Abstract
Determination of the feeding history of polyphagous insect pests, such as noctuid moths (Lepidoptera: Noctuidae), is a critical element in developing population and resistance management strategies for such pests. To identify reliable markers for larval host plant determination and to develop simple extraction and detection methods, a metabolomics approach was implemented after acid hydrolysis of adult moth samples. We identified a derivative from cotton metabolites as a marker in adult moths that were fed cotton tissues as a larval diet, and we propose that the marker is tricycloheliocide H4 based on NMR and mass fragmentation analysis. Using this derivative from cotton metabolites as a marker, a targeted LC-MS/MS method reliably identified cotton as a larval diet in extracts of three noctuid moth species: Helicoverpa zea (cotton bollworm), Chloridea (Heliothis) virescens (tobacco budworm) and Chrysodeixis includens (soybean looper). We are using similar approaches to identify markers for other host plants including soybean.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Knowing patterns of host plant use by polyphagous crop pests is a prerequisite to understanding population dynamics, which, in turn, is necessary for designing appropriate programs to manage and delay pest resistance development. To determine larval host plant use, stable isotope measurement of 13C/12C ratios in moth wing tissues, using isotope ratio-mass spectrometry (IR-MS), has been used to distinguish insects that fed on C3-type plants such as cotton and soybean as larvae from insects that consumed C4-type plants such as corn and sorghum (Gould et al. 2002). While this method can differentiate larvae feeding on C3- or C4-type plants, it cannot differentiate larvae feeding on host plants with the same photosynthetic pathway, such as cotton and soybean (both C3). There is an analytical method that detects gossypol residues that were ingested by a larval feeding on cotton and retained in the adult moth (Orth et al. 2007). That method was developed to estimate the percentage of pest populations using cotton as a host plant for two cotton pests, Helicoverpa zea (cotton bollworm) and Chloridea (Heliothis) virescens (tobacco budworm). In this method, the total gossypol in moth body tissues is extracted as dianilino-gossypol by forming a Schiff base with aniline, and the extraction method is specific to gossypol, not other metabolites. Using data from both 13C/12C ratios and gossypol residues in adult moths, the importance of different hosts for H. zea was assessed (Head et al. 2010). However, it would be valuable to have an analytical method to simultaneously determine multiple feeding sources of lepidopteran pests such as corn, soybeans, and cotton based on the host plant(s) consumed by a larva.
Insect metabolomes can vary depending on the host plants (Riach et al. 2019) or diets (Dong et al. 2017) that the insects consume. There are numerous examples of insects sequestering plant secondary metabolites for their own defense or other purposes (Duffey 1980), and sequestration of plant defense compounds is well known in some specialist insect herbivores such as monarch butterfly, Danaus plexippus (Jones et al. 2019). Although it is not clear whether polyphagous lepidopteran crop pest species sequester plant metabolites actively or passively, one of the major cotton secondary metabolites, gossypol, is present in moth bodies (Orth et al. 2007). Therefore, it is possible for plant-specific metabolites to be present in adult moth tissues following larval feeding. However, identification of new metabolite markers in adult moths is challenging due to variation in the levels of marker metabolites in host plants, which depend on plant developmental stage and tissue type, and on the stability of plant metabolites inside the insect body during development. Moreover, the optimization of extraction and detection methods for different classes of metabolites in a moth sample would require a rigorous validation effort.
To identify multiple markers for host plant determination and to develop a simple extraction and detection method, we used a metabolomics approach after an acid hydrolysis of moth samples. We identified a cotton marker in adult moths that were fed cotton tissues as a larval diet. The derivative from cotton metabolites as a marker was produced during an acid hydrolysis in the presence of methanol, and we propose the structure as tricycloheliocide H4 based on NMR and mass fragmentation analysis. The targeted LC-MS/MS method identified cotton as a larval diet source in the extract of three lepidopteran moth species—H. zea, C. virescens and Chrysodeixis includens (soybean looper)—with high accuracy.
Methods and Materials
Extraction of Adult Moths and Plant Tissue Samples
Larvae of cotton bollworm (Helicoverpa zea; CBW), tobacco budworm (Chloridea (Heliothis) virescens; TBW) and soybean looper (Chrysodeixis includens; SBL) were reared from larva to adult by the Bayer Plant Biotech Entomology group in Chesterfield, MO, USA. For a larval diet, cotton bolls or cotton leaves (Gossypium hirsutum) and soybean pods or soybean leaves (Glycine max) were collected from plants that were grown in a growth chamber (Chesterfield, MO, USA) whereas edamame (Glycine max) and corn kernels (Zea mays) were purchased at Trader Joe’s and Schnucks (Chesterfield, MO, USA), respectively. Adult moths were lyophilized for three days, the wings were removed, and the moth body including head, thorax and abdomen was analyzed. Plant tissues used in larval diets, were lyophilized for five days and analyzed. For extraction from moth bodies, a single moth body was transferred into a 3.1-ml glass vial (Xpertek 3.1 ml Hi Recovery Clear Glass Vial, 15 × 45 mm; P.J. Cobert Associates, Saint Louis, MO, USA), and two glass beads were added (4-mm Glass Beads; VWR, Radnor, PA, USA). The body tissues were ground for 2 min at 1100 rpm using a mega-grinder that was similar to a commercial paint shaker and manufactured in-house. For extraction from plant tissues, 10 ± 1 mg of ground and lyophilized tissue was transferred into a glass vial (Xpertek 3.1 ml).
