Abstract
A major part of chemical conversions is carried out in the fluid phase, where an accurate modeling of the involved reactions requires to also take into account solvation effects. Implicit solvation models often cover these effects with sufficient accuracy but can fail drastically when specific solvent–solute interactions are important. In those cases, microsolvation, i.e., the explicit inclusion of one or more solvent molecules, is a commonly used strategy. Nevertheless, microsolvation also introduces new challenges—a consistent workflow as well as strategies how to systematically improve prediction performance are not evident. For the COSMO and COSMO-RS solvation models, this work proposes a simple protocol to decide if microsolvation is needed and how the corresponding molecular model has to look like. To demonstrate the improved accuracy of the approach, specific application examples are presented and discussed, i.e., the computation of aqueous pKa values and a mechanistic study of the methanol mediated Morita–Baylis–Hillman reaction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Currently available quantum chemical methods are fairly advanced for treating individual molecules or smaller clusters of molecules in vacuum or an ideal gas phase where it is possible to make predictions within the experimental accuracy (between 0 and 1 kcal/mol) [1,2,3]. However, quantum chemistry is still not able to treat molecules in fluid environments in an equally accurate and yet computationally affordable manner. We estimate that around 90% of industrial chemistry and almost 100% of biochemistry takes place in solution (or in liquid bulk) and solvation effects on reaction thermodynamics and kinetics are of major importance in many cases. Thus, the availability of suitable models to describe these effects constitutes an important (and sometimes underestimated) bottleneck for the practical applicability of quantum chemistry to problems in chemical research.
Currently, there are different approaches to account for solvent effects. A direct insight with high accuracy is obtained by ab initio molecular dynamics [4], which takes into account intermolecular interactions most straightforwardly and in principle in the most appropriate way possible. However, large ensembles of molecules and long simulation times are required to obtain converged values of thermodynamic functions (e.g. Gibbs free energies), which makes ab initio molecular dynamics prohibitive in many cases. A shortcut to this approach is the combination of quantum mechanics (QM) and molecular mechanics (MM): These QM/MM approaches treat the region of interest in the system using quantum chemistry and the remaining part, which often corresponds to the surrounding solvent molecules, with classical molecular mechanics. This approach yields reasonable results if suitable force fields are available but is still computationally quite expensive [5]. In this context, also the Reference Interaction Site Model (RISM) [6] should be mentioned, which represents a cluster expansion method applied to molecular fluids. This approach allows to account for preferred explicit interactions without the necessity of performing long dynamic simulation runs. The original 1D site–site theory by Chandler and Anderson [6] was extended to 3D RISM by several authors [7,8,9,10]. Kast et al. combined 3D RISM with quantum chemical method via an embedded cluster approach (EC-RISM) [11].
Dielectric continuum solvation models enable the much more efficient alternative of static calculations [12] and are therefore widely used by the quantum chemical community. These approaches are based on the assumption of a continuous polarizable medium around the solute, which is fully characterized by its dielectric constant. This practically means, that the charge distribution of the solute polarizes the dielectric continuum which results in an external potential introduced into the Hamiltonian of the solute. The physics of the solute–solvent system has to obey the Poisson equation as a boundary condition. This approach led to the development of several solvation methods such as PCM [13,14,15], IEFPCM [16], IPCM [17], SMD [18], SM8 [19, 20], COSMO [22], COSMO-RS [25, 26], etc.
The polarizable continuum method, mostly known as PCM, was the first implementation of the dielectric continuum solvation approach and was developed by Tomasi and co-workers almost four decades ago [13]. However, continuum solvation methods lead in best case to qualitative results of how the total energy of a molecule changes upon the transition from gas phase to solution. They are even less suited to predict free energies or entropies in solution as they do not provide access to changes of the molecular partition function upon solvation, which is required for the computation of most thermodynamic functions. Several improvements of the original PCM method have been addressed in different flavors of PCM methods such as COSMO-PCM (CPCM), isodensity-PCM (IPCM), integral equation formalism (IEFPCM) and surface and volume polarization for electrostatic interaction (SVPE). However, all these approaches still exhibit some deficiencies with short range contributions caused by the description of the solvent as continuum with fixed or locally variable dielectric constant. A more recent series of solvation models developed by the Truhlar group and are the so-called SMx models [19, 20], where x is an alphanumeric label to indicate the version. These achieve slightly better results compared with the PCM methods. Further, the solvation model based on density [18] (SMD) has to be mentioned due to its rather good prediction performance for Gibbs free energies in solution [21].
