Abstract
Numerous studies have identified that circular RNAs (circRNAs) functioned as important regulators in tumor initiation, carcinogenesis, drug or radiation resistance. This study aims to reveal the function of circ_0008344 on radiosensitivity in glioma. The quantitative real-time polymerase chain reaction (qRT-PCR) was implemented for detecting the circ_0008344 and microRNA-433-3p (miR-433-3p) levels. Cell survival intensity and apoptosis were analyzed through colony formation assay and flow cytometry respectively. The protein levels were examined via Western blot. Dual-luciferase reporter assay was exploited for the analysis of target combination. Xenograft models were established in mice for circ_0008344 research in vivo. Our data showed that circ_0008344 level was signally increased in radioresistant glioma tissues and its down-regulation facilitated the susceptibility of glioma cells to radiation. Additionally, we found that circ_0008344 could interact with miR-433-3p and regulated radiosensitivity of glioma cells via sponging miR-433-3p. Ring finger protein 2 (RNF2) was proved to be a target of miR-433-3p and it was regulated by circ_0008344/miR-433-3p axis. The promotion of circ_0008344 knockdown on radiosensitivity was counteracted by RNF2 overexpression in glioma cells. Further experiment in vivo indicated that circ_0008344 down-regulation inhibited glioma growth and acted on miR-433-3p/RNF2 axis to enhance the radiosensitivity in glioma. These evidences manifested that knockdown of circ_0008344 exerted the radiosensitivity-promoting effect on glioma via the miR-433-3p-mediated RNF2 down-regulation, identifying circ_0008344 as a novel diagnostic biomarker of radioresistance and therapeutic target in glioma radiotherapy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Glioma originates from (precursor of) glial cells is one of the most common malignant tumors in the central nervous system (CNS) (Lenting et al. 2017). Gliomas are histologically divided into Grades I to IV, oligodendroglioma (Grade II and III), anaplastic astrocytoma (Grade III) and glioblastoma multiform (GBM, Grade IV) are considered as serious (Gladson et al. 2010). Radiotherapy has been used as an effective treatment in the management of glioma (Hu et al. 2016; Tabatabaei et al. 2017), while the radiotherapeutic effect is generally limited because of the acquired radioresistance (Han et al. 2017). Hence, understanding the radioresistance mechanism and discovering the sensitized molecules are especially important.
As a novel type of non-coding RNAs (ncRNAs) with the absence of protein-coding capacity, circular RNAs (circRNAs) have emerged as essential regulators in cancer formation and progression (Yin et al. 2019). For example, Huang et al. reported that circ_0008039 promoted cell proliferation and metastasis in breast cancer (Huang et al. 2020); Wu et al. asserted that circ_0005075 overexpression blocked the carcinogenesis of gastric cancer (Wu et al. 2020). CircRNAs also have great values in diagnosis and treatment of glioma (Sun et al. 2020). A recent study exhibited that circ_VCAN was increased in radioresistant glioma tissues and inhibited the radiosensitivity of glioma as a tumor-promoting factor (Zhu et al. 2020). Circ_0008344 was also up-regulated and acted as an oncogene in glioma (Zhou et al. 2018), but its role in regulating glioma radiosensitivity needs further exploration.
MicroRNAs (miRNAs) are small endogenous regulatory ncRNAs in various biological behaviors via regulating gene expression through combining with 3′ untranslated regions (3′UTRs) of target mRNA (Pu et al. 2019). Sun et al. found that microRNA-433-3p (miR-433-3p) played an anti-tumor role in glioma and promoted chemosensitivity by targeting CREB (Sun et al. 2017). It is unclear whether miR-433-3p exerts the vital effect on radiosensitivity in glioma. Ring finger protein 2 (RNF2) is a fundamental component of polycomb protein family related to histone 2A modification (Chrispijn et al. 2019). Knockdown of RNF2 could increase the radiosensitivity in squamous cell carcinoma (Yang et al. 2019) and glioma (Zhou et al. 2014). However, the relation between miR-433-3p and RNF2 is ill-defined.
In terms of functional mechanism, circRNAs usually function as the ‘miRNA sponges’ to regulate the expression of genes in tumor development (Panda 2018; Qin et al. 2020). Except for the biological function of circ_0008344 in glioma radiosensitivity, we will also elaborate on the relationship among circ_0008344, miR-433-3p and RNF2 to exploit a representative mechanism in radioresistance of glioma.