The acid hydrolysis solution, which included daidzein-d4 (CDN ISOTOPES, Pointe-Claire, Canada) as an internal standard, was prepared in a glass container by adding 30 μl of daidzein-d4 (10 μg ml−1) into 100 ml of 1 N HCl in methanol. One milliliter of the acid hydrolysis solution was added to each vial of tissue, which was then shaken using the mega-grinder for 2 min at 1100 rpm. After centrifuging the vials for 20–30 s (Centrifuge 5804, Eppendorf, Hauppauge, NY, USA) at 3000 rpm, vials were tightly capped and placed in a water bath (Circulating Water Bath; Thermo Fisher, Waltham, MA, USA) at 55 °C for 16 ± 1 h. Then, 70 μl of 28–30% NH4OH (J. T. Baker) was added to each vial, and sample vials were processed in a SpeedVac (Savant SC250EXP; Thermo Fisher) at 45 °C to complete dryness.
To perform the liquid/liquid extraction, 0.8 ml of 0.1% formic acid in H2O was added to each vial followed by 1.5 ml of ethyl acetate, and then vials were shaken using the mega-grinder for 2 min at 1100 rpm. Vials were centrifuged for 10 min at 3000 rpm, and then 1 ml of the ethyl acetate (top layer) was transferred into an Xpertek 2-ml vial (12 × 32 mm, Clear Glass Robotic Screw Thread Vial with Graduated Markings Spot; P.J. Cobert Associates). The Xpertek 2-ml vials were processed in the SpeedVac at 45 °C to partial dryness. The second ethyl acetate extraction was done by adding 1 ml of ethyl acetate to the original 3.1-ml glass vial followed by shaking using the mega-grinder for 2 min at 1100 rpm. Sample vials were centrifuged for 10 min at 3000 rpm. As the second extraction, 1 ml of ethyl acetate (top layer) was transferred into the same Xpertek 2-ml vial containing the partially dried extract, and the Xpertek 2-ml vials were placed in the SpeedVac at 45 °C to complete dryness. The dried extract was reconstituted by adding 200 μl of 0.1% formic acid in methanol, and centrifuging for 2 min at 3000 rpm. A 96-well membrane filter (AcroPrep Advance 96 Filter Plate, 0.45 μm PTFE, 350 μl well; Pall, Port Washington, NY, USA) was placed on top of a 2-ml collector plate (Xpertek 2-ml Deep 96-Well Collector Plate; P.J. Cobert Associates), and the reconstituted sample extract was transferred into the 96-well membrane filter wells. After centrifugation of the filter plate for 2 min at 3000 rpm, each filtrate, which is the final extract, was transferred into an insert (Xpertek 250 μl Insert, Bottom Spring GLR Glass; P.J. Cobert Associates) in a 2-ml vial, and vials were stored at −20 °C until analyzed.
Discovery of Metabolite Marker Candidates Using Untargeted Metabolomics
Untargeted metabolomics was performed to identify metabolite marker candidates that are unique in moths that were fed cotton tissue as larvae (cotton-fed insects) compared to moths fed other diets (i.e. corn or soy tissues) as larvae. A UPLC-QTOF-MS instrument (Xevo G2 QTOF; Waters, Milford, MA, USA) with an electrospray source was connected with a UPLC column (ACQUITY UPLC BEH C18, 1.7 μm, 2.1 × 150 mm; Waters). The flow rate was 300 μl min−1, and the two mobile phases were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The linear gradient was as follows: 5% B at 0 min, 5% to 40% B from 0 to 12 min, 40% to 80% B from 12 to 20 min, 80% B from 20 to 30 min, 80% to 5% B from 30 to 30.1 min, and the initial condition was held until 32 min. The Xevo G2 QTOF mass spectrometer was run in negative mode, and the data were collected from 200 Da to 1200 Da. Source parameters were as follows: capillary voltage, 2.1 kV; sampling cone voltage, 50.0 V; source temperature, 130 °C; desolvation temperature, 350 °C; cone gas flow, 5 L h−1; and desolvation gas flow, 900 L h−1. To ensure mass accuracy, leucine-enkephalin was used as the reference lock mass (m/z 554.2615) with the LockSpray interface. The data were processed using Waters MarkerLynx 4.1 software for the mass signal extraction with the peak alignment. Selection of marker candidates (i.e. mass features) was based on the presence of a mass ion only in cotton-fed insects and cotton tissues, not in corn or soy tissues or in insects fed these tissues. Selected marker candidates were checked manually using extracted ion chromatograms to verify the presence of a peak in cotton-fed insect samples or cotton tissue samples.
To obtain structural information about the cotton marker, a full scan and data-dependent scan acquisition were carried out in an Orbitrap Fusion mass spectrometer (Thermo Fisher) connected to a Vanquish UHPLC system including a Detector, Autosampler, Pump and Column compartment (Thermo Fisher). At least three different LC gradients were used with either an ACQUITY UPLC CSH C18 column (1.7 μm, 2.1 mm × 100 mm) or a BEH C8 column (1.7 μm, 2.1 mm × 150 mm). For positive mode, 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) were used whereas 0.005% formic acid in water (A) and 0.005% formic acid in acetonitrile (B) were implemented in negative mode. The mass spectrometer parameters are provided in Table S1. The MS2 and MS3 fragmentation data were further studied using Mass Frontier 8.0 (Thermo Fisher).