The conductor-like screening model [22], usually abbreviated as COSMO, is another popular solvation method that employs the dielectric continuum solvation approach. Originally developed by Klamt and Schüürmann in 1995, it shows a high degree of similarity to PCM. The novelty of COSMO consists in using scaled conductor boundary conditions instead of the exact dielectric conditions. This modification of the boundary conditions strongly reduces the outlying charge error which leads to an easier to handle and computationally more efficient approach than standard PCM. Both PCM based and COSMO solvation models, have been successfully applied in numerous computational studies of chemical reactions when solvation effects cancel out [23, 24]. Nevertheless, when applying COSMO or PCM to more challenging cases (and many problems of technical relevance fall into this category), several weaknesses of these approach were observed. One example is the inability to distinguish between rather different solvents with similar dielectric constant, e.g., cyclohexane and benzene or methoxyphenol and heptanone. This limitation led to the development of a new strategy called conductor-like screening model for realistic solvation (COSMO-RS). It combines COSMO with a statistical thermodynamics treatment of interacting molecular surface charges [25, 26]. In contrast to simple solvation models, COSMO-RS contains some fitted parameters, but these are not chemistry-dependent, so that it still represents a theory that can in principle be applied to the whole periodic table. With the inclusion of different types of molecular interaction, e.g., using specific parameters to describe hydrogen bonding and dispersive interactions, this approach allows to treat molecular systems in solution at variable temperature and mixtures in a more proper way. This opens the door for many application areas which previously had been inaccessible for continuum models. It should be mentioned that the abovementioned fitted parameters used in the (original) COSMO-RS solvation model are proprietary and unpublished, apart from those of the very first few COSMO-RS versions.
Originally, the COSMO-RS model was rather designed for chemical engineering applications like phase diagram construction or solvent selection. Nevertheless, this theory can as well be used to obtain Gibbs free energies of formation for species in solution, which allows for the treatment of chemical reactivity [27, 28]. Despite its success in describing chemistry in solution for neutral molecules and even ionic systems with moderate ion–solvent interactions [29], the accuracy of COSMO-RS is significantly lowered when used to describe very strong intermolecular interactions such as hydrogen bonding between protic solvents and highly localized charges. One workaround for improving these deficiencies is the inclusion of up to a few explicit solvent molecules into the molecular model of the solute while describing the effects of less strongly interacting further solvent molecules with the same implicit solvation models as discussed before. Such approaches are called cluster-continuum model or microsolvation.
Since more than three decades quantum chemists from time to time have included solvent molecules in their models for a more realistic description of solutes or to obtain converged bulk properties, as e.g., reviewed by Gadre et al. [30]. A major step towards the use of microsolvation for a more predictive access to thermodynamics of ions was the paper of Pliego and Riveros from 2001 [31]. The authors used a “variational principle” not too dissimilar from the approach presented in this work to predict solvation free energies of a number of singly charged ions. With the employed IPCM model, the number of explicit (water) solvent molecules required by their protocol was rather high (in several cases three, although very small cations and anions were considered), but the presented approach was able to reduce the error in solvation free energies by a factor of around 2.5 with respect to the neat continuum model. The same authors also applied microsolvation to pKa prediction [32] as well as for the exploration of a reaction mechanism (basic formamide hydrolysis) [33]. In both cases, a significantly improved agreement with the experiment suggests that microsolvation in fact provided a much better—or more balanced—description of the solutes of interest. Klamt, the inventor of COSMO-RS, together with Eckert and Diedenhofen, also observed that the addition of one or two water molecules (with solvent H atom attached to anion acceptor sites and solvent O atom attached to cation acidic H atoms—in the neutral species the explicit solvent was kept at the same positions) leads to a better pKa prediction for strong acids and bases, i.e., the slope of the correlation line of predicted vs experimental values was closer to 1 [34]. In the same work also the problem of a too high structural flexibility of clusters of water with less strongly interacting species is discussed, showing that there is not the one way towards better results. At the same time, Ho and Coote reported for a set of 55 carboxylic acids, that the addition of one explicit water molecule to both ions and neutral species led to improvements in predicted pKa when using different solvent models [35]. Ho and Ertem also studied the effect of microsolvation by a varying number of solvent (water) molecules on solvation free energies and concluded that—as one would expect for entropic reasons—that there is no “the more—the better” [36]. The same tendency was also found by Cramer and Truhlar when using SMx and SMD solvent models [18, 37] and Basdogan and Keith when investigating the Morita–Baylis–Hillman reaction [38]. This indicates that microsolvation can improve in a similar way the results of any implicit solvation model (if applied correctly). However, the one major drawback of microsolvation has also been the subject of more recent work by Florez, Restreppo et al. The conformational space strongly increases when adding several solvent molecules to a solute, which can make it quite difficult to identify the lowest-energy conformer or, even worse, to end up with a meaningful conformer ensemble [39, 40]. These observations show that although microsolvation clearly represents a way how to treat more complicated solutes in computations, there still is no clear concept how to predictively apply it on a daily basis in projects focusing on diverse chemical problems.