2 Materials and methods
2.1 Patient specimens
This study was approved by the Ethical Committee of Beijing You’an Hospital, Capital Medical University, and all patients have signed the written informed consent. The normal brain tissues (n = 40) were collected from patients who suffered the excision of brain tissues due to other brain diseases except glioma, and glioma samples (n = 40) were acquired from glioma patients received radiotherapy during the surgical resection at Beijing You’an Hospital, Capital Medical University. After radiotherapy for three months, 23 patients were found with local complete response (CR, lesion disappearance) or partial response (PR, baseline lesions shrinkage ≥ 30%) and 17 patients were found with progressive disease (PD, baseline lesions increase ≥ 20%) or stable disease (SD, baseline lesions between PR and PD). These glioma tissues were classified into radiosensitive group (n = 23) and radioresistant group (n = 17) in compliance with the Response Evaluation Criteria In Solid Tumors (RECIST) criteria (Pierson et al. 2018). After tissue collection from patients, the specimens must be immediately preserved in liquid nitrogen.
2.2 Cell culture and X-ray irradiation
Human normal astrocytes NHA and glioma cell lines (U251 and LN229) were acquired from QCHENG BIO (Shanghai, China), then cultured in humid atmosphere at 37°C with 5% CO2. Dulbecco’s modified eagle medium (DMEM; Hyclone, Logan, UT, USA) containing 10% fetal bovine serum (FBS; Gibco, Carlsbad, CA, USA) and 1% Penicillin-Streptomycin solution (10000 unit/mL of penicillin and 10000 µg/mL of streptomycin; Gibco) was used as the complete medium. For radiation treatment, U251 and LN229 cells were exposed to X-ray irradiation using a linear accelerator (Faxitron Bioptics LLC, San Francisco, CA, USA) with the dose rate of 2 Gy/min.
2.3 Cell transfection
Oligonucleotides or the constructed vectors were transfected into U251 and LN229 cells using Lipofectamine3000 reagent (Invitrogen, Carlsbad, CA, USA), following the detailed procedures provided by the manufacture. The used oligonucleotides (RIBIBIO, Guangzhou, China) included small interfering RNA (siRNA) targeting circ_0008344 (si-circ#1: 5′-ACTAGACATGAGCCTTTGAAT-3′, si-circ#2: 5′-AGACACTAGACATGAGCCTTT-3′ and si-circ#3: 5′-AGACATGAGCCTTTGAATACA-3′), short hairpin RNA (shRNA) targeting circ_0008344 (sh-circ: 5′-GCCAAGCCAGTTCCATTAAAT-3′), miR-433-3p mimic and inhibitor (miR-433-3p: 5′-AUCAUGAUGGGCUCCUCGGUGU-3′ and anti-miR-433-3p: 5′-UCAACAUCAGUCUGAUAAGCUA-3′), as well as the respective negative controls (si-NC, sh-NC, miR-NC and anti-NC). The sequences of circ_0008344 and RNF2 were separately cloned into the pCE-RB-Mam (RIBIBIO) and pcDNA (Invitrogen) expression vectors, generating the recombined vectors pCE-RB-Mam-circ_0008344 (circ_0008344) and pcDNA-RNF2 (RNF2).
2.4 Analysis of quantitative real-time polymerase chain reaction (qRT-PCR)
After the isolation of total RNA from tissues and cells by TRIzol™ Reagent (Invitrogen), the complementary DNA (cDNA) was synthesized (2 µg RNA/sample) through ReverTra Ace® qPCR RT Master Mix (Toyobo, Kita-ku, Osaka, Japan) followed by the operation of qRT-PCR via Roche LightCycler 480 system (Roche, Basle, Switzerland) using SYBR® Green Real-time PCR Master Mix (Toyobo). The forward (F) and reverse (R) primer used for qRT-PCR were as follows: circ_0008344: 5′-TGGTCGAAGTCAGCAGACAC-3′ (F) and 5′-GCAGGAGGTAATGACGGAAG-3′ (R); miR-433-3p: 5′-GGAGAAGTACGGTGAGCCTGT-3′ (F) and 5′-GAACACCGAGGAGCCCATCAT-3′ (R); β-actin: 5′-TGGCATCCACGAAACTACC-3′ (F) and 5′-GTGTTGGCGTACAGGTCTT-3′ (R); U6: 5′-CTCGCTTCGGCAGCACA-3′ (F) and 5′- AACGCTTCACGAATTTGCGT-3′ (R). The results were analyzed using the comparative cycle threshold method (2−∆∆Ct) with β-actin (for circ_0008344) and U6 (for miR-433-3p) as the reference genes.
2.5 Colony formation assay
After transfection and treatment with X-ray irradiation for respective 0 min, 1 min, 2 min and 4 min (0 Gy, 2 Gy, 4 Gy and 8 Gy), 3 × 102 harvested cells were transplanted into each well of the 6-well plates to be incubated for 12 d at 37°C. Then 4% paraformaldehyde (Invitrogen) was added into the well-plates to fasten the colonies and 0.1% crystal violet (Thermo Fisher Scientific, Waltham, MA, USA) was used to stain the colonies, followed by the counting of the colonies under a microscope (Thermo Fisher Scientific). Ultimately, the survival fraction intensity was calculated by the ratio of colony number and seed cell number.
