that radiotherapy was widely used as a main adjuvant therapy for the majority of patients with breast cancer in the clinical treatment [6]. Although radiotherapy could contribute to decrease the recurrence risk and increase long-term survival of most breast cancer patients, the curative effect was related to the radioresistance of breast cancer [7]. Therefore,{Huang, 2021 #566} it is urgent to explore the new therapeutic targets for the radioresistance in breast cancer.{Huang, 2021 #566}{Huang, 2021 #566}.
Multiple research studies reported that non-coding RNAs (ncRNAs) played vital roles in the progression of various cancers [8]. Circular RNAs (circRNAs) are a special class of endogenous ncRNAs and featured by the continuous covalently circular structure without 3’-end poly A tail and 5’-end cup [9, 10]. MicroRNAs (miRNAs), the small, short-stranded ncRNAs with the size of approximately 20 nucleotides, exert function through the posttranscriptional degradation of the target mRNA or suppressing the target mRNA translation [11, 12]. Growing evidence revealed that circRNAs with close-loop structure could act as the stable molecular diagnosis and therapy biomarkers in diverse tumors [13]. CircRNAs were proved to serve as the major regulators in cancer biology and correlated with cancer radioresistance [14,15,16]. A previous report showed that there were 37 downregulated circRNAs and 30 upregulated circRNAs in lung cancer cells under irradiation treatment [17]. Currently, accumulating evidence also suggested the regulatory effect of some circRNAs on breast cancer development, while there were few researches on the involvement of circRNAs in breast cancer radiosensitivity. Interestingly, major roles of miRNAs in the pathogenesis of breast cancer and response to radioresistance have been explored [18, 19]. For instance, miR-22 served as an inhibitor in breast cancer, as evidenced by restraining cell proliferation and enhancing cell apoptosis and radiosensitivity [20]. MiR-200c modulated UBQLN1 to repress the radioresistance and autophagy in MDA-MB-231 cells [21]. In addition, it was reported that circTADA2As modulated the miR-203a/SOCS3 axis to restrain the metastasis of breast cancer cells [22]. More importantly, a previous research uncovered that hsa_circ_0008500 (circ_0008500; Position: chr3:196,831,773–196,846,401), a novel circRNA, was found to be upregulated in breast cancer samples [23]. However, the critical role of circ_0008500 and its potential underlying mechanism in the radiosensitivity and development of breast cancer are yet to be studied.
Herein, we determined the level of circ_0008500 in breast cancer tissues and cells. Moreover, we also explored the functional role of circ_0008500 and its molecular mechanism in breast cancer radiosensitivity.
Material and Methods
Clinical Tissue Samples Acquirement
50 breast cancer patients admitted to the Fourth Hospital of Hebei Medical University were recruited to obtain the cancerous tissues and the adjacent normal tissues. The collected fresh tissues were immediately kept at –80˚C for further analysis. The related breast cancer patients agreed to take part in this study and signed the written informed consents. This project received the ratification of the Ethics Committee of the Fourth Hospital of Hebei Medical University.
Cell Culture
MCF-10A cells (normal breast epithelial cell line), and MDA-MB-468 and MCF-7 cells (human breast cancer cell lines) were obtained from Shanghai Academy of life Science (Shanghai, China). 10% fetal bovine serum was added into dulbecco’s modified Eagle’s medium (DMEM; Gibco, Carlsbad, CA, USA) containing 100 units/mL penicillin and 100 μg/mL streptomycin, and the medium was used to maintain the cells in an incubator at 37˚C under the condition with the setting of 5% CO2.
Cell Transfection
MCF-7 and MDA-MB-468 cells (5 × 105 cells/well) were seeded into the 12-well plates and cultured for 24 h. Small interfering RNA (siRNA) specifically against circ_0008500 (si-circ_0008500; 5’-AACCTCTTTCAGGCTTTAATAGA-3’) and its siRNA control (si-NC), microRNA-758-3p (miR-758-3p) mimic and the control (miRNA NC), and miR-758-3p inhibition (miR-758-p inhibitor) and its negative control (inhibitor NC) were synthesized and acquired from Genepharma (Shanghai, China). Then, the abovementioned oligonucleotides (0.5 μg) were transfected into the cells using 0.6 μL of Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA). Profilin 2 (PFN2) overexpression vector (pc-PFN2) was generated by cloning the full sequence of PFN2 into pcDNA3.1 (Invitrogen), with pc-NC as the negative control. Then, the cells were transfected with 0.2 μg of pc-PFN2 or pc-NC using 0.5 μL of Lipofectamine 3000 (Invitrogen).
Irradiation
X-ray irradiation was conducted with the single dose of 0, 2, 4, 6 or 8 Gy using an exposure instrument Cs-137 irradiator (HWMD-2000, Siemens, Germany) at a dose rate of 2.4 Gy/min. For time-course assays, MCF-7 and MDA-MB-468 cells were irradiated with 4 Gy and harvested every 6 h within 24 h post-irradiation.