The reason why an explicit consideration of the solvent is needed, even if in principle advanced solvation models are used, is simply because interactions a chemist would still call “intermolecular” can become as strong or even stronger than actual chemical bonds. Computed gas-phase enthalpies of interaction (obtained by the authors with the same protocol as described below in “Computational Details” apart from optimizing structures in the gas phase and—of course—not adding solvation contributions, either) can give an idea of which interactions a solvation model has to be able to deal with. Enthalpies of hydrogen bond formation between neutral species are in the range of only up to 20 kJ/mol (e.g. HOH…OH2: − 14.9 kJ/mol; HOH⋯NH3: − 20.0 kJ/mol; FH⋯FH: − 16.7 kJ/mol). However, if hydrogen bonding interactions take place with ionic species, interaction enthalpies can be ten times larger (e.g. HOH…OH−: − 142.0 kJ/mol; H2OH+⋯OH2: − 152.6 kJ/mol; FH⋯F: − 207.3 kJ/mol). Consequently, for such systems solvation models have to account for interactions that are as strong as (more labile) chemical bonds (e.g. O2N…NO2: − 52.5 kJ/mol; F⋯F: − 128.9 kJ/mol; HO…OH: − 206.9 kJ/mol). This demonstrates why the incorporation of explicit solvent molecules into a molecular model sometimes is a must to yield a more realistic description of the considered system: Interactions as strong as chemical bonding lead to significant structural relaxation and changes in electron density—and these effects are simply not covered by any implicit solvation model.
This work investigates under which conditions microsolvation should be applied when using COSMO or COSMO-RS as implicit solvation models. Application to the prediction of aqueous pKa values shows a clear improvement when using microsolvation, compared to isolated free ions treated by implicit solvation only. As an even more critical use case, calculations on the mechanism of the Morita–Baylis–Hillman are presented. Because this reaction starts from neutral reactants and involves intermediates with highly localized negative charge in organic media, it represents a major challenge for any solvation treatment. Significant improvements are observed when applying microsolvation, although the results still are not in quantitative agreement with experiments. These results highlight that there is still significant room for improvement in methods and protocols for the computational treatment of such systems.
Computational details
All computations were performed at the density functional theory (DFT) level using the TURBOMOLE 7.3 program package [41, 42]. For all species as well as transition states (with and without explicit solvation molecules) a conformational sampling was performed to identify the energetically lowest conformer. A set of initial conformers was generated by rotating all single bonds (separately) by a defined angle, usually 120°. The investigated systems are rather small, so only terminal symmetrical groups, e.g., CH3 or bonds to DABCO substituents were excluded from that treatment. For transition states additional constraints were made and the bond lengths of the bonds involved in the reaction were kept constant. Any molecular model containing explicit solvent molecules was also subjected to the same generation of initial structures and conformational sampling. More details on the placing of the explicit solvent molecules are given in the section “Computation of acid–base reactions and pKa values”.
In the conformational sampling, the geometries of all conformers were optimized at the TPSS level [43], employing the triple-zeta def2-TZVP (TZ) basis sets [44], the COSMO solvation model [22] with a dielectric constant of infinity, and the D3 dispersion correction [45] with Becke–Johnson (BJ) damping [46, 47]. For each of the optimized conformers, a single-point energy in the gas phase at the TPSS-D3/def2-TZVP level was computed and the solvation free energy was calculated with the COSMO-RS solvation model [25, 26] as implemented in COSMOtherm 2018 [48] employing the 2018 BP86/def-TZVP parametrization (and the older 2015 and 2012 BP86/def-TZVP parametrizations for a few test cases). The sum of electronic energy in the gas phase (ΔE on TPSS-D3/TZ level) and the solvation free energy (ΔδGTsolv, see below) was used to rank the conformers. This sum is related to a “Gibbs energy in solution”, i.e., the electronic energy in solution plus the excess chemical potential.
To obtain COSMO-RS solvation free energies at 25 °C the standard procedure with two single point calculations, one in the gas phase and one in an ideal conductor (with infinite dielectric constant), at the default BP86 [49, 50]/def-TZVP [51] level of theory was performed and used as input for COSMOtherm.
The results presented for the two use cases—pKa prediction and the Morita–Baylis–Hillman reaction—were obtained based on single (lowest-Gibbs-energy) conformers. Given the errors encountered for more challenging cases for solvation models, the error by neglecting conformational entropy was assumed to be rather small.
The energetically lowest conformer for each intermediate and transition state was reoptimized on the TPSSh [43, 52]-D3/def2-TZVP level. Activation (ΔG‡) and reaction (ΔG) Gibbs free energies were obtained via a thermodynamic cycle (see supporting information Figure S.1.) as a sum of three contributions: (1) the gas phase activation/reaction energy (ΔE‡/ΔE) on the B3LYP-D3/def2-QZVP (QZ) or M06-2X/QZ level, (2) the difference in zero-point vibrational energies and thermostatistical contributions (ΔGTtrv) for products and reactants on the TPSSh-D3-COSMO/TZ level at temperature T, and (3) the difference in solvation free energies (ΔδGTsolv(X)) for products and reactants on the COSMO-RS level at temperature T in solvent X: \(\Delta G = \Delta E + \Delta G_{trv}^{T} + \Delta \delta G_{solv}^{T} \left( X \right)\).