Targeted LC-MS/MS Method for Detecting the Cotton Marker
The mass spectrometric response of the cotton marker was measured on a triple quadrupole mass spectrometer (TSQ Vantage; ThermoFisher) equipped with an electrospray ionization (ESI) interface in a negative ionization mode. The MS/MS parameters were as follows: precursor ion of m/z 423.26, product ion of m/z 391.26, and collision energy of 40. An Accela UPLC pump and Autosampler (Thermo Fisher) were connected with the TSQ using an ACQUITY UPLC HSS Cyano (CN) column (100 Å, 1.8 μm, 2.1 mm × 50 mm; Waters). The flow rate was 500 μl min−1 and the two mobile phases were 0.005% formic acid in water (A) and 0.005% formic acid in acetonitrile (B). The linear gradient was as follows: 3% B at 0 min, 5% at 0.5 min, 30% at 3.5 min, 80% at 4 min, 95% min at 4.5 min, 100% at 5 min, 3% at 5.5 min, and the initial condition was held until 6 min. A lower percentage of formic acid (i.e. 0.005%) gave rise to a higher peak intensity compared to 0.1% formic acid in mobile phase in our mass spectrometer conditions. After a sample sequence run was completed, the Processing Method in the Xcalibur software (v2.1.0, Thermo Fisher) was run to determine the peak areas of the chromatograms.
Requirement for Methanol to Produce the Derivative from Cotton Metabolites as a Marker
Water, methanol, ethanol and isopropanol were each used to make 1 N HCl acid hydrolysis solutions to test whether methanol is essential for producing the derivative from cotton metabolites as a marker. Lyophilized and ground cotton boll tissues (10 mg dry weight) were extracted using the same method described above except for the acid hydrolysis process, in which the indicated acid hydrolysis solutions were used.
Heliocide Spike Test
Heliocide standards were generously provided by Drs. Lorraine Puckhaber and Bob Stipanovic at the USDA, ARS, SPARC, Insect Control and Cotton Disease Research Unit, College Station, Texas, USA. Ground moth tissue (10 mg) without a signal of the cotton marker (i.e. reared on corn or soy tissues; referred to as “non-cotton-fed”) was spiked with 100 ng of three different heliocide standard stocks (heliocide H1, a mixture of heliocide H2 and H3, and a mixture of heliocide H1 and H4) in methanol before the acid hydrolysis process described above. The negative controls were (1) samples spiked with 100 ng of gossypol and (2) samples without a spike (only ground moth tissue). A signal of the cotton marker was measured using the targeted LC-MS/MS method.
Nuclear Magnetic Resonance Spectroscopy (NMR) Analysis
For NMR measurements, lyophilized cotton boll tissues were extracted using the extraction method above to purify the derivative from cotton metabolites as a marker. The extract was injected into a Preparative LC system (Agilent 1260 Pump, VWD and Prep ALS, and 1290 Infinity II Fraction Collector, Santa Clara, CA, USA). A Synergi 4 μm Hydro-RP (80 Å) column (50 × 21.2 mm; Phenomenex, Torrance, CA, USA) was used to collect the fraction containing the marker. The flow rate was 40 ml min−1, and the two mobile phases were water (A) and acetonitrile (B). The LC gradient was as follows: 45% B at 0 min, 50% at 7 min, 100% at 8 min, 100% at 9 min, 45% min at 9.5 min, and 45% at 11 min. The marker was collected between 6.25 and 6.5 min. The semi-purified fraction of the marker was subjected to an LC fractionation for a further purification using an Agilent 1100 Series HPLC (Waldbronn, Germany) equipped with a Macherey-Nagel (Düren, Germany) Nucleodur C18 Gravity column (250 × 4.0 mm, 5-μm particle size). The eluents used were 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B). The linear gradient was as follows: 0% B for 5 min, 95% B at 25 min, 95% B at 35 min followed by a re-equilibration at 0% B. The flow rate of 1 ml min−1 was directed to an Agilent 1260 Fraction Collector (Waldbronn, Germany), and a time-based fraction of 6 s was collected. The assessment of the fraction was performed on an Orbitrap Fusion mass spectrometer (Thermo Fisher, Bremen, Germany). The fraction was dried in a vacuum concentrator, and the sample was dissolved and transferred into an NMR tube using 50 μl of deuterated methanol (CD3OD). To obtain structural information, 1H, 13C, 1H,1H-COSY, 13C-1H-HSQC, 1H-13C-HMBC, and 1H,1H-NOESY experiments were conducted using a BRUKER AVANCE NEO 800 MHz NMR spectrometer (Karlsruhe, Germany) equipped with a 1.7 mm-TCI cryoprobe.