2.6 Flow cytometry
FITC Annexin V staining is typically used for apoptosis detection in conjunction with a pivotal dye propidium iodide (PI). After different transfection for 72 h and radiation for 2 min (4 Gy), cells were washed by cold-phosphate buffer solution (PBS; Hyclone) twice. FITC Annexin V and PI staining was then performed via the FITC Annexin V Kit (BD Biosciences, San Diego, CA, USA) following the user’s guideline. After staining for 20 min, a flow cytometer (BD Biosciences) was applied for the identification of apoptotic cells. Specially, the Annexin V+/PI- and Annexin V+/PI+ labelled cells were recognized as the apoptotic cells.
2.7 Western blot
Western blot was conducted for protein expression analysis. Cells (transfection for 48 h and radiation for 2 min) were washed twice using PBS (Hyclone) and collected by centrifugation at 500g for 15 min, followed by protein lysis in RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific) and quantification by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific). Whereafter, 40 µg proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on NuPAGE™ 10% Bis-Tris Midi Protein Gels (Invitrogen) for 2 h, and transferred onto the PVDF Transfer Membrane (0.45 µm; Thermo Fisher Scientific). After the non-specific protein binding was blocked by Membrane Blocking Solution (Invitrogen), the membranes were subjected to the incubation of antibodies (primary antibodies at 4°C overnight and secondary antibody at room temperature for 45 min). These antibodies were bought from Abcam (Cambridge, UK): anti-Cleaved-caspase-3 (anti-Cleaved-casp3; ab2302, 1:1000), anti-phosphorylated H2AX (anti-γH2AX; ab2893, 1:1000), anti-RNF2 (ab101273, 1:2000), anti-β-actin (ab8227, 1:3000) and anti-Rabbit IgG H&L (HRP) (ab205718, 1:5000). Then ECL Substrate Kit (Abcam) was used for the presence of proteins on the washed membranes, and the grey level analysis was performed via ImageLab software version 4.1 (Bio-Rad, Hercules, CA, USA).
2.8 Dual-luciferase reporter assay
Construction of the luciferase plasmids was implemented by cloning the wild-type (WT) and mutant-type (MUT) sequences of circ_0008344 and RNF2 3′UTR into the pMIR-REPORT vector (Promega, Madison, WI, USA). The positive plasmids were named as circ_0008344 WT, circ_0008344 MUT, RNF2 3′UTR WT and RNF2 3′UTR MUT. U251 and LN229 cells were co-transfected with the above constructed each plasmid along with miR-433-3p or miR-NC. Following transfection for 48 h, cells were lysed in Passive Buffer (Promega) and the relative luciferase activity was measured using the dual-luciferase reporter system (Promega). The renilla luciferase activity was used to normalize the firefly luciferase activity.
2.9 In vivo experiment
Male BALB/c nude mice (6-week-old, n = 10) were purchased from Shanghai SIPPR-BK Laboratory Animal Co. Ltd. (Shanghai, China) and arbitrarily divided into two groups with 5 mice/group. 3 × 106 U251 cells transfected with sh-NC or sh-circ via lentiviral vector were subcutaneously injected into the mice. When the tumor volume (length × width2 × 0.5) grew to 100 mm3, mice in sh-NC and sh-circ groups were given radiation exposure for 3 min (6 Gy), followed by the measurement of tumor volume per 5 days. After 30 days, these mice were sacrificed for tumor separation. Tumor weight was measured on an electronic scale, then the associated RNA and protein changes were detected after RNA and protein extraction. All mice were tenderly cared and this assay was approved by the Animal Ethical Committee of Beijing You’an Hospital, Capital Medical University.
2.10 Statistical analysis
Data from three independent experiments were expressed as mean ± standard deviation (SD), then analyzed using SPSS 24.0 (IBM Corp., Armonk, NY, USA) and Graphpad prism 7 (GraphPad Inc., La Jolla, CA, USA). The survival curves of patients were generated by the Kaplan-Meier plot and analyzed by log-rank test. The linear relation between circ_0008344 and miR-433-3p was analyzed via Pearson’s correlation coefficient. Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test was performed for difference analysis. Statistically, P < 0.01 was defined as the significant difference.
3 Results
3.1 Circ_0008344 was up-regulated in radioresistant glioma tissues
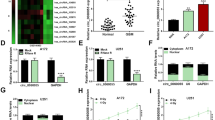
By qRT-PCR analysis and data calculation in figure 1A, we affirmed the up-regulation of circ_0008344 in glioma tissues (n = 40) by comparison with those normal brain tissues (n = 40). Among these glioma tissues, circ_0008344 expression was higher in radioresistant samples (n = 17) than radiosensitive samples (n = 23) (figure 1B). According to the median value (3.036) of circ_0008344 expression, 40 glioma tissues were divided into high expression group (n = 20) and low expression group (n = 20). Log-rank test indicated that the overall survival of high expression group was much lower than that of low expression group (P<0.05), implicating that high circ_0008344 could predict poor prognosis in glioma patients (figure 1C). Relative to NHA cells, circ_0008344 level in U251 and LN229 cells was increased by more than 3-fold changes (figure 1D). High circ_0008344 was related to poor prognosis and radioresistance.