RT-qPCR Analysis
Total RNA was isolated using TRIzol reagent (Invitrogen). Reverse transcription (5 μg RNA) was conducted to synthesize cDNA using First Strand cDNA Synthesis Kit (Takara, Dalian, Liaoning, China). Then, the transcription was performed in a 10 μL reaction mixture, including 5 × PrimeScript Buffer (2 μL), polyadenylated RNA (100 ng), RT primer mixture (1 μL), PrimeScript RT Enzyme Mix I (0.5 μL), and RNase-free water. The quantitative analysis of circ_0008500, DLG1 and PFN2 was conducted using the SYBR® Premix Ex Taq™ (Takara) in 15 μL final mixture harboring 7.5 μL 2 × SYBR Green PCR master mix, 1.5 μL template cDNA and 3 μL of each forward and reverse primers. The cDNA amplification of miR-758-3p was carried out using a mirVana™ real-time RT-PCR microRNA detection kit (Life Technologies, Carlsbad, CA, USA). RT-qPCR analysis was carried out on the StepOnePlus system (Applied Biosystems, CA, USA). The level of circ_0008500, DLG1, miR-758-3p and PFN2 was calculated by the 2 −∆∆Ct method, with the normalization by U6 and GADPH. The primer sequences were represented as below: circ_0008500, F 5’-CTAGTCATGAGCAGGCAGCA-3’, R 5’-CCAAGAAGGAAGATGGGCTA-3’; miR-758-3p, F 5’-GCCGAGTTTGTGACCTGGTCCA-3’, R 5’-GTGCAGGGTCCGAGGT-3’; PFN2, F 5’-GAGACTCTGGGTTCTAGCTGC-3’, R 5’-TTAAGTGTGCCTCCGTGGAC-3’; DLG1, F 5’-ATAGGGAGGTCCTCGTCACC-3’, R 5’-CTTCCGGACCGGCATTTTTC-3’; U6, F 5’-CTCGCTTCGGCAGCACATA-3’, R 5’-CGAATTTGCGTGTCATCCT-3’; GAPDH (qPCR), F 5’-AAGGCTGTGGGCAAGGTCATC-3’, R 5’-GCGTCAAAGGTGGAGGAGTGG-3’; circ_0008500 (Convergent), F 5’-TCTCCAGCCAGATACTCCCC-3’, R 5’-GCAGCTCTGAGGTCAACACT-3’; circ_0008500 (Divergent), F 5’-GGAGATCGTATTATATCGGT-3’, R 5’-CCCTTGTAATTTCATCATCT-3’; GAPDH (PCR), F 5’-GTGACTAACCCTGCGCTCC-3’, R 5’-GGAAAAGCATCACCCGGAGG-3’.
CircRNA Identification
For agarose gel electrophoresis assay, genomic DNA (gDNA) was isolated from the cells using the mammalian genomic DNA extraction kit (Beyotime, Shanghai, China). The products of gDNA and cDNA were determined by 1% TAE running buffer at 160 V for 15 min. The bands were obtained by UV irradiation.
For RNase R digestion assay, total RNA (2 μg) was incubated with or without RNase R (Epicentre, Madison, WI, USA). After cultivation at 37℃ for 30 min, the level of circ_0008500 and DLG1 was determined using RT-qPCR assay.
CCK-8 Assay
Cell viability of breast cancer cells was assessed by the CCK-8 kit (Beyotime). MCF-7 and MDA-MB-468 cells (4000 cells per well) were seeded in the 96-well plates. After the relevant treatment, 10 μL CCK-8 solution was added and incubated with the cells for 2 h. Then, the absorbance at 450 nm was assessed using a microplate reader (Thermo Labsystems, Waltham, MA, USA).
EdU Staining Assay
2 × 104 MDA-MB-468 and MCF-7 cells were seeded in the 12-well plates. MDA-MB-468 and MCF-7 cells irradiated with 4 Gy or transfected with the indicated plasmids and oligonucleotides were cultured with 20 μM EdU (RiboBio, Nanjing, China) for 2 h, and then treated with 10 μL DAPI (Sigma-Aldrich, St. Louis, MO, USA) for 15 min to stain the nuclei. Finally, EdU-positive cells were imaged by a microscope.
Colony Formation Assay
Briefly, MDA-MB-468 and MCF-7 cells were seeded and grown in the 6-well plates at a density of 500 cells per well. Then, the cells were irradiated with 4 Gy or transfected with the indicated plasmids and oligonucleotides. After incubation for 14 d, the colonies were washed with PBS, fixed with methanol for 20 min, and stained with 0.1% crystal violet for 15 min. Finally, the colonies were recorded by an inverted microscope (Carl Zeiss AG, Heidenheim, Germany).
Flow Cytometry
Cell apoptosis assessment was performed using Annexin V-FITC apoptosis detection kit (Beyotime). Briefly, MDA-MB-468 and MCF-7 cells irradiated with 4 Gy or transfected with the indicated plasmids and oligonucleotides were washed, collected and re-suspended in 400 μL 1 × binding buffer (Invitrogen). Then, Annexin V-FITC (5 μL; Beyotime) and PI (10 μL; Beyotime) were used to culture cells away from light for 15 min and finally analyzed by a flow cytometry.
Western Blot
The RIPA buffer (Beyotime) was used to isolate the protein. The extracted proteins (20 μg) were loaded on SDS-PAGE and then transferred on the PVDF membranes. The membranes were blocked with 5% non-fat milk for 2 h. Then, the membranes were incubated with the primary antibodies at 4℃ overnight and the secondary antibody for 1.5 h at room temperature. The antibodies were anti-BAX (1:1000; ab32503, Abcam, Cambridge, UK), anti-BCL2 (1:1000; ab32124, Abcam), anti-PFN2 (1:1000; 60,094–2-Ig, ProteinTech Group, Chicago, IL, USA), anti-GAPDH (1:1000; ab181602, Abcam), and the secondary antibody (goat anti-rabbit, 1:2000; ab6721, Abcam). The protein complexes were presented using the enhanced chemiluminescence (Millipore, Bradford, MA, USA).
Subcellular Localization Assay
Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek Corp., Belmont, MA, USA) was used to measure the localization of circ_0008500 according to the accompanying protocols. In brief, the cells were lysed using Lysis Buffer J. After centrifugation, anhydrous ethanol and Buffer SK were used to incubate the nuclear and cytoplasmic RNA, respectively. Finally, circ_0008500 content in the cytoplasmic and nucleus fractions was measured using RT-qPCR.
Dual-Luciferase Reporter Assay
The wild-type (WT) segments of circ_0008500 or PFN2 harboring the complementary sites of miR-758-3p were individually cloned into the pGL3 vector (Promega, Madison, WI, USA) to construct the WT-circ_0008500 or WT-PFN2 3’UTR luciferase reporter vectors. The mutated segments of circ_0008500 or PFN2 containing the mutant target regions of miR-758-3p were used to form the mutant (MUT) vectors (MUT-circ_0008500 and MUT-PFN2 3’UTR). MiRNA NC or miR-758-3p mimic was co-transfected with each reporter construct into the cells using Lipofectamine 3000 (Invitrogen). The luciferase activity was determined using the Dual-Glo® Luciferase Assay System (Promega).