Results
Discovery and Validation of Cotton Marker Candidates Using Lab-Reared Moths
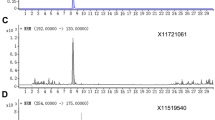
Untargeted metabolomics provided candidate mass features that were detected only in cotton tissue samples or cotton-fed insect samples, not in other plant tissues or non-cotton-fed insects. One of the candidate mass ions detected was m/z 423.218 [M-H]−, corresponding to a molecular formula of C26H31O5. Figure 1 shows that the peak of m/z 423.218 [M-H]− in extracted ion chromatograms was detected at 8.03 min only in an extract from a cotton boll or from a cotton bollworm moth that was reared on cotton boll tissues as a larval diet. There is another peak having the same mass ion (i.e. m/z 423.218) at 8.32 or 8.33 min, and this cotton marker isomer had an additional fragment ion of m/z 393.205 [M-H]− in addition to m/z 391.19 [M-H]− in an MS2 fragmentation study, which is provided in Supplementary data (Fig. S1). In general, signal intensities of the marker were higher in negative mode than in positive mode under our conditions (data not shown).
Extracted ion chromatograms of the cotton marker at 8.03 min (m/z 423.215–423.220 in negative mode) in (a) cotton boll, (b) edamame (soybean), (c) corn kernel and cotton bollworm (CBW) moths reared on cotton boll (CBW-COTTON, d), edamame (CBW-SOY, e) or corn kernel (CBW-CORN KERNEL, f). Y-axis of signal intensity was fixed at 1.5 e8 for plant tissue samples and at 1.8 e6 for moth samples. The peak at 8.32 or 8.33 min is an isomer of the cotton marker (see Fig. S1 for MS2 fragments). A 10-min LC gradient was used in these chromatograms
A targeted LC-MS/MS method was developed to confirm the mass ion of m/z 423.218 as a marker (Fig. S2). A total of 377 moths reared in the lab on different plant tissues were analyzed along with 138 blank extracts (Table 1). Daidzein-d4, which is a stable isotope of one of the legume isoflavones, was used as an internal standard that was spiked into moth samples before extraction. Although daidzein-d4 is not structurally related to the marker, we observed that it was stable during the acid hydrolysis process and showed a similar trend to the marker regarding variability in the extraction process or instrument response. The range of marker ratios (marker peak area response/daidzein-d4 peak area response) in cotton-fed insects of three different species (total 106 moths) was between 11.31 and 410.23 for CBW fed cotton bolls and for TBW and SBL fed cotton leaves. The SBL moths had a lower range of marker ratios (between 13.53 and 68.23) than the other two insect species (Table 1). The distribution of all data points is shown in Fig. S3.
Since a standard for the marker was not available, the extract of cotton-fed moths was mixed with that of non-cotton-fed moths (i.e. moths from soy- or corn-fed larvae) in different ratios and injected into LC-MS/MS to estimate the lower limit of detection, which was 0.2. Moths reared on non-cotton tissues such as soy and corn had marker ratios from 0 up to 3.26, although most of these lower but positive cotton signals appeared to be due to carryover, particularly in non-cotton-fed samples injected after cotton-positive samples with relatively high marker signals (data not shown). No false-negative sample was observed in 106 cotton-fed moths, and the highest marker ratio among non-cotton-fed samples (3.26) was 3.5 times less than the lowest marker ratio among samples reared on cotton tissues (11.31; Table 1 and Fig. S3). Eight out of 138 blank extract samples also showed marker signals. Nonetheless, this targeted LC-MS/MS method determined cotton-fed insects with high accuracy without any false-negative detection, although method optimization would be necessary to further reduce the number of lower, but false-positive signals.
Production of the Derivative from Cotton Metabolites as a Marker during an Acid Hydrolysis Step
The marker peak of m/z 423.218 [M-H]− was not detected in a sample either when water was used instead of methanol in the acid hydrolysis solution or when cotton tissues were extracted using methanol without an acid hydrolysis step (data not shown). This indicates that a methanolysis is essential for producing the marker. When ethanol or isopropanol was used in the acid hydrolysis solution instead of methanol, we detected an ethylated (m/z 437.232 [M-H]−) or isopropylated (m/z 451.248 [M-H]−) mass ion, respectively, rather than a methylated marker, which was determined based on the expected accurate masses with a delayed retention time (Fig. 2).
Extracted ion chromatograms of cotton extracts in the presence of (a) methanol, (b) ethanol or (c) isopropanol during the acid hydrolysis step. The derivative from cotton metabolites as a marker (i.e. m/z 423.218, d) was detected only in a sample extracted using a methanolic hydrolysis solution whereas an ethylated (m/z 437.232, e) or isopropylated (m/z 451.247, f) cotton marker was detected based on the expected accurate masses with a delayed retention time
We identified a cotton secondary metabolite having the same molecular weight as the marker, i.e. heliocide B (C26H32O5, monoistotopic mass of 424.2250). Heliocide B has an additional methyl group compared to heliocide H (C25H30O5, monoistotopic mass of 410.2093), which is abundant in cotton tissues (Opitz et al. 2008) whereas heliocide B is present only in a few cotton species such as G. barbadense (Stipanovic et al. 1977b). We initially hypothesized that heliocide B is the marker compound, produced by a methylation of heliocide H in an acid hydrolysis step. However, the peak of the heliocide B standard was detected at 19.54 min whereas the cotton marker had a retention time of 10.95 min in a 23-min LC gradient method (Fig. S4), leading us to conclude that heliocide B is not the marker.