Circ_0008344 was up-regulated in radioresistant glioma tissues. (A-B) Circ_0008344 level was assayed by qRT-PCR in glioma/normal tissues (A) and radioresistant/radiosensitive tissues (B). (C) The survival curves of high and low circ_0008344 expression groups were generated by Kaplan-Meier plot and analyzed using log-rank test. (D) The qRT-PCR was administrated for circ_0008344 detection in U251, LN229 and NHA cells. **P < 0.01, ***P < 0.001.
3.2 Knockdown of circ_0008344 made glioma cells more sensitive to radiation
Given the abnormal high expression of circ_0008344 in radioresistant glioma tissues, we further studied the potential role of circ_0008344 in radiosensitivity. The siRNAs targeting circ_0008344 (si-circ#1, si-circ#2 and si-circ#3) were severally transfected into U251 and LN229 cells, and their inhibitory efficiencies were all significant in contrast to si-NC control group (figure 2A-B). The most significant si-circ#1 was chosen for the subsequent analysis of biological processes. After exposure to different doses of radiation (0 Gy, 2 Gy, 4 Gy and 8 Gy), colony formation assay revealed that cell survival fraction intensity in si-circ#1 group was prominently decreased compared with si-NC group in U251 (figure 2C) and LN229 (figure 2D) cells with the increasing of radiation dose. Transfection of si-circ#1 was found to increase the apoptotic rate and protein level of the pro-apoptotic Cleaved-caspase in U251 and LN229 cells, contrasted with transfection of si-NC (Supplementary figure 1A-B). Three treatment group were designed (4 Gy, 4 Gy+si-NC and 4 Gy+si-circ#1) for apoptosis detection, which demonstrated that 4 Gy radiation treatment elevated the apoptotic rate of U251 and LN229 cells (compared with control group) while the introduction of si-circ#1 further aggravated the radiation-induced cell apoptosis (figure 2E). In addition, the co-treatment of 4 Gy radiation and si-circ#1 transfection also increased the protein levels of Cleaved-casp3 and γH2AX by comparison with 4 Gy+si-NC group (figure 2F-G). The γH2AX induction is in response to a broad range of DNA lesions (Kopp et al. 2019). Therefore, si-circ#1 promoted the radiation-induced cell growth inhibition as well as enhancement of apoptosis and DNA damage, hinting that glioma cells were more sensitive to radiation after knockdown of circ_0008344.
Knockdown of circ_0008344 made glioma cells more sensitive to radiation. (A-B) The interference efficiencies of si-circ#1, si-circ#2 and si-circ#3 in U251 and LN229 cells were evaluated using qRT-PCR. (C-D) After si-circ#1 or si-NC transfection and radiation treatment (0 Gy, 2 Gy, 4 Gy and 8 Gy), cell survival intensity was determined through colony formation assay. (E) Flow cytometry was implemented to analyze the effects of different treatment groups (control, 4 Gy, 4 Gy+si-NC and 4 Gy+si-circ#1) on cell apoptosis. (F-G) Cleaved-casp-3 and γH2AX protein levels were examined via Western blot. *P < 0.05, **P < 0.01, ***P < 0.001.
3.3 Circ_0008344 was a sponge of miR-433-3p
Circinteractome was used for the bioinformatics analysis between circ_0008344 and miR-433-3p. As the presentation of figure 3A, wild-type circ_0008344 sequence contained the binding sites of miR-433-3p. Then circ_0008344 WT or circ_0008344 MUT plasmid and miR-433-3p or miR-NC were co-transfected into U251 and LN229 cells to perform the relative luciferase detection. Results suggested that miR-433-3p overexpression evoked the luciferase activity inhibition of circ_0008344 WT plasmid, which was not found in circ_0008344 MUT plasmid (figure 3B-C). The qRT-PCR data indicated that miR-433-3p was downregulated in radioresistant tissues compared with radiosensitive tissues (figure 3D). After analysis of Pearson’s correlation coefficient, a negative relation (r = -0.5396, P<0.01) was found between circ_0008344 and miR-433-3p levels in glioma tissues (figure 3E). Obviously, miR-433-3p expression was reduced in U251 and LN229 cells with NHA as normal control cells (figure 3F). With respect to the effect of circ_0008344 on miR-433-3p expression, circ_0008344 down-regulation was shown to increase the expression of miR-433-3p (figure 3G). On the contrary, the overexpression vector circ_0008344 (figure 3H) inhibited the relative miR-433-3p level (figure 3I).The above evidence revealed that circ_0008344 was a valid miR-433-3p sponge in glioma cells.