RIP Assay
A RIP kit (Geneseed, Guangzhou, China) was used to test the correlation between miR-758-3p and circ_0008500 or PFN2. Cell lysates of MCF-7 and MDA-MB-468 cells were acquired using RIP lysis buffer, and then mixed with the magnetic beads conjugated with anti-IgG (1:100; ab190475, Abcam) or anti-Ago2 (1:50; ab186733, Abcam). After immunoprecipitation, the abundance of circ_0008500, miR-758-3p and PFN2 was quantified by RT-qPCR analysis.
Tumor Formation Assay
Cs-137 irradiator (HWMD-2000, Siemens) was utilized for irradiation in tumor formation assay. Beijing Vital River Laboratory Animal Technology (Beijing, China) provided 16 BALB/c nude mice (4-week-old) to establish the murine xenograft model of breast cancer. To stably knockdown circ_0008500, sh-circ_0008500 lentivirus vector was constructed and the lentiviruses were packaged and purified by GenePharma (Shanghai, China). 1 × 106 stably modified (circ_0008500 knockdown) or control (sh-NC) MCF-7 or MDA-MDA-468 cells were injected into the right flank of the nude mice. After injection for 7 d, the mice were treated with or without 4 Gy irradiation every 7 d. The mice were divided into sh-circ_0008500, sh-NC, sh-NC + radiation, and sh-circ_0008500 + radiation groups (n = 4 per group). The tumor volume (length × width2 × 0.5) was recorded weekly. 4 weeks later, the mice were euthanatized and tumor weight was detected. Animal experiments were ratified by the Animal Ethics Committee of the Fourth Hospital of Hebei Medical University.
Immunohistochemistry (IHC)
The tissue sections acquired from xenograft model mice were fixed with formalin (Sigma-Aldrich) and embedded with paraffin. Paraffin-embedded tissues were sliced into 5 µm slides. Then, the sections were incubated with anti-PFN2 (1:50; 60,094–2-Ig, ProteinTech Group) and the secondary antibody. Following the staining using the diaminobenzidine (DAB) kit (Sigma-Aldrich), the positive protein expression of tissue sections was observed and photographed under a microscope.
Statistical Analysis
The assessment data from three replications were analyzed by GraphPad Prism software (La Jolla, CA, USA) and shown as the mean ± standard deviation (SD). The difference was assessed by one-way analysis of variance (ANOVA) or Student’s t test. A value of P less than 0.5 meant the statistically significant difference.
Results
Circ_0008500 Was Downregulated by Radiation Treatment in Breast Cancer Cells
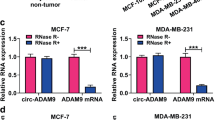
To investigate the involvement of circ_0008500 in breast cancer radiosensitivity, RT-qPCR was performed to gauge the expression of circ_0008500 in breast cancer tissues and cells. The increase of circ_0008500 was observed in breast cancer tissues (n = 50) compared with the adjacent normal tissues (n = 50) (Fig. 1A). Our data showed that circ_0008500 was lowly expressed in triple-negative breast cancer (TNBC; n = 24) tissues compared with the non-TNBC tissues (n = 26) (Fig. 1B). Additionally, the expression of circ_0008500 was highly expressed in ER/PR-positive BC tissues, while there was no remarkable difference between HER2-negative and HER2-positive BC tissues (Fig. 1C–E). Moreover, the expression of circ_0008500 was higher in breast cancer cells (MCF-7 and MDA-MB-468 cells) than that in MCF-10A cells (Fig. 1F). As shown in Fig. 1G, circ_0008500 was consisted of exons 13, 14, and 15 of DLG1. The results of agarose gel electrophoresis assay showed that circ_0008500 was amplified only from cDNA using the divergent primer, while circ_0008500 could not be amplified from gDNA (Fig. 1H). In RNase R treatment assay, we found that the expression of DLG1 was inhibited by RNase R treatment, whereas circ_0008500 was more stable than DLG1 and resistant to RNase R (Fig. 1I, J). As displayed in Fig. 1K– L, 4 Gy radiation induced a significant decrease of circ_0008500 in MCF-7 and MDA-MB-468 cells.
Circ_0008500 was downregulated by radiation treatment in breast cancer cells. (A) The level of circ_0008500 in breast cancer tissues (n = 50) and the adjacent normal tissues (n = 50) was measured by RT-qPCR. (B) RT-qPCR was employed to detect the level of circ_0008500 in TNBC tissues (n = 24) and the non-TNBC tissues (n = 26). (C) RT-qPCR was used to measure the expression of circ_0008500 in ER-negative (n = 27) or ER-positive (n = 23) BC tissues. (D) RT-qPCR was used to measure the expression of circ_0008500 in PR-negative (n = 28) or PR-positive (n = 22) BC tissues. (E) RT-qPCR was used to measure the expression of circ_0008500 in HER2-negative (n = 25) and HER2-positive (n = 25) BC tissues. (F) The expression of circ_0008500 in breast cancer cell lines (MCF-7 and MDA-MB-468 cells) and the normal breast epithelial cell line (MCF-10A) was detected by RT-qPCR. (G) Circ_0008500 was formed by the cyclization of exons 13, 14 and 15. (H) The divergent primers amplified circ_0008500 from cDNA but not from gDNA. (I and J) The expression of circ_0008500 and linear DLG1 was determined by RT-qPCR after treatment with RNase R. (K and L) Circ_0008500 level in MCF-7 and MDA-MB-468 cells after radiation treatment (4 Gy) was measured by RT-qPCR. *P < 0.05. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
Circ_0008500 Knockdown Enhanced Radiosensitivity and Aggravated Radiation-caused Suppression Effect On Cell Proliferation As Well As Promotion Effect On Cell Apoptosis in Breast Cancer Cells
The data of RT-qPCR represented a successful knockdown efficiency of si-circ_0008500 in MCF-7 and MDA-MB-468 cells (Fig. 2A). CCK-8 assay indicated that cell viability was significantly inhibited by circ_0008500 knockdown in breast cancer cells (Fig. 2B). Moreover, circ_0008500 downregulation aggravated radiation treatment caused inhibition on cell proliferation in breast cancer cells (Fig. 2C, D). The results in colony formation assay suggested that circ_0008500 knockdown exacerbated the suppressive impact of radiation treatment on the number of colonies in MCF-7 and MDA-MB-468 cells (Fig. 2E). In addition, the promotion effect of radiation treatment on cell apoptosis was facilitated by circ_0008500 knockdown (Fig. 3A, B). Meanwhile, the enhanced BAX protein expression and the reduced BCL2 protein expression induced by radiation treatment were intensified by transfection of si-circ_0008500 in MCF-7 and MDA-MB-468 cells (Fig. 3C, D).