Final single-point energies in the gas phase were computed with B3LYP [35, 53,54,55]-D3 and M06-2x [56] density functionals using the quadruple-zeta basis sets def2-QZVP (QZ) [51]. For such large basis sets the basis set superposition error almost vanishes and no special treatment, e.g., a computationally demanding counterpoise correction, is required. Computations of the harmonic vibrational frequencies were performed numerically and used to calculate the zero-point vibrational energy as well as the statistical thermodynamics corrections to obtain a Gibbs free energy at finite temperature (25 °C). The vibrational frequencies were used unscaled. Some of the small molecules in the acid–base reactions are symmetric and the corresponding symmetry number was considered when calculating the external-rotor entropy. DABCO, the catalyst in case of the MBH reaction, has a symmetry number 6, which was considered when calculating the free rotor entropy. Solvation free energies were computed with COSMO-RS as described above.
In all cases the resolution-of-identity (RI) approximation for Coulomb integrals was applied using matching default auxiliary basis sets [57, 58]. For the integration of the exchange–correlation contribution, the numerical quadrature grids m4 (for geometry optimizations and BP86 single-point energies) and m5 (for B3LYP and M06-2x single-point energies) were employed [59].
For all Gibbs free energies reported in the present work, a mixed thermodynamic reference state is defined as follows: The reference state of all solutes is c = 1 mol/l. If a solvent molecule (water or methanol) is involved in a reaction (including association reactions), its reference state is that of the pure phase, i.e., c = 55.4 mol/l for water (for acid–base reactions) and c = 24.7 mol/l for methanol (for MBH reaction) at a temperature of 25 °C. This mixed reference state is the same as in the experimental study of the Morita–Baylis–Hillman reaction [60], so that computed and experimental Gibbs free energies of activation and reaction can be directly compared to each other. For more details how the reference state is obtained in our computations see the supporting information section S.1.2 and the general discussion about molecular and solvation thermodynamics by Jensen [61].
Initial guess structures for the transition states (TS) were either build manually or obtained using the molecular growing string method (MGSM) [62,63,64]. These structures were then optimized using eigenvector following as implemented in TURBOMOLE. The resulting TS geometries were subjected to a conformational search with frozen TS bonds (as described above). The energetically lowest conformer was again optimized using eigenvector following. All minimum and TS geometries were confirmed as such by the vibrational frequencies.
Pictures of the computed molecular geometries were generated using CYLview [65]. Pictures of the COSMO surfaces with screening charge densities were created with COSMOthermX [48].
General strategy for the application of microsolvation
In order to apply microsolvation in predictive computational protocols, it is necessary to have clear computational criteria when (and how many) additional solvent molecules have to be added to a solute of interest. The criterion applied within this work is the Gibbs free energy ∆Ga for the association of a solvent molecule to a solute to form an explicit solute–solvent couple when applying already implicit solvation to all species:
Let us assume that the implicit solvent model already provides a “good solvation” treatment, which is defined in the following as being able to correctly take into account weaker interactions, typically up to hydrogen bonding of neutral species. If such implicit solvation treatment is at work, for the first of these “quasi-reactions”, the following results can be expected:
-
(a)
If solvent and solute have no clearly preferred points of interaction, the obtained ∆Ga should be moderately positive: Localizing solvent and solute in specific positions and directions with respect to each other always means a loss of entropy with respect to freely mobile species that are able to orient more or less randomly with respect to each other.
-
(b)
If, in contrast, there is one clearly preferred intermolecular interaction geometry, e.g., due to a hydrogen bond, ∆Ga should be close to zero, if both the explicit quantum chemical description and the implicit solvation description correctly take into account intermolecular interactions. However, in order to reach this, apart from having a “good solvation” model, also from the quantum chemical side the computations have to be rather accurate. Both energy and entropy of this step must be computed rather carefully. In reactions which make one species out of two, vibrational and conformational entropic contributions clearly matter and omitting them leads for both to a stronger preference of isolated species, i.e., a more positive ∆Ga. In addition, if there are several equally preferred interaction sites (i.e., oligomeric structures of equal energy), also a permutational entropic term must be added.
-
(c)
Even for “good solvation” models, there will be interactions that are simply too strong to fall in the application range, i.e., there is a point where the description as an aggregate resulting from a “chemical reaction” becomes mandatory. Very strong hydrogen bonds involving ionic species are examples for such interactions. Here, ∆Ga of the quasi-reaction between solute and solvent will be negative, indicating that the solvation model fails to reproduce the full extent of structural reorganization upon the interaction and the resulting relaxation of electronic wave functions.
Assuming that ∆Ga1 is negative, i.e., that the added solvent is thermodynamically favored (case c)), at least one explicit solvent should be included in the molecular model for the solute. The question if more explicit solvents are needed, is answered by looking at ∆Ga2, ∆Ga3 and so on: As long as negative ∆Ga are obtained, further explicit solvents should be added. Luckily, with the solvation model COSMO-RS typically also for ionic species only up to one explicit solvent molecule is required, unless highly charged species (or highly localized charges) are considered. Furthermore, also case b) represents an interesting way to challenge the consistency of solvation models when studying the solvent itself as solute (at least, if it exhibits the abovementioned preferred point(s) of interaction). If values for bulk solvent interaction are at least close to zero, this indicates that case c) will also be identified rather reliably, so that the above-mentioned criteria when to use microsolvation represent a reasonable choice.
