Abstract
Background
Circular RNA (circRNA) has been proven to play a critical role in breast cancer progression. Therefore, this study was designed to clarify the role and underlying molecular mechanisms of circ-disintegrin and metalloproteinase 9 (circ-ADAM9) in breast cancer.
Methods
A quantitative real-time polymerase chain reaction (RT-qPCR) was conducted to assess the expression levels of circ-ADAM9, microRNA-383-5p (miR-383-5p), and profilin 2 (PFN2). Cellular growth curves of breast cancer cells were determined by colony-forming assay. Cell viability and apoptosis were measured by MTT and flow cytometry, respectively. The protein expression level was analyzed by western blot. Cell migration and invasion were evaluated by wound healing and Transwell assays. A xenograft experiment was established to clarify the functional role of circ-ADAM9 inhibition in vivo. The interactions among circ-ADAM9, miR-383-5p, and PFN2 were analyzed by dual-luciferase reporter, RNA immunoprecipitation (RIP), and RNA pull-down assays.
Results
We found that circ-ADAM9 was upregulated in breast cancer tissues and cells compared to controls. Inhibition of circ-ADAM9 expression impaired proliferation, migration, and invasion, but increased radiosensitivity and apoptosis in breast cancer cells; besides, radiotherapy combined with circ-ADAM9 inhibition showed significant inhibitory effects on tumor growth. The functional effects of circ-ADAM9 were related to miR-383-5p, a target of circ-ADAM9. Overexpression of miR-383-5p-mediated malignant behaviors and radiosensitivity of breast cancer cells were dependent on PFN2.
Conclusion
Circ-ADAM9 was found to participate in breast cancer progression through targeting the miR-383-5p/PFN2 axis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breast cancer, a female malignancy, is a common cause of cancer-related deaths worldwide, the incidence of which has shown a marked upward tendency in recent years [1, 2]. Despite decades of research on its pathophysiology, effective approaches to treatment of breast cancer are still limited to date [3]. Current treatments include chemotherapy, radiotherapy, and surgery, but patients have a high risk of recurrence after treatment [4]. Therefore, developing novel therapies is imperative.
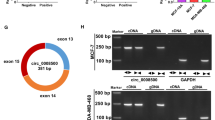
Circular RNAs (circRNAs) are covalently closed RNAs which form through a process named back-splicing that links the end of a 3′ exon to an upstream 5′ exon and represent a new class of long non-coding RNAs [5]. Circ-ADAM9 (has_circ_0001791) is derived from a disintegrin and metalloproteinase 9 (ADAM9) gene and is located on chromosome 8 (38879161-38880844). A previous study has highlighted the carcinogenic effect of circ-ADAM9 in pancreatic cancer [6]. Recently, circ-ADAM9 was documented to be highly regulated in breast cancer and served as a diagnostic biomarker for breast cancer detection [7], which attracted much attention. However, the molecular determinants underlying the regulatory function of circ-ADAM9 largely remain to be clarified in breast cancer.
As a type of non-coding RNA, microRNAs (miRNAs) are the most prevalent and functionally diverse class [8]. MiR-383-5p has been implicated in the pathogenesis of multiple tumors, including cervical cancer [9], gastric cancer [10], and oral squamous cell carcinoma [11]. Additionally, Zhang et al. reported that miR-383-5p was a possible target for breast cancer treatment [12]. It remains unexplored whether miR-383-5p mediates molecular effects protecting against radioresistance in breast cancer.
Recent studies have demonstrated that profilin 2 (PFN2) is involved in regulating actin polymerization and endocytosis [13, 14]. Although the molecular roles of PFN2 have not been entirely clarified, it was commonly reported that dysregulation of PFN2 was able to regulate the development of certain cancer types [15, 16].
Therefore, in this study, we focused on investigating the roles of circ-ADAM9 in breast cancer progression. We demonstrated that circ-ADAM9 functioned as an upstream regulator of the miR-383-5p/PFN2 axis, and that this regulatory pathway was associated with breast cancer progression by regulating malignant phenotypes of breast cancer cells.
Materials and methods
Tissue sample collection
Tissue samples of breast cancer (N = 45) and adjacent noncancerous tissues (N = 45) were obtained from breast cancer patients undergoing a surgical procedure at Heping Hospital Affiliated to Changzhi Medical College. No patients with breast cancer had received radiotherapy or chemotherapy, and all patients provided written informed consent prior to surgery. All tissues were promptly frozen in liquid nitrogen and then transferred to −80 °C. The entire investigation was approved and supervised by the Ethics Committee of Heping Hospital Affiliated to Changzhi Medical College.
Cell lines and cell culture
The breast cancer cell lines (MCF‑7, MDA-MB-468, and MDA-MB-231) and human normal breast epithelial cells (MCF-10A) were purchased from Nanjing Key Gen Biotech (Nanjing, China) and then cultured in Dulbecco’s modified Eagle medium (GIBCO BRL, Grand Island, NY, USA) supplemented with 10% (v/v) fetal bovine serum (FBS; GIBCO BRL) at 37 °C with 5% CO2.