Circ_0008500 knockdown aggravated radiation-caused suppression effect on cell proliferation in breast cancer cells. (A) The knockdown efficiency of circ_0008500 was evaluated by RT-qPCR. (B) Cell viability in MCF-7 and MDA-MB-468 cells transfected with sh-NC or si-circ_0008500 was detected by CCK-8 assay. (C–E) MCF-7 and MDA-MB-468 cells were treated without radiation (0 Gy) or with radiation (4 Gy), and 4 Gy + si-NC, or 4 Gy + si-circ_0008500. (C and D) The EdU-stained MCF-7 and MDA-MB-468 cells were quantified, and the scale bar was 50 μm. (E) The number of colonies was detected by colony formation assay. *P < 0.05. Three independent experiments were performed. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
Circ_0008500 downregulation enhanced radiation-induced promotion effect on cell apoptosis in breast cancer cells. (A–D) MCF-7 and MDA-MB-468 cells were treated without radiation (0 Gy) or with radiation (4 Gy), and 4 Gy + si-NC, or 4 Gy + si-circ_0008500. (A and B) Cell apoptosis was assessed by flow cytometry. (C and D) The protein expression of BAX and BCL2 was detected by western blot. *P < 0.05. Three independent experiments were performed. One-way ANOVA followed by Tukey’s test was used to determine statistical significance
Circ_0008500 Interacted with miR-758-3p
The results showed that circ_0008500 was mainly enriched in the cytoplasm (Fig. 4A, B). Venn diagram displayed that one overlapped miRNA was predicted by circBank and circinteractome (Fig. 4C). CircBank predicted the binding sites between circ_0008500 and miR-758-3p (Fig. 4D). RT-qPCR analysis represented a successful overexpression efficiency of miR-758-3p mimic in MCF-7 and MDA-MB-468 cells (Fig. 4E). Next, dual-luciferase reporter and RIP assays were performed to determine the correlation between circ_0008500 and miR-758-3p. The results of dual-luciferase reporter assay revealed that miR-758-3p mimic remarkably reduced the luciferase activity in WT-circ_0008500 group, while the luciferase activity in MUT-circ_0008500 group was not impacted (Fig. 4F, G). RIP assay showed that circ_0008500 and miR-758-3p were enriched in anti-Ago2 group in comparison to anti-IgG group (Fig. 4H, I). Moreover, the data disclosed that miR-758-3p was significantly downregulated in breast cancer tissues (Fig. 4J). The results in Fig. 4K suggested that miR-758-3p was elevated in TNBC tissues compared with the non-TNBC tissues. Furthermore, miR-758-3p was significantly decreased in ER/PR-positive BC tissues, but not in HER2-positive BC tissues (Fig. 4L–N). In addition, our data showed that miR-758-3p was lower in MCF-7 and MDA-MB-468 cells relative to that in MCF-10A cells (Fig. 4O). As described in Fig. 4P, the expression of miR-758-3p was upregulated by circ_0008500 knockdown in MCF-7 and MDA-MB-468 cells.
Circ_0008500 interacted with miR-758-3p. (A and B) Subcellular fraction assay was used to reveal the subcellular location of circ_0008500 in MCF-7 and MDA-MB-468 cells. (C) Venn diagram was utilized to select the overlapping downstream miRNAs of circ_0008500 through the intersection of circinteractome and circBank databases. (D) The binding sequences between circ_0008500 and miR-758-3p were predicted by circBank, and the mutant sequences of circ_0008500 were constructed. (E) The level of miR-758-3p was detected by RT-qPCR. (F and G) Dual-luciferase reporter assay was devoted to determine the relationship between circ_0008500 and miR-758-3p in breast cancer cells. (H and I) The enrichment of circ_0008500 and miR-758-3p was detected by RIP assay. (J) The expression of miR-758-3p in breast cancer tissues (n = 50) and normal tissues (n = 50) was assessed by RT-qPCR. (K) The expression of miR-758-3p was measured in non-TNBC (n = 26) and TNBC tissues (n = 24) using RT-qPCR. (L) RT-qPCR was used to measure the expression of miR-758-3p in ER-negative (n = 27) or ER-positive (n = 23) BC tissues. (M) RT-qPCR was used to measure the expression of miR-758-3p in PR-negative (n = 28) or PR-positive (n = 22) BC tissues. (N) RT-qPCR was used to measure the expression of miR-758-3p in HER2-negative (n = 25) and HER2-positive (n = 25) BC tissues. (O) RT-qPCR analysis was used to measure the level of miR-758-3p in MCF-10A, MCF-7 and MDA-MB-468 cells. (P) MiR-758-3p expression in MCF-7 and MDA-MB-468 cells transfected with si-NC or si-circ_0008500 was assessed by RT-qPCR analysis. *P < 0.05. Three independent experiments were performed. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
Circ_0008500 Downregulation Repressed Cell Proliferation and Elevated Cell Apoptosis by Targeting miR-758-3p in Breast Cancer Cells
Based on the above results, we further investigated whether miR-758-3p was involved in the function of circ_0008500 in breast cancer development. The data of RT-qPCR analysis indicated that miR-758-3p inhibitor markedly suppressed the expression of miR-758-3p in MCF-7 and MDA-MB-468 cells (Fig. 5A). Further analysis displayed that miR-758-3p inhibitor strikingly counteracted the inhibitory effect on cell viability caused by circ_0008500 absence in breast cancer cells (Fig. 5B). EdU and colony formation assays demonstrated that the inhibition of miR-758-3p significantly blocked circ_0008500 downregulation-induced suppressive impact on cell proliferation (Figs. 5C, D and S1C, D). Besides, circ_0008500 knockdown led to the enhanced cell apoptosis, which was harbored by miR-758-3p inhibitor (Figs. 5E and S1E). Furtherly, upregulation of BAX and downregulation of BCL2 protein expression caused by circ_0008500 knockdown were rescued by inhibition of miR-758-3p in MCF-7 and MDA-MB-468 cells (Fig. 5F, G).