Circ_0008344 was a sponge of miR-433-3p. (A) Circinteractome was exploited for predicting the binding sites between circ_0008344 and miR-433-3p. (B-C) Dual-luciferase reporter assay was employed to analyze whether circ_0008344 could bind to miR-433-3p in glioma cells. (D) The qRT-PCR was performed for miR-433-3p analysis in radiosensitive and radioresistant glioma tissues. (E) The potential linear correlation between circ_0008344 and miR-433-3p was analyzed via Pearson’s correlation coefficient. (F) The miR-433-3p level was measured by qRT-PCR in glioma cells and normal NHA cells. (G) The effect of si-circ#1 on miR-433-3p was assessed by qRT-PCR with si-NC as control group. (H-I) The qRT-PCR was adopted to measure the relative expression of circ_0008344 (H) and miR-433-3p (I) in U251 and LN229 cells transfected with circ_0008344 or vector. **P < 0.01, ***P < 0.001.
3.4 Circ_0008344 knockdown-mediated radiosensitivity promotion was dependent on the up-regulation of miR-433-3p in glioma cells
After anti-miR-433-3p transfection, miR-433-3p level was decreased by 50% in U251 and LN229 cells (figure 4A). Colony formation assay manifested that the si-circ#1-induced inhibitory effect on cell survival intensity was returned by miR-433-3p inhibitor in U251 and LN229 cells treated with radiation (figure 4B-C). The apoptotic rates increased by knockdown of circ_0008344 in 4 Gy-treated U251 and LN229 cells were also lessened after miR-433-3p was down-regulated (figure 4D-E). As the consequences of miR-433-3p inhibition, the stimulative effects of si-circ#1 on the radiation-induced Cleaved-casp-3 and γH2AX protein levels were abrogated for the most part (figure 4F). Thus, miR-433-3p up-regulation was responsible for the promotive radiosensitivity caused by circ_0008344 knockdown in glioma cells.
Circ_0008344 knockdown-mediated radiosensitivity promotion was dependent on the up-regulation of miR-433-3p in glioma cells. (A) The transfection efficiency of anti-miR-433-3p was determined by qRT-PCR. (B-C) Colony formation assay was conducted to assess cell survival intensity in U251 and LN229 cells after transfection (si-NC+anti-NC, si-circ#1+anti-NC and si-circ#1+anti-miR-433-3p) and radiation treatment (0 Gy, 2 Gy, 4 Gy and 8 Gy). (D-E) The apoptotic rate was detected using flow cytometry in U251 and LN229 cells of the following groups: si-NC+anti-NC, 4 Gy+si-NC+anti-NC, 4 Gy+si-circ#1+anti-NC and 4Gy+si-circ#1+anti-miR-433-3p. (F) The protein analyses of Cleaved-casp-3 and γH2AX were carried out by Western blot. *P < 0.05, **P < 0.01, ***P < 0.001.
3.5 Circ_0008344 increased RNF2 expression by sequestering miR-433-3p
Starbase exhibited the binding sites of miR-433-3p on the RNF2 3′UTR sequence (figure 5A), and their mutual effect was verified by miR-433-3p-induced inhibition of luciferase signal in RNF2 3′UTR WT group (figure 5B-C). Western blot displayed the high expression of RNF2 in radioresistant tissues (figure 5D) and glioma cells (figure 5E), by comparison with radiosensitive tissues and NHA cells. The miR-433-3p level was up-regulated by 10-fold changes in miR-433-3p-transfected U251 and LN229 cells relative to miR-NC-transfected cells, showing the great overexpression efficiency of miR-433-3p in glioma cells (figure 5F). Moreover, there was a repressive effect of miR-433-3p overexpression on RNF2 protein level while this effect was abolished after circ_0008344 level was upregulated (figure 5G-H). Altogether, RNF2 was the target of miR-433-3p and circ_0008344 could sponge miR-433-3p to up-regulate the RNF2 expression.
Circ_0008344 increased RNF2 expression by sequestering miR-433-3p. (A) The bioinformatics analysis between miR-433-3p and RNF2 was performed by Starbase. (B-C) The combination between miR-433-3p and RNF2 was analyzed using dual-luciferase reporter assay. (D-E) RNF2 protein expression was measured by Western blot in radiosensitive/radioresistant tissues (D) and glioma/normal cells (E). (F) The overexpression effect of miR-433-3p mimic on miR-433-3p level was assessed using qRT-PCR. (G-H) Western blot was employed for RNF2 determination after transfection of miR-NC, miR-433-3p, miR-433-3p+vector and miR-433-3p+circ_0008344 in U251 (G) and LN229 (H) cells. **P < 0.01, ***P < 0.001.