MCF‑7 and MDA-MB-231 cells received radiation using a linear accelerator (Varian2300EX; Varian, Palo Alto, CA, USA). The setting conditions were as follows: 160 kV, 6.3 mA, for 1.1, 2.3, 3.5, 4.6, and 5.7 min, at a dose of 0, 2, 4, 6, and 8 Gy, respectively.
Quantitative real-time polymerase chain reaction
Total RNA was extracted by Trizol reagent (Invitrogen, Carlsbad, CA, USA) and the quantity of total RNA was measured under the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA). Complementary DNA (cDNA) was synthesized by PrimeScript™ RT reagent kit (Takara, Dalian, China). Real-time PCR (RT-qPCR) was conducted using SYBR Select Master Mix (Applied Biosystems, Carlsbad, CA, USA) under the IQTM5 Multicolour Real-Time PCR detection system (Bio-Rad, Hercules, CA, USA). All target genes were standardized to the endogenous reference glyceraldehyde-3-phosphate dehydrogenase (GAPDH) by the 2−∆∆Ct method. Hairpin-it TM miRNAs qPCR Quantitation Kit (Genepharma, Shanghai, China) was used to measure the level of miR-383-5p, with nuclear RNA U6 as control. Three independent experiments were included in this assay, and three duplicated wells were included for each sample. The primers were circ-ADAM9, upstream 5′-CGTCGGAGACATGACAGTGC-3′ and downstream 5′-TCCAAGTAGTTTGCCAGGAGAA-3′; ADAM9, upstream 5′- TCTTGCCACAGACCCGGTAT-3′ and downstream 5′-ATCTCCAGTCCAACTAGCACA-3′; miR-383-5p, upstream 5′-GCGCGAGATCAGAAGGTGATT-3′ and downstream 5′-AGTGCAGGGTCCGAGGTATT-3′; PFN2, upstream 5′-ATGATTGTAGGAAAAGACCGGGA-3′ and downstream 5′-GCAGTCACCATCGACGTATAGAC-3′; U6, upstream 5′-ATCCTTACGCACCCAGTCCA-3′ and downstream 5′-GAACGCTTCACGAATTTGC-3′; GAPDH, upstream 5′-TGAACCATGAGAAGTATGAC-3′ and downstream 5′-TCTTACTCCTTGGAGGCCA-3′.
RNase R and actinomycin D experiments
The partial extracted total RNA was treated with or without 3 U/mg RNase R (Geneseed, Guangzhou, China) for 15 min at 37 °C. Actinomycin D (2 mg/mL) was purchased from Sigma-Aldrich (Louis, Missouri, USA) and then added into Dulbecco’s modified Eagle medium to assess RNA stability. Three independent experiments were included in this assay.
Transfection assay
The short hairpin RNA targeting circ-ADAM9 (sh-circ-ADAM9) and matched control (sh-NC), circ-ADAM9-overexpression vector (circ-ADAM9) and negative control (pCD-ciR), and PFN2-overexpression vector (PFN2) and negative control (pcDNA3.1) were designed by RiboBio (Guangzhou, China). The mimics of miR-383-5p and negative control (miR-383-5p and NC mimics), and inhibitor of miR-383-5p and negative control (miR-383-5p inhibitor and NC inhibitor) were purchased from Genomeditech (Shanghai, China). Transfections of shRNA and miR-383-5p mimics/inhibitor were performed using RNAiMAX (Invitrogen) at a final concentration of 20 nM. Besides, lipofectamine 2000 (Sigma-Aldrich) was used for transfection of vector, and the concentrations of plasmids were 1 μg.
Colony-forming and 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-2H-tetrazol-3-ium bromide (MTT) assays
MCF‑7 and MDA-MB-231 cells were transfected for 48 h and then sowed into 6‑well plates (600 cells/well); the following incubation was at 37 °C with 5% CO2. Two weeks later, the numbers of visible colonies were measured under the inverted microscope (Leica Microsystems GmbH, Wetzlar, Germany) after staining with 0.1% crystal violet (Beyotime, Shanghai, China). A colony is defined by a cell number ≥ 50 cells. Plating efficiency (PE) = the number of clones/the number of plated cells × 100%, and survival fraction (SF) = PE the experimental group/PE the control group × 100%.
Cell proliferation was assessed by MTT assay following the manufacturer’s protocol. In brief, approximately 3000 cells/well were plated into 96-well plates and incubated for the indicated times. 20 μL of MTT (Beyotime) was used to incubate MCF‑7 and MDA-MB-231 cells for 4 h. The cell proliferation curves were plotted by measuring absorbance at 490 nm under the microplate reader (Bio-Rad). Three independent experiments were included in this assay, and three duplicated wells were included for each sample.
Flow cytometry assay
Transfected MCF‑7 and MDA-MB-231 cells were washed with cold phosphate buffered saline to remove residual medium. After digestion by trypsin and centrifugation, single-cell suspension (1 × 106/mL) was incubated with annexin V (Invitrogen) labeled with fluorescein isothiocyanate (FITC; Invitrogen) and propidium iodide (Invitrogen) for 30 min in dark conditions. The apoptotic cells were sorted using an Attune™ Flow Cytometer (Applied Biosystems). Three independent experiments were included in this assay.