Circ_0008500 downregulation repressed cell proliferation and elevated cell apoptosis by targeting miR-758-3p in breast cancer cells. (A) The level of miR-758-3p in MCF-7 and MDA-MB-468 cells transfected with inhibitor NC or miR-758-3p inhibitor was detected by RT-qPCR. (B-G) MCF-7 and MDA-MB-468 cells were transfected with si-NC, si-circ_0008500, si-circ_0008500 + inhibitor NC, or si-circ_0008500 + miR-758-3p inhibitor. (B) Cell viability was measured by CCK-8 assay. (C) Cell proliferation was detected by EdU assay. (D) The number of colonies was assessed by colony formation assay. (E) Flow cytometry was used to measure cell apoptosis. (F and G) Western blot was utilized to detect the protein expression of BAX and BCL2. *P < 0.05. Three independent experiments were performed. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
PFN2 Was a Target of miR-758-3p and circ_0008500 Regulated PFN2 Expression Through Targeting miR-758-3p
Next, we further studied the downstream regulator of miR-758-3p in the development of breast cancer. Starbase predicted the potential binding sites between miR-758-3p and PFN2 (Fig. 6A). Dual-luciferase reporter assay indicated that the luciferase activity was remarkably reduced by miR-758-3p mimic in WT-PFN2 3’UTR group, but not in MUT-PFN2 3’UTR group (Fig. 6B, C). Moreover, the relationship between miR-758-3p and PFN2 was also verified by RIP assay (Fig. 6D, E). RT-qPCR analysis revealed that PFN2 was highly expressed in breast cancer tissues (n = 50) compared with the normal tissues (n = 50) (Fig. 6F). Moreover, PFN2 was decreased in TNBC tissues in comparison to the non-TNBC tissues (Fig. 6G). The upregulated PFN2 expression was observed in ER/PR-positive BC tissues, while the expression of PFN2 showed no significant difference between HER2-positive and HER2-negative BC tissues (Fig. 6H–J). Western blot showed that the protein expression of PFN2 was significantly increased in breast cancer tissues and cells (Fig. 6K, L). The data in Fig. 6M suggested that miR-758-3p mimic markedly repressed PFN2 protein expression in MCF-7 and MDA-MB-468 cells. Additionally, circ_0008500 knockdown suppressed the protein expression of PFN2 by targeting miR-758-3p (Fig. 6N).
PFN2 was a target of miR-758-3p and circ_0008500 regulated PFN2 expression through targeting miR-758-3p. (A) Starbase predicted the putative complementary sites between miR-758-3p and PFN2. (B-E) Dual-luciferase reporter (B and C) and RIP (D and E) assays were performed to verify the relationship between miR-758-3p and PFN2. (F) The expression of PFN2 in breast cancer tissues (n = 50) and normal tissues (n = 50) were measured by RT-qPCR. (G) The level of PFN2 was determined in non-TNBC (n = 26) and TNBC (n = 24) tissues using RT-qPCR. (H) RT-qPCR was used to measure the expression of PFN2 in ER-negative (n = 27) or ER-positive (n = 23) BC tissues. (I) RT-qPCR was used to measure the expression of PFN2 in PR-negative (n = 28) or PR-positive (n = 22) BC tissues. (J) RT-qPCR was used to measure the expression of PFN2 in HER2-negative (n = 25) and HER2-positive (n = 25) BC tissues. (K) The protein expression of PFN2 in breast cancer tissues and normal tissues were measured by western blot. (L) Western blot was utilized to evaluate the protein expression of PFN2 in MCF-10A, MCF-7 and MDA-MB-468 cells. (M) PFN2 protein expression was detected by western blot in MCF-7 and MDA-MB-468 cells transfected with miRNA NC or miR-758-3p mimic. (N) PFN2 protein expression was assessed by western blot in MCF-7 and MDA-MB-468 cells with transfection of si-NC, si-circ_0008500, si-circ_0008500 + inhibitor NC, or si-circ_0008500 + miR-758-3p inhibitor. *P < 0.05. Three independent experiments were performed. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
MiR-758-3p Inhibited Cell Proliferation, and Enhanced Cell Apoptosis by Regulating PFN2 in Breast Cancer Cells
The results from western blot showed that the protein expression of PFN2 was significantly upregulated in MCF-7 and MDA-MB-468 cells transfected with pc-PFN2 (Fig. 7A). Functionally, overexpression of PFN2 strikingly blocked miR-758-3p mimic-induced repression on cell viability (Fig. 7B) and proliferation (Figs. 7C, D, and S2C, D) in MCF-7 and MDA-MB-468 cells. In addition, miR-758-3p mimic-caused promotion on cell apoptosis was reversed by PFN2 overexpression (Figs. 7E and S2E), as evidenced by the decreased BAX and the increased BCL2 (Fig. 7F, G).