Next, we tested whether heliocide compounds are a precursor of the cotton marker. We spiked 100 ng of heliocide H1, a heliocide H1 + H4 mix, a heliocide H2 + H3 mix or gossypol (negative control) into ground moths that were reared on non-cotton diets such as corn kernel. Heliocides H2, H3 and H4 were not available alone, only in mixtures with other heliocides (i.e. H2 + H3 or H1 + H4). After an extraction, we used a targeted LC-MS/MS method with a 6-min gradient to detect the cotton marker, and Fig. 3 shows that the marker peak was detected in samples spiked with either heliocide H1 or the mixture of heliocides H1 and H4. The marker peak areas in samples spiked with the mixture of heliocides H1 + H4 were approximately 2-fold those in samples spiked with heliocide H1 standard alone. Relatively smaller peaks were detected in samples spiked with the mixture of heliocides H2 + H3, and the mean peak area of the marker was approximately 11% of that in samples spiked with the mixture of heliocides H1 + H4. One of three replicate samples spiked with gossypol showed a small marker peak, but the mean area of the peak was approximately 8% of that in samples spiked with the mixture of heliocides H1 + H4. No peak was detected when moth samples were extracted without a spike of heliocides. When three different amounts of the mixture of heliocides H1 and H4 were spiked (200, 500 or 2000 ng), the marker ratios were between 5.5 and 32.9 with a linearity of R2 = 0.9913 (Fig. S5). Therefore, the results of heliocide spike experiments suggested that the origin of the cotton marker is heliocide H1, heliocide H4 or a combination of the two.
Chromatograms of the cotton marker after three different heliocide standard stocks (i.e. H1, H2 + H3 or H1 + H4) or gossypol (100 ng each per sample) were spiked into a non-cotton-fed ground moth sample prior to extraction. A targeted LC-MS/MS method with a 6-min LC gradient was used to detect the derivative from cotton metabolites as a marker, and chromatograms of all three replications are shown except for the ‘No spike’ control (n = 2). Y-axis indicates peak intensity, and the highest value was set as 7000 in all chromatograms. X-axis shows retention time (minute)
NMR Analysis
The purified marker from cotton boll tissues was analyzed using a BRUKER AVANCE NEO 800 MHz NMR spectrometer (Table 2; Figs. S6–S11). The data were analyzed using ACD/Structure Elucidator Suite (ACD/Labs, Toronto, Ontario, Canada) for a structure elucidation, and the proposed structure is shown in Fig. 4.
NMR data analysis revealed the presence of several structural features more reminiscent of heliocide H4 than heliocide H1. These features included the presence of one vinylic proton at C-18 (δH,C 5.5, 121.2, CH) next to a methyl group (C-20, δH,C 1.7, 23.0, CH3) attached to C-17 (δC 135.8), an aromatic ring fully substituted and carrying two hydroxy functions at C-6 (δC 145.7, OH) and C-7 (δC 147.4, OH), an isopropyl group corresponding to C-12 (δH,C 4.3, 27.6, CH), C-13 (δH,C 1.3, 20.6, CH3) and C-14 (δH,C 1.3, 20.8, CH3) attached at the aromatic C-8 (δC 135.7) (Figs. S6, S7 and S11). However, the dimethyl-7-hemigossypolone portion was likewise altered, i.e. there was no longer an aldehydic function at C-11 (δH,C 5.4, 77.6, CH), which normally substitutes the aromatic C-5 (δC 123.5), and only one carbonyl function was detected (C-1, δC 204.0 ppm). In addition, the 2-methylbut-2-ene sidechain at C-16 (δH,C 2.0, 45.7, CH) was distorted, hinting at a new compound, even though the methyl group (C-15, δH,C 1.2, 21.0) at C-2 (δC 50.7, C) remained intact (Fig. S6). The basic skeleton of heliocide H4—made of three rings—was clearly supported by 2D-NMR analysis and all the correlations observed.
The resonance frequencies of the methine at position C-11 and the HMBC correlations of H-11 to a quaternary carbon at δC 85.2 ppm (C-4) indicated the presence of a dihydrobenzofuran ring, originating seemingly from the former carbonyl and aldehyde functions in heliocide H4 (Fig. S7). Moreover, the spectral assignment revealed the presence of a six-membered ring, made of a methylene (C-25, δH,C 2.0 & 2.3, 41.7, CH2), a quaternary carbon (C-23, δC 76.6), and a methine (C-22, δH,C 2.3, 41.0, CH) adjacent to the dihydrofuran ring, and all the expected 2D correlations were observed (Figs. S6, S7, S8 and S10). A methyl rest (C-24, δH,C 0.3, 24.2, CH3) and a methoxy group (C-26, δH,C 3.1, 48.7, CH3O) were found attached to C-23 (Fig. S6), as supported by the HMBC correlations from the methyl protons to C-23, C-22 and C-25, and those from the methoxy to C-23 (Fig. S7). The extreme shielding of the methyl group in the 1H spectrum hinted at a particular spatial arrangement. To form this tetrahydropyran ring, the methylene and the methine rests were attached to the five-membered heterocycle at C-11 and C-4, respectively. This was proved by the HMBC correlations from the diastereotopic protons at C-11 to C-5, and those from the methine in position 22 to C-4, C-10 (δC 144.6, C) and C-3 (δH,C 2.1, 44.7, CH).