Wound healing and Transwell assay
Transfected MCF‑7 and MDA-MB-231 cells were incubated overnight in the 6‑well plates. Thereafter, wounded areas were generated by a 200 μL pipette tip and then reported under the inverted microscope (Leica Microsystems GmbH). After incubation for 24 h, images were reported again. Cell invasion properties were detected using a 24-well Transwell chamber with Matrigel (Becton Dickinson, San Jose, CA, USA). In the top compartment, 2 × 105 MCF‑7 and MDA-MB-231 cells in 200 μL of serum-free medium were seeded, while the lower chamber was filled with complete medium. Following incubation at 37 °C for 24 h, the invading cells were imaged under the inverted microscope (Leica Microsystems GmbH) at 100 × amplification. Three independent experiments were included in this assay, and three duplicated wells were included for each sample.
In vivo experiment
BALB/c nude mice (females, 4 weeks of age) were purchased from Vital River Laboratory (Beijing, China) and fed according to the institutional guide. All nude mice studies were permitted by the Institutional Animal Care and Use Committee of Heping Hospital Affiliated to Changzhi Medical College. In all experiments, 5 nude mice per group were used and nude mice were randomized before injection of breast cancer cells. A volume of 200 μL FBS-free culture medium containing 2 × 106 MCF-7 cells stably transfected with sh-circ-ADAM9 was hypodermically inoculated into the left back near the forelimb, with sh-NC as control. Tumor volume was calculated regularly using volume = 1/2 (length × width2). BALB/c nude mice in irradiation groups were irradiated with 2 Gy every other day (total 10 Gy) when tumor volume > 150 mm3. At the time of sacrifice 25 d after injection, tumor tissues were collected for weight detection and RNA extraction.
Dual-luciferase reporter assay
Circinteractome (https://circinteractome.nia.nih) was used for prediction of potential binding miRNAs of circ-ADAM9. The target protein of miR-383-5p, PFN2, was predicted by Starbase (http://starbase.sysu.edu.cn/). Partial sequences of circ-ADAM9 or the PFN2 3′-untranslated region (UTR) containing putative miR-383-5p complementary sequences were synthesized and inserted into pmirGLO vector (Millipore, Billerica, MA, USA). After co-transfection with the indicated luciferase reporter plasmids and miR-383-5p mimics or NC mimics, MCF‑7 and MDA-MB-231 cells were incubated at 37 °C for 24 h. The relative firefly luciferase activity was analyzed with Dual-Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) and then normalized to Renilla luciferase activity. Three independent experiments were included in this assay.
RNA immunoprecipitation (RIP) and RNA pull-down assays
The RIP assay was conducted by using Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore). In brief, transfected MCF‑7 and MDA-MB-231 cells were collected and lysed by RIP buffer. Cell lysates were then treated with magnetic beads embracing Ago2 or IgG antibodies at 4 °C for 24 h, with the input group as a positive control. After proteinase K treatment, immunoprecipitated RNA was subjected to RT-qPCR assay. For RNA pull-down assay, 1 × 107 breast cancer cells were harvested, lysed, and then sonicated. The 200 μL of cell lysates were incubated with 3′-end biotinylated miR-383-5p (Bio-miR-383-5p; GenePharma) or control for 3 h at 4 °C. After pull-down by streptavidin magnetic beads (Life Technologies, Carlsbad, CA, USA), the RNA complexes were extracted for RT-qPCR analysis. Three independent experiments were included in two assays.
Western blot assay
Cultured MCF‑7 and MDA-MB-231 cells or tumor tissues were washed with cold phosphate buffered saline and homogenized in RIPA lysis buffer (Beyotime). Equivalent protein samples (50 µg per lane) were loaded onto 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and then transferred onto nitrocellulose membranes (Bio-Rad). Rabbit primary antibodies were used to incubate membranes overnight at 4 °C. After washing, membranes were incubated with HRP-conjugated secondary antibody (#7074S; 1:2000 dilution; Cell Signaling Technology, Danvers, MA, USA). Enhanced chemiluminescence (Life Technologies) was used to visualize immunoreactive bands. The following antibodies were used: Proliferating Cell Nuclear Antigen (PCNA; #13110S; 1:1500 dilution; Cell Signaling Technology), B‑cell lymphoma‑2 (Bcl‑2; #4223S; 1:1500 dilution; Cell Signaling Technology), Cleaved-caspase‑3 (c-caspase 3; #9664S; 1:1500 dilution; Cell Signaling Technology), caspase 3 (#14220S; 1:1500 dilution; Cell Signaling Technology), GAPDH (#2118S; 1:2000 dilution; Cell Signaling Technology), and PFN2 (#8875S; 1:1500 dilution; Cell Signaling Technology). Three independent experiments were included in this assay.
Statistical analysis
Quantitative data were expressed as mean ± standard deviation. Statistical analysis was conducted by Student’s t-test or analysis of variance followed by post hoc Bonferroni’s using GraphPad Prism 7 (GraphPad, La Jolla, CA, USA), and P < 0.05 indicated statistical significance. Pearson’s correlation analysis was applied for correlation analysis.