Table 1 summarizes the results for the ∆Ga1 of solvent–solvent interaction (dimerization) at the COSMO and COSMO-RS levels for six species: hydrogen cyanide, water, ethyl amine, methanol, ethanol, and hydrogen fluoride. The values suggest that on average COSMO-RS yields a balanced description between explicit intramolecular and implicit intermolecular interactions. The scattering of values of course indicates certain deficiencies, but overall values are much closer to zero than for COSMO. However, it should be noted, that also for COSMO, all ∆Ga1 are quite similar (close to + 10 kJ/mol), which hints that also here systematic criteria might be derived when to apply microsolvation.
Computation of acid–base reactions and pKa values
As a first application, the computation of Gibbs free energies for several acid–base reactions in aqueous solution and the resulting pKa values of the acids are discussed. We deliberately chose a variety of examples with pKa values ranging from 0 (H3O+) to 16 (EtOH).
As outlined above, for each solute the required degree of microsolvation needs to be determined. In order to decide for which atom(s) of which species the necessity of microsolvation should be checked at all, criteria are needed. These are most probably related to electronic structure and can in principle be based on any electron density-dependent descriptor. Population analyses yielding partial charges could be one option. If COSMO-RS is used as solvation model, a COSMO calculation for the ideal conductor has to be performed anyway. Thus, the maximum screening charges (or rather the screening charge densities) of segments for the atoms of interest seem a convenient property. COSMO theory refers to the solvent and thus, provides the screening charge density, which is typically of opposite sign to the molecular screened surface charge, i.e., screening charge densities for negatively charged atoms are positive and vice versa. Figure 1 shows the cases of ethanol/ethoxide and acetic acid/acetate as example. The first column provides the screening charge density on the COSMO surface of the plain molecules: The darker the shade of red, the more positive the screening charge density is; the darker the shade of blue, the more negative it is. For the ethoxide anion the negative charge is localized on the oxygen atom and the maximal screening charge density is + 0.031 e/Å2. In case of acetate, the negative charge is delocalized, and the maximal screening charge density is + 0.022 e/Å2. For ethanol and acetic acid the maximal screening charge densities are below ± 0.020 e/Å2.
Screening charge densities on COSMO surfaces of ethanol/ethoxide and acetic acid/acetate and the best conformers (see section on computational details) of their complexes with explicit water molecules. The association free energies (∆Ga) were computed at the M06-2x/QZ + COSMO-RS levels and are given in kJ/mol. The unit of the maximal screening charge density is e/Å2
If the maximal screening charge density is positive, the explicit water molecule acts as hydrogen bond donor and stabilizes the negative charge on the atom or the functional group. If it is negative, mostly the case for hydrogen atoms, the explicit solvent molecule acts as hydrogen bond acceptor. For molecules like alcohols or acids usually both options, hydrogen bond donor and acceptor positions have to be tested. For the ethanol/ethoxide and acetic acid/acetate these cases are shown in Fig. 1. Here, the explicit water molecules were placed manually in a typical hydrogen bonding arrangement. These initial solute-water complexes were subjected to a conformational sampling by rotating all single bonds as described in the section “Computational Details” to obtain a reasonable structure with minimal Gibbs free energy. In an automated workflow e.g., the “ssc_strength” option of COSMOtherm could be used to construct the most strongly interacting solute–solvent pairs.
Other automated approaches to place solvent molecules have of course been developed. Kildgaard et al. published a stochastic algorithm for the generation of hydration clusters by placing water molecules in selected orientations around the solute in an iterative fashion [66, 67]. This way, they obtained the conformation with the lowest free energy. Reiher and coworkers presented a stochastic generation of solvation clusters which include conformational sampling [68]. Open sites on solvent accessible surface of solute and solvent are randomly chosen and placed opposite each other at a certain distance and random angle that avoid atom clashes. The geometries of the resulting solute–solvent cluster are then optimized with standard quantum chemical methods. The addition of more solvent molecules starts from the previous solute–solvent complex. The random selection of sites is not optimal when solute or solvent feature charged or polar functional groups, as it is the case for many of the examples presented here. Our approach, in contrast, places the solvent molecules only at a few sites on the solvent accessible surface, but where the interaction is expected to be strongest (within the framework of the COSMO-RS solvation model).
After identification of the possible positions of explicit solvent molecules, the necessity to include them in the computation of the acid–base reactions is checked via the Gibbs free energy of association (ΔGa) of the water molecules. As discussed above, we assume that whenever the association Gibbs free energy ΔGa is negative, the explicit water molecule must be included. For ethanol and acetate, the Gibbs free energy of association for the first water molecule is thermoneutral or endergonic, which means that no explicit water is needed. For acetic acid ΔGa is slightly negative (− 3 kJ/mol) and one explicit water is included. For the ethoxide anion the association of the first water molecule is strongly exergonic (− 24 kJ/mol) and even the association of a second water is still exergonic (− 9 kJ/mol) on the M06-2x/QZ + COSMO-RS level. Therefore, the recommendation is to use two explicit water molecules.