3.6 RNF2 overexpression neutralized the enhancement of radiosensitivity caused by circ_0008344 inhibition in glioma cells
The RNF2 vector was constructed for ascertaining the function of RNF2 on radiosensitivity, and the transfection efficiency was successful in U251 and LN229 cells (figure 6A). Then the rescued experiment demonstrated that RNF2 transfection evidently relieved the si-circ#1-caused survival inhibition (figure 6B) and apoptosis elevation (figure 6C) in U251 and LN229 cells exposed to radiation. Also, the protein expression levels of Cleaved-casp-3 and γH2AX were lower in 4Gy+si-circ#1+RNF2 treatment group than these in 4Gy+si-circ#1+pcDNA group in U251 and LN229 cells (figure 6D). In a word, the increased radiosensitivity by the inhibition of circ_0008344 was counteracted following the overexpression of RNF2.
RNF2 overexpression neutralized the enhancement of radiosensitivity caused by circ_0008344 inhibition in glioma cells. (A) The protein level of RNF2 was assayed via Western blot in pcDNA and RNF2 transfection groups. (B) Following transfection (si-NC+pcDNA, si-circ#1+pcDNA and si-circ#1+RNF2) and radiation treatment (0 Gy, 2 Gy, 4 Gy and 8 Gy), the analysis of cell survival intensity was implemented by colony formation assay. (C-D) After treatment of si-NC+pcDNA, 4 Gy+si-NC+pcDNA, 4 Gy+si-circ#1+pcDNA and 4 Gy+si-circ#1+RNF2, cell apoptosis (C) and protein expression (Cleaved-casp-3 and γH2AX) (D) were severally examined by flow cytometry and Western blot. **P < 0.01, ***P < 0.001.
3.7 Repression of circ_0008344 sensitized glioma to radiation by the miR-433-3p/RNF2 axis in vivo
U251 cells with stable expression of sh-NC or sh-circ were injected into mice. Tumor volume was measured weekly and mice were scarified when tumor volume was reach about 800 mm3. The tumor volume curve and tumor weight indicated that tumor growth was markedly inhibited in sh-circ group by comparison with sh-NC group (supplementary figure 1C–D). In addition, the effect of circ_0008344 on the radiosensitivity of glioma was investigated. Mice were exposed to 6 Gy irradiation after tumor volume of sh-NC or sh-circ group was 100 mm3, and then tumor volume was determined every 5 days. Tumor volume of sh-circ group was notably reduced contrasted with sh-NC group in the late period of observation (from 20 to 30 days) (figure 7A). After 30 days, tumors were excised and tumor weight was repressed in sh-circ group relative to sh-NC group (figure 7B). The circ_0008344, miR-433-3p and associated protein levels were detected in the tumor tissues from mice. The results of qRT-PCR showed that circ_0008344 was lower (figure 7C) and the miR-433-3p level was higher (figure 7D) in sh-circ group than these in sh-NC group. Western blot revealed that knockdown of circ_0008344 distinctly suppressed the RNF2 protein expression but facilitated the protein levels of Cleaved-casp-3 and γH2AX (figure 7E). These results implied that circ_0008344 downregulation decreased glioma growth in vivo and contributed to the radiosensitivity of glioma via regulating the miR-433-3p/RNF2 axis.
Repression of circ_0008344 sensitized glioma to radiation by the miR-433-3p/RNF2 axis in vivo. (A) Tumor volume was measured per 5 d after radiation treatment in mice for 5 d. (B) Tumors were weighed at 30 d. (C-D) Circ_0008344 (C) and miR-433-3p (D) levels were examined through qRT-PCR. (E) Western blot was administrated for the determination of RNF2, Cleaved-casp-3 and γH2AX. **P < 0.01, ***P < 0.001.
4 Discussion
Radiotherapy is considered as an important therapeutic strategy for glioma patients, but patients are at the risk of local recurrence and poor prognosis after the occurrence of radioresistance. Our research clarified that circ_0008344 knockdown targeted the miR-433-3p/RNF2 axis to enhance the sensitization of glioma to radiation, which might be beneficial for addressing the issue of radioresistance in glioma.
As a novel class of regulatory molecules in recent years, circRNAs have been found to participate in the radiosensitive regulation in cancers. Many circRNAs have been found to be differentially expressed in radioresistant esophageal cancer cells (Su et al. 2016) and radiation-treated cervical cancer cells (Yu et al. 2018), implying that the dysregulation of circRNAs was correlated to radioresistance in cancers. Upregulation of circATRNL1 could sensitize oral squamous cell carcinoma to irradiation (Chen et al. 2020), while the radiosensitivity was promoted by the down-regulation of circ_0001313 in colon cancer cells (Wang et al. 2019). A study of glioma clarified that circATP8B4 aggravated cell radioresistance (Zhao et al. 2019). As for the effect of circ_0008344 on radiotherapy in glioma, we found it was highly expressed in radioresistant tissues and speculated that circ_0008344 was associated with radioresistance in glioma. Expectedly, knockdown of circ_0008344 repressed colony survival intensity but enhanced apoptosis and DNA damage in glioma cells under radiation. These results showed that the low expression of circ_0008344 induced the enhancement of radiosensitivity in glioma.