Results
Circ-ADAM9 is overexpressed in breast cancer tissues and cells
Herein, we measured the expression level of circ-ADAM9 in breast cancer tissues and adjacent non-tumor tissues and found that circ-ADAM9 was upregulated in breast cancer tissues compared with paired neighboring normal tissues (Fig. 1a). Consistent with these findings, circ-ADAM9 was strongly expressed in MCF‑7, MDA-MB-231, and MDA-MB-468 cells relative to control MCF-10A cells (Fig. 1b). Given that circ-ADAM9 expression was relatively higher in MCF‑7 and MDA-MB-231 cells compared to MDA-MB-468 cells, MCF‑7 and MDA-MB-231 cells were selected to explore the role of circ-ADAM9. Additionally, RNase R and actinomycin D were used to determine the existence and stability of circ-ADAM9. Compared to the linear form of ADAM9 mRNA, circ-ADAM9 was resistant to RNase R digestion and hardly degraded by actinomycin D (Fig. 1c,d). Our observations raised the possibility that circ-ADAM9 was closely tied to breast cancer progression.
Expression level of circ-ADAM9 in breast cancer tissues and cells. a Quantification of circ-ADAM9 was performed by RT-qPCR in breast cancer tissues (n = 45) and paired non-tumor tissues (n = 45), paired t-test. b RT-qPCR was used to examine circ-ADAM9 level in breast cancer cells and control MCF-10A cells, one-way ANOVA. c, d Relative levels of circ-ADAM9 and ADAM9 mRNA were determined by RT-qPCR after treating with RNase R or actinomycin D, unpaired t-test and one-way ANOVA (**P < 0.01; ***P < 0.001)
Depletion of circ-ADAM9 expression impaired proliferation, migration, and invasion but increased radiosensitivity and apoptosis in breast cancer cells
To systematically identify biologically relevant circ-ADAM9 in breast cancer, we performed loss-of-function assays using shRNA-mediated targeting of circ-ADAM9 in breast cancer cells. As compared to the sh-NC group, circ-ADAM9 was obviously downregulated in the sh-circ-ADAM9 group (Fig. 2a). In vitro depletion of circ-ADAM9 significantly repressed the survival of breast cancer cells in all test groups; simultaneously, circ-ADAM9 inhibition enhanced radiosensitivity in MCF‑7 and MDA-MB-231 cells (Fig. 2b). Next, 4 Gy of radiation was chosen to examine cell survival number. Sh-circ-ADAM9-transfected MCF‑7 and MDA-MB-231 cells also exhibited decreased cell proliferation relative to sh-NC-transfected cells, no matter whether with or without radiation (Fig. 2c). Furthermore, circ-ADAM9 knockdown promoted radiation-induced apoptosis in MCF‑7 and MDA-MB-231 cells (Fig. 2d). As shown in Fig. 2e, depletion of circ-ADAM9 increased the rate of c‑caspase 3/caspase 3 and decreased PCNA and Bcl‑2 levels in MCF‑7 and MDA-MB-231 cells, which was further enhanced after treatment with radiation. Cell migration was lower in the sh-circ-ADAM9 group than in the sh-NC group (Fig. 2f). Consistently, invasion inhibition was observed in MCF‑7 and MDA-MB-231 cells after silencing of circ-ADAM9 (Fig. 2g). Together, these findings demonstrated that suppression of circ-ADAM9 inhibited the malignant phenotypes of breast cancer cells.
Effects of circ-ADAM9 inhibition on radiosensitivity, proliferation, apoptosis, migration, and invasion in breast cancer cells. a Interference efficiency of sh-circ-ADAM9 was assessed by RT-qPCR in MCF‑7 and MDA-MB-231 cells, unpaired t-test. b–g MCF‑7 and MDA-MB-231 cells transfected with sh-NC or sh-circ-ADAM9 were exposed to an increasing dose of Gy. b Cellular growth curves of MCF‑7 and MDA-MB-231 cells were determined by colony-forming assay, two-way ANOVA. c Cell viability was measured by MTT assay, two-way ANOVA. d Apoptosis rate was measured by flow cytometry, one-way ANOVA. e Representative images of western blot analysis and protein expression levels of PCNA, Bcl‑2, and rate of c‑caspase 3/caspase 3 normalized to control GAPDH, one-way ANOVA. f, g Migration and invasion assay histograms show the migrated and invaded cell count from MCF‑7 and MDA-MB-231 cells determined by wound healing and Transwell assays, unpaired t-test (***P < 0.001)
Circ-ADAM9 inhibition impeded tumor growth following irradiation
The functional effect of circ-ADAM9 inhibition on tumor growth was assessed by in vivo xenograft experiments. The shortage of circ-ADAM9 reduced the xenograft volume compared with control; after irradiation treatment, nude mice bearing sh-circ-ADAM9 tumors were the smallest in all test groups (Fig. 3a). Consistent with a delay in tumor growth, circ-ADAM9 knockdown combined with irradiation could reduce tumor weight (Fig. 3b,c). The expression of circ-ADAM9 was downregulated in the sh-circ-ADAM9 group and the IR+sh-circ-ADAM9 group compared with matched controls (Fig. 3d). Thus, circ-ADAM9 was a key regulator of tumor growth and radiosensitivity.