Furthermore, the COSY spectrum (Fig. S8) showed a correlation between the methine at C-22 and a methylene group (C-21, δH,C 1.1 & 1.6, 27.5, CH2)—belonging to another cyclic spin system—which itself was further attached to the methine at C-16. The mutual HMBC correlations from the two methine groups at positions 22 and 16 and to C-3, combined with the HMBC correlation series from the diastereotopic protons at C-22 to the quaternary carbons C-4, C-10 and C-23, clearly demonstrated the presence of a further six-membered ring. The HMBC, COSY and NOESY experiments fully agreed with this assignment featuring a contiguous hexacyclic system, carrying a heliocide-like scaffold, as displayed in Fig. 4.
The novel compound possesses seven stereocenters, i.e. four protonated and three quaternary carbons, which further increase the difficulty for the elucidation of the full stereochemistry. However, the chain made by C-21, C-22, C-23 and C-25 confers rigidity to the core carrying all these stereocenters. The compactness of this scaffold would make the stereocenters so interdependent that the determination of one could be enough to progressively reveal the others. For this purpose, the NOESY interactions were carefully analyzed in order to deduce the relative stereochemical orientation (Fig. S9).
The ‘decisive’ methyl attached at C-2 showed a NOESY interaction with H-3, and this kind of interaction is typical of most natural heliocide sesquiterpenes carrying a 2,3-trans relative configuration between C-2 and C-3 (Stipanovic et al. 1977a). In addition, the NOESY experiment revealed the same trans arrangement with C-16 by the interaction from Me-15 to H-16, which forced Me-15 to be in an opposite plane from the octahydro-chromene portion. The lack of NOEs between H-16 and H-22, despite the differentiated interactions with the diastereotopic protons at C-21, indicated their relative trans orientation. The observed NOEs between one of the diastereotopic protons at C-19 and H-22 corroborated this 16,22-trans orientation deduction. The strong NOESY interaction arrays between one of the diastereotopic protons at C-25 with the highly shielded Me-24, while the other interacted with the MeO-26, combined with the correlation between Me-24 and the two H-21 indicated the 22,23-trans-relative configuration. This assignment implied only one possible helicity left to the dihydrofuran ring, namely, with the oxygen atom in an opposite plane from the two contiguous six-membered rings.
Modeling tricycloheliocide H4 showed that the C-24 methyl group is held in a rigid position above the plane of the phenyl ring, and hence in the shielding cone. Thus, it is not surprising that the C-24 methyl protons are shifted upfield (i.e. δH 0.3) due to the strong shielding effect of the phenyl ring. On the other hand, the strongly shielded 1H shift of Me-24 (i.e. δH 0.3) would be an indication of the huge steric compression—caused by the three condensed rings—which corroborate this spatial arrangement (Hansen and Spanget-Larsen 2017; Kleinpeter et al. 2011; Prashad et al. 1980). Processing all the gathered relative stereochemical information together, the number of possible stereoisomers for this rigid structure drastically shrinks from 27 stereoisomers to two enantiomers, 2S,3R,4R,11S,16S,22S,23R or 2R,3S,4S,11R,16R,22R,23S. Considering its most probable origin from heliocide H4, the configuration of the derivative from cotton metabolites that we put forward as a marker is proposed as 2S,3R,4R,11S,16S,22S,23R. Therefore, we named the marker compound as tricycloheliocide H4.
MS Fragmentation Analysis
To better understand the marker structure, mass fragmentation data were processed using Mass Frontier 8.0, in which the ‘Fragments & Mechanisms Module’ automatically generated fragments, detailed fragmentation, and rearrangement mechanisms from a chemical structure. In positive mode, structures of most major fragment ions in MS2 and MS3 acquisitions were generated by Mass Frontier 8.0 with a mass error of <0.05 Da (Figs. S12–S15). The loss of CH3OH or CH4 at C-26 (Fig. 4) appears to be an initial fragmentation reaction of tricycloheliocide H4. In negative mode, fragment structures were not generated by Mass Frontier 8.0, but the neutral losses such as CH3OH showed a similar pattern with those observed in positive mode (Figs. S16–S18).
Discussion
Determination of the feeding history of insect pests is a critical element for developing insect refuge and resistance management strategies for transgenic crops (Head et al. 2010). It is desirable to have a method that would readily identify the feeding source(s) of target insect pests, particularly polyphagous lepidopteran species such as CBW, TBW and SBL, which are key pests of major crops such as corn, soybean, and cotton in the USA and other geographic locations.
Previously, IR-MS (Isotope Ratio-Mass Spectrometry) was implemented successfully to identify an insect feeding source as a C3- or C4-type host plant (Gould et al. 2002). Although the δ13C composition in a moth characteristic of either a C3- or C4-type host cannot distinguish individual host plant species within the same type, this provides valuable information for understanding the makeup of a moth population when used with an additional method based on plant-specific metabolites such as gossypol in cotton plants (Head et al. 2010). Orth et al. (2007) developed a gossypol residue assay that was based on the formation of a Schiff base with aniline followed by a solid-phase extraction (SPE) to ensure high selectivity as well as sensitivity. To cover a broader range of plant secondary metabolite classes, we used an acid hydrolysis followed by a liquid/liquid extraction to identify metabolite markers for major crops such as cotton or soybean in a moth that utilized that host plant during the larval stage. An acid hydrolysis process would increase the concentration of some of secondary metabolites that are present as a glycoside by removing glycoside moieties from the major structural backbone such as a variety of isoflavone glycosides in soybean (Chiang et al. 2001). However, some metabolites can be labile during extraction due to the acidic condition and heat treatment (55 °C for 16 h under our conditions).