MiR-758-3p inhibited cell proliferation and accelerated cell apoptosis by regulating PFN2 in breast cancer cells. (A) The protein expression of PFN2 was detected by western blot in MCF-7 and MDA-MB-468 cells transfected with pc-NC or pc-PFN2. (B-G) MCF-7 and MDA-MB-468 cells were transfected with miRNA NC, miR-758-3p mimic, miR-758-3p mimic + pc-NC, or miR-758-3p mimic + pc-PFN2. (B) CCK-8 assay was used to measure cell viability. (C and D) Cell proliferation was measured by EdU (C) and colony formation (D) assays. (E) Flow cytometry was employed to determine cell apoptosis. (F and G) The protein expression of BAX and BCL2 was assessed by western blot. *P < 0.05. Three independent experiments were performed. Student’s t test and one-way ANOVA followed by Tukey’s test were used to determine statistical significance
Circ_0008500 Knockdown Inhibited Tumor Growth and Elevated the Radiosensitivity in Breast Cancer in Vivo
To further verify the functional role of circ_0008500 in breast cancer in vivo, the xenograft tumor assay was employed. As displayed in Figs. 8A, B and S3A, B, tumor volume and weight were significantly repressed by circ_0008500 downregulation or radiation treatment, and a more remarkable suppression of tumor volume and weight was found in sh-circ_0008500 transduction + radiation treatment group compared with the radiation treatment group. Moreover, the data indicated that circ_0008500 knockdown enhanced miR-758-3p expression and inhibited circ_0008500 and PFN2 expression in the tissues extracted from the mice. Additionally, circ_0008500 knockdown aggravated the inhibition effect of radiation treatment on circ_0008500 and PFN2 expression, as well as promotion effect on miR-758-3p expression (Figs. 8C–F and S3C-F). Furthermore, IHC assay suggested that PFN2 was reduced in sh-circ_0008500 or radiation treatment group, and circ_0008500 knockdown enhanced the inhibition impact of radiation treatment on PFN2 expression (Figs. 8G and S3G).
Circ_0008500 knockdown inhibited tumor growth and elevated radiosensitivity in breast cancer in vivo. (A–F) The mice were injected with 1 × 106 stably modified (circ_0008500 knockdown) or control (sh-NC) MCF-7 cells and divided into four groups: sh-NC group, sh-circ_0008500 group, sh-NC + radiation group, and sh-circ_0008500 + radiation group. (A and B) Tumor volume (A) and weight (B) were detected. (C and D) The expression of circ_0008500 (C) and miR-758-3p (D) was measured using RT-qPCR. (E–F) The mRNA and protein expression of PFN2 was determined by RT-qPCR and western blot, respectively. (G) Expressed PFN2 in tumor tissues from mice was examined by IHC assay, and the scale bar was 100 μm. *P < 0.05. Three independent experiments were performed. One-way ANOVA followed by Tukey’s test was used to determine statistical significance
Discussion
CircRNAs, a new type of ncRNAs, are regarded as the aberrant splicing byproducts and exert function by regulating gene expression [24]. Moreover, circRNAs acted as the major regulators in the biological behaviors of human cancers, such as cell multiplication, metastasis and apoptosis [25]. Accumulating evidence revealed that circRNAs could modulate the sensitivity to irradiation treatment in various cancers [26, 27]. A recent research reported that circ_0008500 was a apparently upregulated circRNA in breast cancer [23]. However, the impacts of circ_0008500 on the biological behaviors and radiosensitivity in breast cancer have not been investigated. In our research, we demonstrated that circ_0008500 regulated the sensitivity to radiation treatment and development in breast cancer through the miR-758-3p/PFN2 axis.
Previous researches displayed that the aberrantly expressed circRNAs played major roles in the development of various malignancies, including breast cancer [28, 29]. It was reported that the radioresistance of human malignancies could be modulated by circRNAs, such as hsa_0009035 [30], circRNA Cyclin B2 [31], and circ_0086720 [16]. A recent study showed that exosome-transmitted circ_IFT80 boosted the radioresistance and tumorigenesis in colorectal cancer by increasing MSI1 expression via sponging miR-296-5p [32]. Circ_0001313, a remarkably increased circRNA in radio-resistant colon cancer, repressed the radiosensitivity in colon cancer through regulating the expression of tumor inhibitor miR-338-3p [33]. Moreover, a few circRNAs were proved to modulate breast cancer development, like hsa_0053063 [34], hsa_circ_0072995 [35], hsa_circ_0008039 [36]. Here, the role of circ_0008500 in modulating the development and radiosensitivity in breast cancer was revealed. Our data suggested that circ_0008500 was markedly upregulated in breast cancer tissues and cells, consistent with the previous results [23]. Furthermore, we observed that radiation treatment decreased circ_0008500 level in breast cancer. However, the regulatory mechanism of irradiation-mediated circ_0008500 downregulation is unclear. We speculated that irradiation might inhibit the transcription of genes or regulate the expression of the upstream factors, thereby reducing the level of circ_0008500. Moreover, our study demonstrated that circ_0008500 knockdown inhibited cell multiplication and accelerated cell apoptosis and radiosensitivity in breast cancer. Additionally, our results presented that knockdown of circ_0008500 elevated radiation-induced inhibitory impact on tumor growth in vivo. These findings explained that circ_0008500 functioned as a promoter in breast cancer development and knockdown of circ_0008500 improved the sensitivity to radiation treatment in breast cancer.
CircRNAs exerted action through different ways, like acting as the competing endogenous RNAs for miRNAs [37]. In the present research, circBank predicted the potential complementary sites between miR-758-3p and circ_0008500, and the correlation between them was demonstrated. MiR-758-3p was verified as an inhibitor in some human cancers, such as cervical cancer and ovarian cancer [38, 39]. By targeting NOTCH2, cell metastasis and the proliferative ability were dramatically restrained by miR-758-3p in bladder cancer cells [40]. In clear cell renal cell carcinoma, a recent study disclosed that miR-758-3p predicted the great prognosis and suppressed cell growth and metastasis [41]. MiR-758-3p, as a downstream gene of lncRNA DANCR, was lowly expressed and weakened cell autophagy and apoptosis in breast cancer [42]. Consistently, we also found that miR-758-3p was decreased in breast cancer, and circ_0008500 directly interacted with miR-758-3p, thereby modulating its expression. In addition, circ_0008500 knockdown caused inhibition impact on cell proliferation and promotion impact on cell apoptosis and radiosensitivity by upregulating miR-758-3p in vitro.