The maximal screening charge densities of all neutral species and the association free energies on the B3LYP-D3/QZ + COSMO-RS and M06-2x/QZ + COSMO-RS levels are given in Table 2. The results for the charged species are presented in Table 3. The corresponding results for B3LYP-D3/QZ and M06-2x/QZ with the COSMO solvation model are collected in the supporting information Tables S.3 and S.4.
As expected, neutral species have slightly lower absolute values of (maximal) surface screening charge densities. In automated workflows, a check on the necessity of microsolvation could, e.g., be performed if absolute values exceed ± 0.020 e/Å2, although making the threshold atom type dependent could lead also to more precise predictions how to computationally treat solute species.
For the neutral molecules (Table 2) ΔGa is usually slightly positive or slightly negative, i.e., close to thermoneutral, and thus, explicit water molecules are needed in some cases only. The exceptions with a more negative ΔGa are water, sulfuric acid, and phosphoric acid which have ΔGa of − 12, − 23, and − 10 kJ/mol, respectively, for the association of the first water molecule on the M06-2x/QZ + COSMO-RS level. The association of a second water molecule is slightly endergonic for water and phosphoric acid, but still exergonic for sulfuric acid (− 8 kJ/mol). Thus, sulfuric acid is the only neutral molecule which requires two explicit water molecules. Overall, with the COSMO-RS solvation model ten neutral molecules need one explicit water and H2SO4 requires two. With the COSMO model only two neutral solutes need explicit solvation: H3PO4 needs one and H2SO4 requires two explicit water molecules.
For ionic molecules (Table 3) there are even more cases where COSMO-RS clearly underestimates the intermolecular hydrogen bonding interactions. This is particularly severe where there is no or only little charge stabilization, like in the ethoxide anion. If the charge is stabilized by resonance effects, like in a phenoxy anion or acetate, the association Gibbs free energy is close to zero. For the ions without charge stabilization, microsolvation is clearly needed, requiring in five cases (H3O+, NMe3H+, PhNH3+, pyridine-H+, imidazole-H+) only one explicit water molecules, but in four cases (EtO−, CF3CH2O−, CH3CHNO2−, PhO−) two explicit water molecules are needed. When COSMO is employed, a beneficial stabilization is obtained with one explicit water molecule for PhO− and with two explicit water molecules for H3O+, OH−, EtO−, and CF3CH2O−.
In a next step, Gibbs free energies of reaction for the aqueous acid–base reactions were performed, both with and without microsolvation. As a reference base imidazole was used. It exhibits a pKa close to 7 (6.95 was used here as suggested by Bruice et al. [68]), i.e., it represents a reference in the center of the aqueous pH scale. Furthermore, the positive charge in the cation is quite delocalized so that deficiencies in predicted ∆G can be rather safely traced back to the acid–base pair of interest. Reaction free energies for the reactions
were computed and the resulting Gibbs free energy was transferred to pKa units to obtain the desired pKa value of the acid.
Calculated (M06-2x) and experimental ΔG of acid–base reactions involving cationic, neutral and anionic acids and the corresponding pKa values of the acids are given Table 4. The analogous results for B3LYP are provided the in the supporting information in Table S.5. The reaction free energies are a bit lower on the B3LYP-D3/QZ level, but the overall performance of the two functionals is similar and we focus here on the M06-2x results. As described in the computational details, the thermodynamic reference state is the following: water molecules are treated as pure water with c = 55.4 mol/l (at 25 °C) and all solutes with c = 1 mol/l.
The reaction free energies of the acid–base reaction are reasonable with an implicit solvation model only in those cases in which the reaction involves neutral species and ions stabilized by resonance effects. The mean absolute error (MAE) is 33 kJ/mol for M06-2x/QZ + COSMO and 15 kJ/mol in case of M06-2x/QZ + COSMO-RS. This corresponds to a mean absolute error of 5.8 and 2.7 on the pKa scale. However, for reactions which involve ionic molecules that exhibit a non-stabilized charge on specific isolated atoms, the results are sometimes far from the experimental values. The maximal error (MAX) is observed for H3O+ (69 kJ/mol, 12.0 on pKa scale) in case of COSMO and for EtOH (51 kJ/mol, 9.0 on pKa scale) in case of COMO-RS. For these systems, microsolvation significantly improves the agreement between theoretical and experimental data, e.g., the error for EtOH decreases to 19 kJ/mol which corresponds to an error of 3.3 on pKa scale.