Silencing of circ-ADAM9 repressed tumor growth in vivo. a Xenograft volume was measured regularly, two-way ANOVA. b, c Representative images and tumor weight after tumor dissection from nude mice, one-way ANOVA. d Expression level of circ-ADAM9 was estimated with RT-qPCR in dissected tumor tissues from different groups, one-way ANOVA (***P < 0.001)
MiR-383-5p is a direct target of circ-ADAM9
Circinteractome predicted that 9 miRNAs had binding sites on circ-ADAM9. Only 4 miRNAs, including miR-1236-3p, miR-383-5p, miR-622, and miR-634, which were downregulated in breast cancer and can exert cancer-suppressive effects in previous studies, were screened and validated. Only miR-383-5p expression was inversely regulated by circ-ADAM9 (Fig. S1a, b). We also found that miR-383-5p was obviously downregulated in breast cancer compared to matched non-tumor tissues (Fig. 4a). MiR-383-5p was also identified as having a significant correlation with circ-ADAM9 in breast cancer tissues (Fig. 4b). The prediction algorithm Circinteractome was used to predict target miRNA of circ-ADAM9, and we displayed the miR-383-5p-binding sites on circ-ADAM9 (Fig. 4c). To validate the putative target of circ-ADAM9, we performed dual-luciferase reporter assay. MiR-383-5p ectopic expression decreased luciferase reporter activity of wt-circ-ADAM9, while it was not decreased in MCF‑7 and MDA-MB-231 cells transfected with MUT-circ-ADAM9 (Fig. 4d). Furthermore, RIP assay suggested that miR-383-5p and circ-ADAM9 were enriched in the Ago2-immunoprecipitated group when compared to the IgG group, indicating that circ-ADAM9 could bind to miR-383-5p in an Ago2-dependent manner (Fig. 4e). Consistently, miR-383-5p expression was lower in MCF‑7, MDA-MB-231, and MDA-MB-468 cells compared to MCF-10A cells (Fig. 4f). As compared to the pCD-ciR group, circ-ADAM9 was obviously upregulated in MCF‑7 and MDA-MB-231 cells transfected with circ-ADAM9 (Fig. 4g). Overexpression of circ-ADAM9 inhibited miR-383-5p expression; conversely, loss of circ-ADAM9 increased miR-383-5p expression (Fig. 4h,i). Collectively, these findings showed that miR-383-5p is a functional target of circ-ADAM9.
Circ-ADAM9 targeted miR-383-5p in breast cancer cells. a Expression level of miR-383-5p was examined by RT-qPCR in breast cancer tissues and paired non-tumor tissues, paired t-test. b Correlation between miR-383-5p and circ-ADAM9 was analyzed by Pearson’s correlation analysis. c Graphic representation showed miR-383-5p binding sites on circ-ADAM9, two-way ANOVA. d Dual-luciferase reporter assay was used to confirm the association between miR-383-5p and circ-ADAM9, two-way ANOVA. e RIP assay was performed in MCF‑7 and MDA-MB-231 cells, one-way ANOVA. f Expression of miR-383-5p was examined by RT-qPCR in breast cancer cells and control MCF-10A cells, one-way ANOVA. g RT-qPCR was used to test circ-ADAM9 expression in MCF‑7 and MDA-MB-231 cells transfected with pCD-ciR or circ-ADAM9, unpaired t-test. h, i Expression of miR-383-5p was assessed by RT-qPCR in MCF‑7 and MDA-MB-231 cells transfected with pCD-ciR, circ-ADAM9, sh-NC, or sh-circ-ADAM9, unpaired t-test (***P < 0.001)
PFN2 is a direct target of miR-383-5p
Considering that miRNA could medicate gene expression by binding to mRNAs, we hypothesized that miR-383-5p might act as a regulator in breast cancer through binding to a certain mRNA. Starbase predicted that there were 1927 genes with binding sites for miR-383-5p, and we reviewed the relevant literature to screen three genes that were upregulated in breast cancer and were associated with radioresistance, i.e., FSTL1, RAD51, and PFN2. RNA pull-down experiments showed that a large amount of PFN2 could bind to miR-383-5p (Fig. S1c, d). Interesting, PFN2 was obviously overexpressed in breast cancer compared to matched non-tumor tissues (Fig. 5a,b), and we also confirmed a negative correlation relationship between miR-383-5p and PFN2 (Fig. 5c). Using the bioinformatics prediction tool Starbase, miR-383-5p was also identified to have complementary sequences on PFN2 (Fig. 5d). Dual-luciferase reporter manifested that only upregulation of miR-383-5p decreased the luciferase activity of the wt-PFN2 3′-UTR group (Fig. 5e). Biotinylated miR-383-5p also enhanced the pull-down efficiency of PFN2 mRNA (Fig. 5f). According to the western blot analysis, PFN2 was upregulated in MCF‑7, MDA-MB-231, and MDA-MB-468 cells compared to MCF-10A cells (Fig. 5g). Transfection with miR-383-5p mimics dramatically increased the expression of miR-383-5p in MCF‑7 and MDA-MB-231 cells; besides, miR-383-5p was decreased in cells transfected with miR-383-5p inhibitor (Fig. 5h,i). Additionally, we found that PFN2 protein expression in MCF‑7 and MDA-MB-231 cells was decreased under transfection of miR-383-5p mimics, while PFN2 was stably increased through miR-383-5p inhibitor (Fig. 5j,k). In view of the above findings, PFN2 was a direct target of miR-383-5p in breast cancer cells.