MiR-433-3p is an inhibitor in the malignant progression of some tumors, such as renal carcinoma (Cai et al. 2020), osteosarcoma (Hou and Mao 2020), and esophageal squamous cell carcinoma (Shi et al. 2018). However, the role of miR-433-3p in cancer radiation has not been fully investigated. Our data demonstrated that miR-433-3p could motivate the radiation susceptibility of glioma cells. In consideration of the sponge mechanism of circRNAs to miRNAs (Hao et al. 2019; Sunet al. 2020), we performed the target analysis between circ_0008344 and miR-433-3p. The results exhibited that circ_0008344 contained the binding sites for miR-433-3p and exerted the sponge effect on miR-433-3p in glioma cells. As a matter of course, the function of circ_0008344 in regulating radiosensitivity was dependent on sponging miR-433-3p.
In addition, we identified RNF2 as a downstream gene of miR-433-3p. Furthermore, circ_0008344 was shown to increase the RNF2 expression by acting as a miR-433-3p sponge and RNF2 overexpression rescued the radiosensitivity-promoting impact of circ_0008344 down-regulation on glioma cells. Liu et al. have illuminated that silencing circ_100367 accelerated the radiosensitivity via the miR-217/Wnt3 axis in esophageal squamous cell carcinoma (Liu et al. 2019) and circ_000543 knockdown exerted the same stimulative function on radiosensitivity in nasopharyngeal carcinoma via targeting the miR-9/PDGFRB axis (Chen et al. 2019). In conformity with these studies, we concluded that knockdown of circ_0008344 promoted the radiation sensitivity of glioma cells through the miR-433-3p/RNF2 axis. This conclusion was also affirmed by our assay in vivo.
To summarize, knockdown of circ_0008344 generated the promotive effect on the response to radiation in glioma via regulating the miR-433-3p/RNF2 signal axis. This study afforded the reliable evidence for the involvement of the circ_0008344/miR-433-3p/RNF2 network in regulating radiosensitivity of glioma cells. It is expected to improve the radiotherapeutic efficacy for glioma patients by down-regulating the level of circ_0008344
References
Cai X, Zhang X, Mo L, Zhu J and Yu H 2020 LncRNA PCGEM1 promotes renal carcinoma progression by targeting miR-433-3p to regulate FGF2 expression. Cancer Biomark 27 493–504
Chen G, Li Y, He Y, Zeng B, Yi C, Wang C, Zhang X, Zhao W, et al. 2020 Upregulation of circular RNA circATRNL1 to sensitize oral squamous cell carcinoma to irradiation. Mol. Ther. Nucleic Acids 19 961–973
Chen L, Zhou H and Guan Z 2019 CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR-9/platelet-derived growth factor receptor B axis. Biochem. Biophys. Res. Commun. 512 786–792
Chrispijn ND, Elurbe DM, Mickoleit M, Aben M, de Bakker DEM, Andralojc KM, Huisken J, Bakkers J, et al. 2019 Loss of the Polycomb group protein Rnf2 results in derepression of tbx-transcription factors and defects in embryonic and cardiac development. Sci. Rep. 9 4327
Gladson CL, Prayson RA and Liu WM 2010 The pathobiology of glioma tumors. Annu. Rev. Pathol. 5 33–50
Han X, Xue X, Zhou H and Zhang G 2017 A molecular view of the radioresistance of gliomas. Oncotarget 8 100931–100941
Hao Z, Hu S, Liu Z, Song W, Zhao Y and Li M 2019 Circular RNAs: functions and prospects in glioma. J. Mol. Neurosci. 67 72–81
Hou XK and Mao JS 2020 Long noncoding RNA SNHG14 promotes osteosarcoma progression via miR-433-3p/FBXO22 axis. Biochem. Biophys. Res. Commun. 523 766–772
Hu X, Fang Y, Hui X, Jv Y and You C 2016 Radiotherapy for diffuse brainstem glioma in children and young adults. Cochrane Database Syst. Rev. CD010439
Huang FJ, Dang JQ, Zhang S and Cheng ZY 2020 Circular RNA hsa_circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur. Rev. Med. Pharmaco.l Sci. 24 1887–1898
Kopp B, Khoury L and Audebert M 2019 Validation of the gammaH2AX biomarker for genotoxicity assessment: a review. Arch. Toxicol. 93 2103–2114
Lenting K, Verhaak R, Ter Laan M, Wesseling P and Leenders W 2017 Glioma: experimental models and reality. Acta Neuropathol. 133 263–282
Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, Gu H, Wang X, et al. 2019 CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging 11 12412–12427