When microsolvation is included, the MAE decreases to 12 kJ/mol (2.2 on pKa scale) and the maximal error drops to 35 kJ/mol (7.2 on pKa scale) for M06-2x/QZ + COSMO-RS. The largest errors for COSMO-RS + microsolvation are observed for H2O (error of 6.2 on pKa scale), H2SO4− (error of 5.8), PhOH (error of 4.0), CF3CH2OH (error of 3.6) and H3O+ (error of 3.6). Only in the cases of H2O and H3O+ the pure COSMO-RS actually performs better, which most likely is due to the special treatment of water within the COSMO-RS model (version 2018 was used here). Surprisingly, when using COSMO-RS the hydroxide anion does not require an explicit water molecule, although the maximal screening charge density is even higher (+ 0.033 e/Å2) than for ethoxide anion (+ 0.031 e/Å2). This again, is probably related to the special treatment of water within the COSMO-RS model. We repeated the computation of association free energies using M06-2x and COSMO-RS with the BP86/def-TZVP parametrizations from 2015 and 2012. The Gibbs free energy of association of a water molecule to the hydroxide anion is 26 kJ/mol, 31 kJ/mol, and 16 kJ/mol when using the COSMO-RS parameters from 2018, 2015, and 2012. In case of the ethoxide anion ΔGa is -24 to -25 kJ/mol for all three parameter versions tested. This indicates that the special treatment of water within the COSMO-RS model changes from version to version. When the COSMO solvation model is employed, the hydroxide anion does require explicit solvation, as one would expect. Nevertheless, the errors for M06-2x/QZ + COSMO are larger than for M06-2x/QZ + COSMO-RS when including microsolvation. The MAE for M06-2x/QZ + COSMO + MS amounts to 26 kJ/mol (4.6 on pKa scale) and the maximal error is 61 kJ/mol (10.7 on pKa scale).
Overall, the results obtained for acid–base reactions suggest that the presented protocol generally works and that it provides a possibility for designing an automated workflow to decide whether microsolvation is needed in combination with the COSMO or COSMO-RS solvation model. Figure 2 summarizes our microsolvation protocol and provides a rough idea how such an automated workflow might look like. In the first step, the maximal screening charge densities are used to identify possible positions of the solvent molecules and in the second step, the association Gibbs free energy is used to decide if formation of the respective solute–solvent cluster is energetically favored. Next, we applied this microsolvation protocol to the challenging use case of an organic reaction mechanism.
Investigation of the alcohol-mediated Morita–Baylis–Hillman reaction
To highlight the importance of microsolvation for chemical reactivity studies and investigations of reaction mechanisms and to test our protocol for microsolvation, we chose the alcohol-mediated Morita-Baylis–Hillman (MBH) reaction as an exemplary case study. Its mechanism has been extensively studied by many groups using computational and experimental methods [69,70,71,72,73,74,75,76,77,78,79,80], but has remained elusive until the very detailed experimental work of Plata, Singleton and coworkers in 2015. Plata and Singleton also demonstrated that computational studies using a simple continuum solvation model fail spectacularly in describing the mechanism of the MBH reaction [60]. This is no surprise, because all intermediates are either zwitterionic or charged and thus, any computational method that utilizes a simple continuum solvation model like COSMO is expected to yield large errors. Two years later in 2017, Harvey, Sunoj and coworkers showed that by employing high-level energy computations on the coupled-cluster level (DLPNO-CCSD(T)) in combination with an explicit solvation treatment based on molecular dynamics they can achieve a good agreement with experiment with an error of 3–20 kJ/mol [81]. Basdogan and Keith showed in 2018, that the required degree of microsolvation can be studied systematically with a static approach [38]. For all intermediates, they identified the low-energy geometries of the solute–solvent clusters with different numbers of methanol molecules (1–5 and 10) with a stochastic computational filtering procedure using a global optimization protocol. Subsequently, they explored the reaction pathways by systematic single-ended growing string method (GSM) [62,63,64] computations. They found that gradually adding more solvent models does not improve the agreement with experiment (five is best) and that once a complete reaction pathway is found, high-level energy computations on the coupled-cluster level only yield a marginal gain.
Compared to these two studies our approach is quite simple and requires neither dynamics simulations nor a global optimization procedure to determine the degree of microsolvation. We start from the reaction mechanism without explicit solvent molecules and have clear criteria if adding more solvent molecules is necessary. Our intention here is not to present a benchmark of DFT functionals and/or solvation models. We simply chose a reasonable combination of computational methods that are widely used, i.e., TPSSh-D3-COSMO/TZ for geometry optimizations, and M06-2x/QZ and B3LYP-D3/QZ single-point energies together with the COSMO or COSMO-RS solvation models to obtain Gibbs free energies of reaction and activation. We use Plata’s and Singleton’s carefully determined experimental thermodynamics and kinetic data for main and side reactions as reference throughout. As discussed in the “computational details” section, the chosen reference state for the presented Gibbs free energies is identical to the reference state used by Plata and Singleton: methanol molecules are treated as pure methanol with c = 24.7 mol/l (at 25 °C), all solutes have c = 1 mol/l. Therefore, the experimental results and our computed values are directly comparable.