Cotton contains a vast array of secondary metabolites including cotton-specific terpenoid aldehydes such as heliocides and gossypol (Benedict et al. 2004; Bell et al. 1978; Bezemer et al. 2004; Liu et al. 1999; McCormick 1982; Stipanovic et al. 1977a, 1977b), which can also be induced by physical wounding (Park et al. 2019). Heliocides H1, H4, B1 and B4 are the products of a Diels–Alder reaction of hemigossypolone and β-ocimene, whereas H2, H3, B2 and B3 require hemigossypolone-7-methyl ether with myrcene (Stipanovic et al. 1977a). The amount of heliocides in leaves was higher than that of gossypol (Opitz et al. 2008), but the toxicity of heliocide compounds against larvae was lower than that of gossypol (Elliger et al. 1978), although a hormetic effect of gossypol on the generalist herbivore Helicoverpa armigera was observed (Celorio-Mancera et al. 2011). When larvae of the three species tested consume cotton tissues, these terpenoid aldehydes would be sequestered, degraded, or detoxified in the body at different rates. The cotton marker was detected in SBL moths at relatively lower amounts than in the other two species (Table 1, Fig. S3), indicating that species-dependent variability exists in the accumulation of cotton secondary metabolites.
Heliocide H4 in cotton plants consumed by lepidopteran larvae appears to be converted into tricycloheliocide H4 (Fig. 4) during an acid hydrolysis in the presence of methanol. Based on NMR analysis, a mechanism of the production of tricycloheliocide H4 is proposed in Fig. 5. An acid-catalyzed carbonyl ene reaction of heliocide H4 would give structure a, and a nucleophilic addition of the newly formed tertiary alcohol with the neighboring aldehyde moiety would give hemi-acetal b. Then, acid-catalyzed dehydration of b would generate oxonium ion c, and a nucleophilic addition of the neighboring alkene to oxonium ion c would give rise to tertiary carbocation d. Finally, methanolysis of carbocation d would lead to tricycloheliocide H4. Heliocide H1 is expected to go through a similar mechanism during acid hydrolysis, and the peak that came out at 8.32 or 8.33 min, after the tricycloheliocide H4 peak at 8.03 min (Fig. 1), could be a product of heliocide H1 although we did not confirm it. These two peaks can come out together without a baseline separation in the targeted LC-MS/MS method, which had a relatively short 6-min LC gradient. That could be why we observed a peak of tricycloheliocide H4 when heliocide H1 was spiked into non-cotton-fed moth tissues; alternatively, there might be heliocide H4 contamination of the purified heliocide H1 sample.
The targeted LC-MS/MS method to detect tricycloheliocide H4 in a moth extract gave no false negatives (Table 1), although relatively weak marker signals were detected in several non-cotton-fed insect samples and blank extracts. We are working to reduce carryover from previous injections by optimizing the current LC-MS/MS method. Nevertheless, the high accuracy of detection of the cotton marker in moth extracts indicates that this LC-MS/MS method can be used as a new screening tool to identify cotton as a moth host plant. We are currently validating a soy marker that was recently discovered using an untargeted lipidomics approach. If soybean, cotton and other plant species of interest could be identified by an “one extraction for all target metabolites with one injection” method, it would provide better information for developing a refuge strategy as well as for assessing cross-crop resistance risk.
Data Availability
All data were captured within the manuscript with supplementary materials.