MiR-383-5p targeted PFN2 in breast cancer cells. a, b Expression levels of PFN2 were measured by RT-qPCR and western blot in breast cancer tissues and control, paired t-test. c Correlation between miR-383-5p and PFN2 was analyzed. d Schematic illustration shows miR-383-5p binding sites on PFN2. e, f Dual-luciferase reporter and RNA pull-down assays were performed to confirm the association between miR-383-5p and PFN2, one-way and two-way ANOVA. g Protein level of PFN2 was assessed by western blot in breast cancer cells and control, one-way ANOVA. h–k Expression of miR-383-5p and PFN2 was determined by RT-qPCR in MCF‑7 and MDA-MB-231 cells transfected with miR-383-5p mimics, NC mimics, miR-383-5p inhibitor, or NC inhibitor, unpaired t-test (**P < 0.01; ***P < 0.001)
MiR-383-5p regulates breast cancer cell function via downregulating PFN2
To characterize the cellular effects of miR-383-5p and PFN2 overexpression, we determined radiosensitivity, proliferation, apoptosis, migration, and invasion in breast cancer cells. PFN2 overexpression was found in MCF‑7 and MDA-MB-231 cells after transfection with PFN2 (Fig. 6a). Importantly, transfection with PFN2 rescued the downregulation of PFN2 in MCF‑7 and MDA-MB-231 cells induced by miR-383-5p mimics (Fig. 6b). As expected, miR-383-5p overexpression by miR-383-5p mimics reduced the colony-forming and proliferative abilities of breast cancer cells no matter with or without radiation, which was reversed by re-expression of PFN2 (Fig. 6c,d). MiR-383-5p overexpression also increased cell apoptosis, which was enhanced after treatment with radiation, and the role of miR-383-5p overexpression was counteracted by PFN2 overexpression (Fig. 6e,f). PFN2 upregulation counteracted miR-383-5p mimics-induced decrease of PCNA and Bcl‑2 and increased the rate of c‑caspase 3/caspase 3 in MCF‑7 and MDA-MB-231 cells under radiation (Fig. 6g,h). The suppressive effects on migration and invasion in miR-383-5p mimics-transfected cells were largely abrogated by PFN2 overexpression (Fig. 6i,j). The above findings suggest that miR-383-5p weakened the malignant behaviors of breast cancer cells via targeting PFN2.
Overexpression of miR-383-5p-mediated effects on radiosensitivity, proliferation, apoptosis, migration, and invasion of breast cancer cells were dependent on PFN2. a Overexpression efficiency of PFN2 was assessed by RT-qPCR in MCF‑7 and MDA-MB-231 cells, unpaired t-test. b–j MCF‑7 and MDA-MB-231 cells were transfected with NC mimics, miR-383-5p mimics, miR-383-5p mimics+pcDNA3.1, or miR-383-5p mimics+PFN2. b Western blot analysis was used to quantify PFN2 level, one-way ANOVA. c, d Cellular growth curves and cell viability of MCF‑7 and MDA-MB-231 cells were determined by colony-forming and MTT assay, respectively, one-way ANOVA. e, f Apoptosis was analyzed by flow cytometry, one-way ANOVA. g Protein expression levels of PCNA, Bcl‑2, and rate of c‑caspase 3/caspase 3 were assessed by western blot analysis, one-way ANOVA. h, i Migration and invasion assays were performed by wound healing and Transwell assays, respectively, one-way ANOVA (**P < 0.01; ***P < 0.001)
Inhibition of circ-ADAM9-mediated effects on breast cancer cells via upregulating miR-383-5p
To investigate whether circ-ADAM9 could regulate breast cancer cell function via miR-383-5p, MCF‑7 and MDA-MB-231 cells were transfected with sh-circ-ADAM9 and miR-383-5p inhibitor. MiR-383-5p was downregulated in MCF‑7 and MDA-MB-231 cells co-transfected with sh-circ-ADAM9 and miR-383-5p inhibitor compared to cells transfected with sh-circ-ADAM9 only (Fig. 7a). Moreover, downregulation of miR-383-5p obviously rescued the inhibitory expression of PFN2 in sh-circ-ADAM9-transfected cells (Fig. 7b). The decreased colony-forming and proliferative abilities in circ-ADAM9-silenced MCF‑7 and MDA-MB-231 cells were also rescued by miR-383-5p inhibitor (Fig. 7c,d). Conversely, downregulation of circ-ADAM9 markedly enhanced cell apoptosis, which was abolished by silencing of miR-383-5p (Fig. 7e). Suppression of circ-ADAM9 increased the rete of c‑caspase 3/caspase 3 and decreased PCNA and Bcl‑2 levels in MCF‑7 and MDA-MB-231 cells, whereas these effects were largely abrogated by concomitant inhibition of miR-383-5p (Fig. 7f). Wound healing and Transwell assays indicated that the inhibitive effects on migration and invasion induced by circ-ADAM9 silencing were overturned by miR-383-5p inhibition (Fig. 7g,h). Circ-ADAM9 plays a cancerogenic role in breast cancer cells via regulating miR-383-5p.