Panda AC 2018 Circular RNAs Act as miRNA Sponges. Adv. Exp. Med. Biol. 1087 67–79
Pierson C, Grinchak T, Sokolovic C, Holland B, Parent T, Bowling M, Arastu H, Walker P, et al. 2018 Response criteria in solid tumors (PERCIST/RECIST) and SUVmax in early-stage non-small cell lung cancer patients treated with stereotactic body radiotherapy. Radiat. Oncol. 13 34
Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y and Ren J 2019 Regulatory network of miRNA on its target: coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol. Life Sci. 76 441–451
Qin H, Liu J, Du ZH, Hu R, Yu YK and Wang QA 2020 Circular RNA hsa_circ_0012673 facilitates lung cancer cell proliferation and invasion via miR-320a/LIMK18521 axis. Eur. Rev. Med. Pharmacol. Sci. 24 1841–1852
Shi Q, Wang Y, Mu Y, Wang X and Fan Q 2018 MiR-433-3p Inhibits proliferation and invasion of esophageal squamous cell carcinoma by targeting GRB2. Cell Physiol. Biochem. 46 2187–2196
Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, et al. 2016 Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J. Transl. Med. 14 225
Sun J, Li B, Shu C, Ma Q and Wang J 2020 Functions and clinical significance of circular RNAs in glioma. Mol. Cancer 19 34
Sun S, Wang X, Xu X, Di H, Du J, Xu B, Wang Q and Wang J 2017 MiR-433-3p suppresses cell growth and enhances chemosensitivity by targeting CREB in human glioma. Oncotarget 8 5057–5068
Tabatabaei P, Visse E, Bergstrom P, Brannstrom T, Siesjo P and Bergenheim AT 2017 Radiotherapy induces an immediate inflammatory reaction in malignant glioma: a clinical microdialysis study. J. Neurooncol. 131 83–92
Wang L, Peng X, Lu X, Wei Q, Chen M and Liu L 2019 Inhibition of hsa_circ_0001313 (circCCDC66) induction enhances the radio-sensitivity of colon cancer cells via tumor suppressor miR-338-3p: Effects of cicr_0001313 on colon cancer radio-sensitivity. Pathol. Res. Pract. 215 689–696
Wu J, Chen Z, Song Y, Zhu Y, Dou G, Shen X, Zhou Y, Jiang H, et al. 2020 CircRNA_0005075 suppresses carcinogenesis via regulating miR-431/p53/epithelial-mesenchymal transition axis in gastric cancer. Cell Biochem. Funct. 38 932–942
Yang J, Yu F, Guan J, Wang T, Liu C, Wang Y, Liu G and Zhu S 2019 Knockdown of RNF2 enhances the radiosensitivity of squamous cell carcinoma in lung. Biochem. Cell Biol. 97 589–599
Yin Y, Long J, He Q, Li Y, Liao Y, He P and Zhu W 2019 Emerging roles of circRNA in formation and progression of cancer. J. Cancer 10 5015–5021
Yu D, Li Y, Ming Z, Wang H, Dong Z, Qiu L and Wang T 2018 Comprehensive circular RNA expression profile in radiation-treated HeLa cells and analysis of radioresistance-related circRNAs. PeerJ. 6 e5011
Zhao M, Xu J, Zhong S, Liu Y, Xiao H, Geng L and Liu H 2019 Expression profiles and potential functions of circular RNAs in extracellular vesicles isolated from radioresistant glioma cells. Onco.l Re.p 41 1893–1900
Zhou C, Yang F, Xi W, Wei M, Zheng G, Wang W, Yang A, Zhang J, et al. 2014 Effect of RNF2 knockdown on apoptosis and radiosensitivity in glioma U87 cells. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi 30 471–475
Zhou J, Wang H, Chu J, Huang Q, Li G, Yan Y, Xu T, Chen J, et al. 2018 Circular RNA hsa_circ_0008344 regulates glioblastoma cell proliferation, migration, invasion, and apoptosis. J. Clin. Lab. Anal. 32 e22454
Zhu C, Mao X and Zhao H 2020 The circ_VCAN with radioresistance contributes to the carcinogenesis of glioma by regulating microRNA-1183. Medicine 99 e19171
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the ethical review committee of Beijing You’an Hospital, Capital Medical University. Written informed consent was obtained from all enrolled patients.
Additional information
Communicated by BJ Rao.
Corresponding editor: BJ Rao
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Di, L., Zhao, X. & Ding, J. Knockdown of circ_0008344 contributes to radiosensitization in glioma via miR-433-3p/RNF2 axis. J Biosci 46, 82 (2021). https://doi.org/10.1007/s12038-021-00198-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12038-021-00198-8