References
Benedict C, Martin G, Liu J, Puckhaber L, Magill C (2004) Terpenoid aldehyde formation and lysigenous gland storage sites in cotton: variant with mature glands but suppressed levels of terpenoid aldehydes. Phytochemistry 65:1351–1359
Bell A, Stipanovic R, O’Brien D, Fryxell P (1978) Sesquiterpenoid aldehyde quinines and derivatives in pigment glands of Gossypium. Phytochemistry 17:1297–1305
Bezemer T, Wagenaar R, Van Dam N, Van Der Putten W, Wackers F (2004) Above- and below-ground terpenoid aldehyde induction in cotton, Gossypium herbaceum, following root and leaf injury. J Chem Ecol 30:53–67
Celorio-Mancera M, Aha S, Vogel H, Heckel D (2011) Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genomics 12:575
Chiang W-D, Shih C-J, Chu Y-H (2001) Optimization of acid hydrolysis conditions for total isoflavones analysis in soybean hypocotyls by using RSM. Food Chem 72:499–503
Dong H-L, Zhang S-X, Tao H, Chen Z-H, Li X, Qiu J-F, Cui W-Z, Sima Y-H, Cui W-Z, Xu S-Q (2017) Metabolomics differences between silkworms (Bombyx mori) reared on fresh mulberry (Morus) leaves or artificial diets. Sci Rep 7:10972
Duffey S (1980) Sequestration of plant natural products by insects. Annu Rev Entomol 25:447–477
Elliger C, Chan B, Waiss A (1978) Relative toxicity of minor cotton terpenoids compared to gossypol. J Econ Entomol 71:161–164
Gould F, Blair N, Reid M, Rennie T, Lopez J, Micinski S (2002) Bacillus thuringiensis-toxin resistance management: stable isotope assessment of alternate host use by Helicoverpa zea. Proc Natl Acad Sci U S A 99:16581–16586
Hansen PE, Spanget-Larsen J (2017) NMR and IR investigations of strong intramolecular hydrogen bonds. Molecules 22:552
Head G, Jackson RE, Adamczyk J, Bradley JR, Van Duyn J, Gore J, Hardee DD, Leonard BR, Luttrell R, Ruberson J, Mullins JW, Orth RG, Sivasupramaniam S, Voth R (2010) Spatial and temporal variability in host use by Helicoverpa zea as measured by analyses of stable carbon isotope ratios and gossypol residues. J Appl Ecol 47:583–592
Jones P, Petschenka G, Flacht L, Agrawal AA (2019) Cardenolide intake, sequestration, and excretion by the monarch butterfly along gradients of plant toxicity and larval ontogeny. J Chem Ecol 45:264–277
Kleinpeter E, Lämmermann A, Kühn H (2011) The anisotropic effect of functional groups in 1H NMR spectra is the molecular response property of spatial nucleus independent chemical shifts (NICS)—conformational equilibria of exo/endotetrahydrodicyclopentadiene derivatives. Org Biomol Chem 9:1098–1111
Liu J, Benedict CR, Stipanovic RD, Bell AA (1999) Purification and characterization of S-adenosyl-L-methionine: Desoxyhemigossypol-6-O-methyltransferase from cotton plants. An enzyme capable of methylating the defense terpenoids of cotton. Plant Physiol 121:1017–1024
McCormick JP (1982) Secondary metabolites of Gossypium: a biogenetic analysis. In: Montalvo J (ed) Cotton dust. ACS symposium series, Vol. 189. American Chemical Society, Washington, DC, pp. 275–300
Opitz S, Kunert G, Gershenzon J (2008) Increased terpenoid accumulation in cotton (Gossypium hirsutum) foliage is a general wound response. J Chem Ecol 34:508–522
Orth R, Head G, Mierkowski M (2007) Determining larval host plant use by a polyphagous lepidopteran through analysis of adult moths for plant secondary metabolites. J Chem Ecol 33:1131–1148
Park S-H, Scheffler J, Scheffler B, Cantrell C, Pauli CS (2019) Chemical defense responses of upland cotton, Gossypium hirsutum L. to physical wounding. Plant direct 3:e00141
Prashad M, Seth M, Bhaduri AP, Banerji A (1980) 13C NMR studies: effect of steric compression on chemical shifts of carbons in diastereoisomers. Org Magn Reson 14:74–75
Riach AC, Perera MVL, Florance HV, Robinson LA, Penfield SD, Hill JK (2019) Metabolic fingerprints reveal how an insect metabolome is affected by different larval host plant species. Arthropod Plant Interact 13:571–579
Stipanovic R, Bell A, O’Brian D, Lukefahr M (1977a) Heliocide H2: an insecticidal sesterterpenoid from cotton (Gossypium). Tetrahedron Letter 6:567–570
Stipanovic RD, Bell AA, Lukefahr MJ (1977b) Natural insecticides from cotton (Gossypium). In: Hedin PA (ed) Host plant resistance to pests. ACS symposium series, Vol. 62. American Chemical Society, Washington, DC, pp. 197–214
Acknowledgments
We appreciate an early effort on structure elucidation by Lloyd Sumner (University of Missouri), Martin Ruebelt and Bosong Xiang (Bayer US Crop Science), and insect rearing by the Plant Biotech Entomology group (Bayer US Crop Science). We also are grateful to Paula Price and Ben Yorke for reviewing the manuscript.
Funding
All authors were employees of Bayer Crop Science, and the work was funded by Bayer Crop Science.
Author information
Authors and Affiliations
Contributions
Jae Hak Kim and Graham Head designed and coordinated the project. Jae Hak Kim, Andreas Lagojda, Dieudonné Tshitenge Tshitenge, Daniel P. Walker, Graham Head wrote the manuscript. Jae Hak Kim performed metabolomics and LC-MS work while Andreas Lagojda, Dirk Kuehne and Dieudonné Tshitenge Tshitenge generated and interpreted NMR data and structure elucidation. Swapan Chaudhuri and Daniel P. Walker contributed to the interpretation of NMR data and a mechanism of the marker production.
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
All authors consented to a participation in this project.
Consent for Publication
All authors reviewed and approved the final version of the manuscript.
Code Availability
Not applicable.
Electronic supplementary material
ESM 1
(DOCX 7790 kb)
Rights and permissions
About this article
Cite this article
Kim, J.H., Lagojda, A., Kuehne, D. et al. Determination of Cotton as a Larval Feeding Source for Lepidopteran Moths Using a Derivative from Cotton Metabolites as a Marker by LC-MS/MS Method. J Chem Ecol 46, 956–966 (2020). https://doi.org/10.1007/s10886-020-01219-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10886-020-01219-w