Inhibition of circ-ADAM9-mediated effects on breast cancer cells were dependent on miR-383-5p. a–h MCF‑7 and MDA-MB-231 cells were transfected with sh-NC, sh-circ-ADAM9, sh-circ-ADAM9+NC inhibitor, or sh-circ-ADAM9+miR-383-5p inhibitor. a, b Levels of miR-383-5p and PFN2 were quantified by RT-qPCR and western blot assays, one-way ANOVA. c, d colony-forming and MTT assays were used to assess growth curves and cell viability of MCF‑7 and MDA-MB-231 cells, two-way ANOVA. e Apoptosis was examined by flow cytometry, one-way ANOVA. f Western blot analysis was conducted to test the protein levels of PCNA, Bcl‑2, and rate of c‑caspase 3/caspase 3, one-way ANOVA. g, h Wound healing and Transwell assays were used to analyze migration and invasion, respectively, one-way ANOVA (**P < 0.01; ***P < 0.001)
Discussion
Breast cancer is a common gynecological malignancy in females [1, 2]. We found that circ-ADAM9 could regulate the miR-383-5p/PFN2 axis to promote breast cancer progression. Considering their stability and sequence conservation, circRNAs have huge potential application value for diagnosis and treatment of human cancers [17]. For example, Tran et al. proposed that circRNA has an extremely high application value for diagnosis and treatment of female reproductive system tumors in the future [18]. Understanding of the crosstalk between circRNA and miRNA is rapidly expanding [19]. Indeed, recent reports have identified that circ-ADAM9 exerted miRNA-sponging functions in human disease by regulating the function of miR-20a-5p and miR-217 [6, 20]. Our results proposed that the function of circ-ADAM9 was dependent on sponging of miR-383-5p.
Work from the past decade has established miRNAs as critical regulators in malignant tumors through base pairing with 3′-UTRs of their target genes and thus affecting the function of target mRNAs [21]. Previous research has suggested that miR-383-5p could function as a tumor inhibitor in breast cancer progression through directly targeting oncogenes, such as programmed death 1, RNA-binding motif protein 3, and lactate dehydrogenase, and miR-379 might be considered as an effective strategy for breast cancer treatment [22,23,24]. Importantly, aberrant expression of miR-383-5p has been demonstrated to be implicated in chemoresistance of tumor cells [25, 26]. Consistent with miR-383-5p having a role in the pathological development of breast cancer, in this paper, miR-383-5p has been shown to impact breast cancer progression via tumor-suppressive roles through PFN2.
Altered PFN2 expression was linked to tumorigenesis, we also found that PFN2-mediated malignant behaviors and radiosensitivity of breast cancer cells were regulated by miR-383-5p. In addition, a report in breast cancer implied that miR-223-3p overexpression reduced cell proliferation, glycolysis, and colony formation, while improving radiosensitivity in breast cancer cells by regulating PFN2 [27]. Importantly, Kim et al. reported that PFN2-associated adhesion molecules could be a target for radiosensitivity [28]. Not surprisingly, upregulation of PFN2 indicated a poor prognosis of patients with breast cancer, proposing PFN2 as a therapeutic target [29, 30]. As expected, PFN2 was also verified as an oncogene in breast cancer progression.
Together, circ-ADAM9 could competitively bind to miR-383-5p to increase PFN2 expression, which facilitated breast cancer progression and radioresistance. These findings have expanded the functional repertoire of circ-ADAM9, identifying circ-ADAM9 as a key driver of breast cancer progression which is dependent on the miR-383-5p/PFN2 axis.
Conclusion
Our findings showed that circ-ADAM9 inhibition impaired proliferation, migration, and invasion, while increasing radiosensitivity and apoptosis in breast cancer cells through the miR-383-5p/PFN2 axis, suggesting the potential value of radiotherapy combined with circ-ADAM9-targeted adjuvant therapy for breast cancer patients.