Accumulating evidence indicated that miRNAs degrade the expression of the target mRNA through directly binding to the mRNA 3’UTR, thereby exerting actions [43]. PFN2, a actin-binding protein, is a member of profilins that participate in regulating cytoskeletal dynamics [44]. Recent evidence revealed that PFN2 functioned as the potential prognosis biomarker and therapeutic target in several cancers, including breast cancer [45, 46]. Due to the oncogenic property of PFN2 in a variety of cancers, including osteosarcoma as well as head and neck squamous cell carcinoma [47, 48], PFN2 might be a potential downstream effector of miR-758-3p function. Our data showed that miR-758-3p was verified to target PFN2, and PFN2 was a vital mediator of miR-758-3p function in breast cancer cells. Former literature suggested that overexpression of PFN2 promoted cell proliferation, epithelial-to-mesenchymal transition and metastasis in triple negative breast cancer cells in vitro and showed stronger tumorigenicity in vivo [49]. PFN2, as a downstream target of miR-223-3p, was proved to aggravate the radioresistance and progression of breast cancer through enhancing glycolysis [50]. Furthermore, a previous study showed PFN2 was related to the radiosensitivity in NCI-60 cells [51]. In this research, we observed that PFN2 was elevated in breast cancer. In addition, miR-758-3p played the suppressive role in the development of breast cancer by targeting PFN2.
To summarize, our findings verified the upregulation of circ_0008500 and PFN2, as well as the downregulation of miR-758-3p in breast cancer, and demonstrated that circ_0008500 knockdown downregulated PFN2 by targeting miR-758-3p to enhance breast cancer radiosensitivity and inhibit breast cancer development, indicating that the circ_0008500/miR-758-3p/axis regulatory pathway might contribute to improve the therapy methods of breast cancer.
References
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, et al. Breast cancer Nat Rev Dis Primers. 2019;5(1):66.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Benvenuto M, Focaccetti C, Izzi V, Masuelli L, Modesti A, Bei R. Tumor antigens heterogeneity and immune response-targeting neoantigens in breast cancer. Semin Cancer Biol. 2021;72:65–75.
DeSantis CE, Ma J, Gaudet MM, Newman LA, Miller KD, Goding Sauer A, et al. (2019) Breast cancer statistics. CA Cancer J Clin. 2019;69(6):438–51.
Mahvi DA, Liu R, Grinstaff MW, Colson YL, Raut CP. Local Cancer Recurrence: The Realities, Challenges, and Opportunities for New Therapies. CA Cancer J Clin. 2018;68(6):488–505.
Zheng R, Yao Q, Xie G, Du S, Ren C, Wang Y, et al. TAT-ODD-p53 enhances the radiosensitivity of hypoxic breast cancer cells by inhibiting Parkin-mediated mitophagy. Oncotarget. 2015;6(19):17417–29.
Sio TT, Joliat GR, Jrebi N. Multidisciplinary approach to uncommon, widely metastatic breast cancer. Eur Rev Med Pharmacol Sci. 2014;18(6):846–50.
Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer. 2018;18(1):5–18.
Yin Y, Long J, He Q, Li Y, Liao Y, He P, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–21.
Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–8.
Wang J, Du Y, Liu X, Cho WC, Yang Y. MicroRNAs as regulator of signaling networks in metastatic colon cancer. Biomed Res Int. 2015:823620.
Hammond SM. An overview of microRNAs. Adv Drug Deliv Rev. 2015;87:3–14.
Memczak S, Papavasileiou P, Peters O, Rajewsky N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One. 2015;10(10):e0141214.
Wu D, Jia H, Zhang Z, Li S. Circ-PRMT5 promotes breast cancer by the miR-509–3p/TCF7L2 axis activating the PI3K/AKT pathway. J Gene Med 2021;23(2):e3300.
Du S, Zhang P, Ren W, Yang F, Du C. Circ-ZNF609 Accelerates the Radioresistance of Prostate Cancer Cells by Promoting the Glycolytic Metabolism Through miR-501-3p/HK2 Axis. Cancer Manag Res. 2020;12:7487–99.
Jin Y, Su Z, Sheng H, Li K, Yang B, Li S. Circ_0086720 knockdown strengthens the radiosensitivity of non-small cell lung cancer via mediating the miR-375/SPIN1 axis. Neoplasma. 2021;68(1):96–107.
Zhang T, Wu DM, Deng SH, Han R, Liu T, Li J, et al. RNAseq profiling of circRNA expression in radiation-treated A549 cells and bioinformatics analysis of radiation-related circRNA-miRNA networks. Oncol Lett. 2020;20(2):1557–66.
Bakhtari N, Mozdarani H, Salimi M, Omranipour R. Association study of miR-22 and miR-335 expression levels and G2 assay related inherent radiosensitivity in peripheral blood of ductal carcinoma breast cancer patients. Neoplasma. 2021;68(1):190–9.
Perez-Anorve IX, Gonzalez-De la Rosa CH, Soto-Reyes E, Beltran-Anaya FO, Del Moral-Hernandez O, Salgado-Albarran M, et al. New insights into radioresistance in breast cancer identify a dual function of miR-122 as a tumor suppressor and oncomiR. Mol Oncol. 2019;13(5):1249–67.
Zhang X, Li Y, Wang D, Wei X. miR-22 suppresses tumorigenesis and improves radiosensitivity of breast cancer cells by targeting Sirt1. Biol Res. 2017;50(1):27.
Sun Q, Liu T, Yuan Y, Guo Z, Xie G, Du S, et al. MiR-200c inhibits autophagy and enhances radiosensitivity in breast cancer cells by targeting UBQLN1. Int J Cancer. 2015;136(5):1003–12.
Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175.
Hu Y, Song Q, Zhao J, Ruan J, He F, Yang X, et al. Identification of plasma hsa_circ_0008673 expression as a potential biomarker and tumor regulator of breast cancer. J Clin Lab Anal. 2020;34(9):e23393.
Xu Z, Li P, Fan L, Wu M. The Potential Role of circRNA in Tumor Immunity Regulation and Immunotherapy. Front Immunol. 2018;9:9.
Yu CY, Kuo HC. The emerging roles and functions of circular RNAs and their generation. J Biomed Sci. 2019;26(1):29.
Yu D, Li Y, Ming Z, Wang H, Dong Z, Qiu L, et al. Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. PeerJ 2018;6:e5011.
Chen Y, Yuan B, Wu Z, Dong Y, Zhang L, Zeng Z. Microarray profiling of circular RNAs and the potential regulatory role of hsa_circ_0071410 in the activated human hepatic stellate cell induced by irradiation. Gene. 2017;629:35–42.