References
Woolston C (2015) Breast cancer. Nature 527(7578):S101
Anastasiadi Z et al (2017) Breast cancer in young women: an overview. Updates Surg 69(3):313–317
Fahad Ullah M (2019) Breast cancer: current perspectives on the disease status. Adv Exp Med Biol 1152:51–64
Wormann B (2017) Breast cancer: basics, screening, diagnostics and treatment. Med Monatsschr Pharm 40(2):55–64
Patop IL, Wust S, Kadener S (2019) Past, present, and future of circRNAs. EMBO J 38(16):e100836
Xing C et al (2019) Circular RNA ADAM9 facilitates the malignant behaviours of pancreatic cancer by sponging miR-217 and upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol 47(1):3920–3928
Ameli-Mojarad M et al (2021) Circular RNA hsa_circ_0005046 and hsa_circ_0001791 may become diagnostic biomarkers for breast cancer early detection. J Oncol 2021:2303946
Liu B, Li J, Cairns MJ (2014) Identifying miRNAs, targets and functions. Brief Bioinform 15(1):1–19
Hu Y et al (2019) LINC01128 expedites cervical cancer progression by regulating miR-383-5p/SFN axis. BMC Cancer 19(1):1157
Wei C, Gao JJ (2019) Downregulated miR-383-5p contributes to the proliferation and migration of gastric cancer cells and is associated with poor prognosis. PeerJ 7:e7882
Shao B et al (2020) RP11-284F21.9 promotes oral squamous cell carcinoma development via the miR-383-5p/MAL2 axis. J Oral Pathol Med 49(1):21–29
Zhang Z et al (2020) MiR-891a-5p as a prognostic marker and therapeutic target for hormone receptor-positive breast cancer. J Cancer 11(13):3771–3782
Schluter K, Jockusch BM, Rothkegel M (1997) Profilins as regulators of actin dynamics. Biochim Biophys Acta 1359(2):97–109
Gareus R et al (2006) Mouse profilin 2 regulates endocytosis and competes with SH3 ligand binding to dynamin 1. J Biol Chem 281(5):2803–2811
Cui XB et al (2016) PFN2, a novel marker of unfavorable prognosis, is a potential therapeutic target involved in esophageal squamous cell carcinoma. J Transl Med 14(1):137
Kim MJ et al (2015) Profilin 2 promotes migration, invasion, and stemness of HT29 human colorectal cancer stem cells. Biosci Biotechnol Biochem 79(9):1438–1446
Patop IL, Kadener S (2018) circRNAs in cancer. Curr Opin Genet Dev 48:121–127
Tran AM et al (2020) A new world of biomarkers and therapeutics for female reproductive system and breast cancers: circular RNAs. Front Cell Dev Biol 8:50
Abdollahzadeh R et al (2019) Competing endogenous RNA (ceRNA) cross talk and language in ceRNA regulatory networks: a new look at hallmarks of breast cancer. J Cell Physiol 234(7):10080–10100
Tian D et al (2020) Circ-ADAM9 targeting PTEN and ATG7 promotes autophagy and apoptosis of diabetic endothelial progenitor cells by sponging mir-20a-5p. Cell Death Dis 11(7):526
Towler BP, Jones CI, Newbury SF (2015) Mechanisms of regulation of mature miRNAs. Biochem Soc Trans 43(6):1208–1214
Azarbarzin S et al (2020) MicroRNA-383-5p restrains the proliferation and migration of breast cancer cells and promotes apoptosis via inhibition of PD-L1. Life Sci 267:118939
Tian Y et al (2019) LINC00096 promotes the proliferation and invasion by sponging miR-383-5p and regulating RBM3 expression in triple-negative breast cancer. Onco Targets Ther 12:10569–10578
Zhang J et al (2020) MicroRNA-383-5p acts as a potential prognostic biomarker and an inhibitor of tumor cell proliferation, migration, and invasion in breast cancer. Cancer Biomark 27(4):423–432
Sun X et al (2020) Expression level of miR-383-5p in colorectal cancer patients following neoadjuvant chemotherapy and its clinical significance in the prognosis. Minerva Med. https://doi.org/10.23736/S0026-4806.20.06656-2
Jiang J et al (2019) Up-regulation of miR-383-5p suppresses proliferation and enhances chemosensitivity in ovarian cancer cells by targeting TRIM27. Biomed Pharmacother 109:595–601
Zhao Y et al (2021) Circle RNA circABCB10 modulates PFN2 to promote breast cancer progression, as well as aggravate radioresistance through facilitating glycolytic metabolism via miR-223-3p. Cancer Biother Radiopharm 36(6):477–490
Kim HS et al (2012) Identification of a radiosensitivity signature using integrative metaanalysis of published microarray data for NCI-60 cancer cells. BMC Genomics 13:348
Jiang M et al (2019) Long noncoding RNA FOXD2AS1/miR1505p/PFN2 axis regulates breast cancer malignancy and tumorigenesis. Int J Oncol 54(3):1043–1052
Ling Y et al (2021) Profilin 2 (PFN2) promotes the proliferation, migration, invasion and epithelial-to-mesenchymal transition of triple negative breast cancer cells. Breast Cancer 28(2):368–378
Author information
Authors and Affiliations
Contributions
P. Song was responsible for drafting the manuscript. P. Song, J. Wu, and J. Chen contributed to the analysis and interpretation of data. P. Song, F. Wang, G. Wang, and J. Chen contributed to data collection. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
P. Song, J. Wu, J. Chen, F. Wang, J. Chen, and G. Wang declare that they have no competing interests.
Ethical standards
All procedures performed in studies involving human participants or on human tissue were in accordance with the ethical standards of the institutional and/or national research committee and with the 1975 Helsinki declaration and its later amendments or comparable ethical standards. Written informed consent was obtained from patients with approval by the Institutional Review Board in Heping Hospital Affiliated to Changzhi Medical College.
Additional information
Consent for publication
Not applicable.
Availability of data and materials
Please contact the corresponding author for the data request.
Supplementary Information
Fig. S1
Screening of miR-383-5p and PFN2. a, b Expression of miR-1236-3p, miR-383-5p, miR-622, and miR-634 in MCF‑7 and MDA-MB-231 cells with sh-circ-ADAM9 or sh-NC transfection checked by RT-qPCR, unpaired t-test. c, d Screening of PFN2 was conducted using RNA pull-down assay, unpaired t-test (***P < 0.00)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Song, P., Wu, J., Chen, J. et al. Knockdown of circ-ADAM9 inhibits malignant phenotype and enhances radiosensitivity in breast cancer cells via acting as a sponge for miR-383-5p. Strahlenther Onkol 199, 78–89 (2023). https://doi.org/10.1007/s00066-022-02006-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00066-022-02006-0