Pan G, Mao A, Liu J, Lu J, Ding J, Liu W. Circular RNA hsa_circ_0061825 (circ-TFF1) contributes to breast cancer progression through targeting miR-326/TFF1 signalling. Cell Prolif. 2020;53(2):e12720.
Zhang XY, Mao L. Circular RNA Circ_0000442 acts as a sponge of MiR-148b-3p to suppress breast cancer via PTEN/PI3K/Akt signaling pathway. Gene. 2021;766:145113.
Zhao X, Dong W, Luo G, Xie J, Liu J, Yu F. Silencing of hsa_circ_0009035 Suppresses Cervical Cancer Progression and Enhances Radiosensitivity through MicroRNA 889–3p-Dependent Regulation of HOXB7. Mol Cell Biol. 2021;41(6):e0063120.
Cai F, Li J, Zhang J, Huang S. Knockdown of Circ_CCNB2 Sensitizes Prostate Cancer to Radiation Through Repressing Autophagy by the miR-30b-5p/KIF18A Axis. Cancer Biother Radiopharm. 2020. https://doi.org/10.1089/cbr.2019.3538.
Li L, Jiang Z, Zou X, Hao T. Exosomal circ_IFT80 Enhances Tumorigenesis and Suppresses Radiosensitivity in Colorectal Cancer by Regulating miR-296-5p/MSI1 Axis. Cancer Manag Res. 2021;13:1929–41.
Wang L, Peng X, Lu X, Wei Q, Chen M, Liu L. Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol Res Pract. 2019;215(4):689–96.
Ji C, Hu J, Wang X, Zheng W, Deng X, Song H, et al. Hsa_circ_0053063 inhibits breast cancer cell proliferation via hsa_circ_0053063/hsa-miR-330-3p/PDCD4 axis. Aging (Albany NY). 2021;13(7):9627–45.
Zhang HD, Jiang LH, Hou JC, Zhou SY, Zhong SL, Zhu LP, et al. Circular RNA hsa_circ_0072995 promotes breast cancer cell migration and invasion through sponge for miR-30c-2-3p. Epigenomics. 2018;10(9):1229–42.
Liu Y, Lu C, Zhou Y, Zhang Z, Sun L. Circular RNA hsa_circ_0008039 promotes breast cancer cell proliferation and migration by regulating miR-432-5p/E2F3 axis. Biochem Biophys Res Commun. 2018;502(3):358–63.
Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–8.
Xie H, Wang J, Wang B. Circular RNA Circ_0003221 Promotes Cervical Cancer Progression by Regulating miR-758-3p/CPEB4 Axis. Cancer Manag Res. 2021;13:5337–50.
Hu X, Li Y, Kong D, Hu L, Liu D, Wu J. Long noncoding RNA CASC9 promotes LIN7A expression via miR-758-3p to facilitate the malignancy of ovarian cancer. J Cell Physiol. 2019;234(7):10800–8.
Wu X, Chen B, Shi H, Zhou J, Zhou F, Cao J, et al. miR-758-3p suppresses human bladder cancer cell proliferation, migration and invasion by targeting NOTCH2. Exp Ther Med. 2019;17(5):4273–8.
Wu Y, Liu Y. miR-758-3p Inhibits Proliferation, Migration, and Invasion of Clear Cell Renal Cell Carcinoma and Predicts Favorable Prognosis. Cancer Manag Res. 2020;12:9285–95.
Zhang XH, Li BF, Ding J, Shi L, Ren HM, Liu K, et al. LncRNA DANCR-miR-758-3p-PAX6 Molecular Network Regulates Apoptosis and Autophagy of Breast Cancer Cells. Cancer Manag Res. 2020;12:4073–84.
Cai Y, Yu X, Hu S, Yu J. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–54.
Pandey DK, Chaudhary B. Evolutionary expansion and structural functionalism of the ancient family of profilin proteins. Gene. 2017;626:70–86.
Cui XB, Zhang SM, Xu YX, Dang HW, Liu CX, Wang LH, et al. PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Transl Med. 2016;14(1):137.
Jiang M, Qiu N, Xia H, Liang H, Li H, Ao X. Long noncoding RNA FOXD2AS1/miR1505p/PFN2 axis regulates breast cancer malignancy and tumorigenesis. Int J Oncol. 2019;54(3):1043–52.
Liu J, Wu Y, Wang Q, Liu X, Liao X, Pan J. Bioinformatic analysis of PFN2 dysregulation and its prognostic value in head and neck squamous carcinoma. Future Oncol. 2018;14(5):449–59.
Zhao ZY, Zhao YC, Liu W. Long non-coding RNA TUG1 regulates the progression and metastasis of osteosarcoma cells via miR-140-5p/PFN2 axis. Eur Rev Med Pharmacol Sci. 2019;23(22):9781–92.
Ling Y, Cao Q, Liu Y, Zhao J, Zhao Y, Li K, et al. Profilin 2 (PFN2) promotes the proliferation, migration, invasion and epithelial-to-mesenchymal transition of triple negative breast cancer cells. Breast Cancer. 2021;28(2):368–78.
Zhao Y, Zhong R, Deng C, Zhou Z. Circle RNA circABCB10 Modulates PFN2 to Promote Breast Cancer Progression, as Well as Aggravate Radioresistance Through Facilitating Glycolytic Metabolism Via miR-223-3p. Cancer Biother Radiopharm. 2021;36(6):477–90.
Kim HS, Kim SC, Kim SJ, Park CH, Jeung HC, Kim YB, et al. Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells. BMC Genomics. 2012;13:348.
Funding
This study was supported by Scientific Research Fund Project of Hebei Provincial Health and Family Planning Commission. (No. 20201103).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no financial conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kong, D., Shen, D., Liu, Z. et al. Circ_0008500 Knockdown Improves Radiosensitivity and Inhibits Tumorigenesis in Breast Cancer Through the miR-758-3p/PFN2 Axis. J Mammary Gland Biol Neoplasia 27, 37–52 (2022). https://doi.org/10.1007/s10911-022-09514-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10911-022-09514-w