Figure 3 shows an overview of the investigated reaction steps of the prototypical MBH reaction of methyl acrylate (MA, 1) and p-nitrobenzaldehyde (4) catalyzed by DABCO (2). First, DABCO adds to MA to yield the zwitterionic intermediate 3, which then undergoes the aldol step with aldehyde 4 to form the zwitterionic intermediate 5. Furthermore, the C-protonation of 3 to yield cationic adduct 8 was also investigated. The conversion from intermediates 5 to 6 was subject of debate for a long time. Many computations favor a proton-shuttle mechanism involving one MeOH [69,70,71,72, 75] but Plata and Singleton showed experimentally that the mechanism follows the acid/base route [58]. The last step is the elimination of DABCO to yield the product 7. Except for the barriers of the initial addition and final elimination step, Plata and Singleton were able to determine accurate reaction (ΔG) and activation (ΔG‡) free energies from their experiments.
Overview of the investigated reaction steps of the alcohol mediated Morita–Baylis–Hillman reaction, analogous to the steps given in Ref. [58]
First, we compare the computed overall thermodynamics of the MBH reaction with experimental results. The overall reaction Gibbs free energy was determined experimentally as − 16 kJ/mol [58]. B3LYP-D3/QZ in combination with both solvent models does not yield an exergonic reaction. The overall reaction Gibbs free energy is 19 kJ/mol with the COSMO and 6 kJ/mol with the COSMO-RS solvation model. B3LYP is known to be a functional that usually does neither provide the best nor the worst results for main group thermochemistry and kinetics [82] but was also found to underestimate the energies of C–C single bonds compared to C–C double bonds [83]. M06-2x/QZ together with the COSMO solvation model yields a slightly exergonic reaction with ΔG of -2 kJ/mol. The reaction Gibbs free energy at the M06-2x/QZ + COSMO-RS level of − 15 kJ/mol, which agrees with experiments quantitatively. The average errors of the M06-2x/QZ computations for all the steps of the MBH reaction are similar compared to B3LYP-D3/QZ (see Table 5) and all observed trends that will be discussed are the same. The M06-x2 functional is in general a good choice for charged systems [29] and we will mainly focus on the M06-2x/QZ results in the following.
Next, we will discuss the computations and solvation effects with respect to the following points: (1) The overall error of Gibbs free energies of activation and reaction compared to experiment, (2) the difference in barrier heights for the aldol-step and the C-protonation of 8, (3) the difference in barrier heights for the proton shuttle and acid/base mechanisms for conversion from 5 to 6, and (4) the overall rate-limiting step.
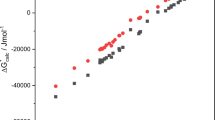
First, we discuss the results obtained with a simple (semi-)continuum solvation model. Figure 4 shows the reaction profile for the M06-2x/QZ computations with COSMO and COSMO-RS solvation. The analogous depiction for B3LYP-D3/QZ can be found in the supporting information Figure S.2. Table 5 provides the Gibbs free energies of activation and reaction for all individual steps (relative to the corresponding reactants) on both the M06-2x and B3LYP levels. The errors compared to experimental values are significant for both solvation models. The mean absolute error (MAE) for ΔG compared to experiment is 20 and 18 kJ/mol for M06-2x/QZ + COSMO and M06-2x/QZ + COSMO-RS, respectively. The maximal absolute error (MAX) of ΔG results in 43 kJ/mol for M06-2x/QZ + COSMO and 46 kJ/mol for M06-2x/QZ + COSMO-RS for the deprotonation step of 9 to 6. For the reaction barriers only three values are known from experiment (see Table 5) and the MAE was omitted. The barrier of the aldol step (MAX) is off by 28 and 12 kJ/mol for M06-2x/QZ + COSMO and M06-2x/QZ + COSMO-RS, respectively.

taken from Ref. [58]
Experimental and computed free energies for the MBH reaction on the M06-2x/QZ level with COSMO and COSMO-RS solvation. Experimental values are
From experiment it was determined that the aldol step 3 + 4 and the C-protonation of 3 have similar barriers of 85 and 91 kJ/mol, respectively. Thus, the difference is only 6 kJ/mol. A very small difference is also obtained from the computations, but the order of the two reaction pathways is wrong. M06-2x/QZ combined with both solvation models yields a lower barrier for the C-protonation of intermediate 3 (115 kJ/mol for both, COSMO and COSMO-RS) than for the aldol step (125 and 116 kJ/mol for COSMO and COSMO-RS).
The conversion from 5 to 6 was experimentally identified as acid/base mechanism via intermediate 9. We computed the acid/base and proton shuttle mechanisms, and both M06-2x/QZ + COSMO and M06-2x/QZ + COSMO-RS fail to yield the acid/base reaction as favored pathway. The barrier for the shuttle mechanism is lower by 16 and 23 kJ/mol, respectively. The experimentally determined rate limiting step of the MBH reaction is the deprotonation of cationic intermediate 9 by MeO− to yield 6, which has a barrier of 89 kJ/mol relative to the starting materials. The aldol step has a slightly lower relative barrier of 85 kJ/mol. Again, M06-2x/QZ in combination with both solvation models fails and indicates the aldol step as the rate-limiting step with an activation barrier of 125 kJ/mol and 116 kJ/mol, respectively.
Thus, with respect to the four discussion points M06-2x/QZ and B3LYP-D3/QZ in combination with the simple COSMO and the more advanced COMO-RS solvation model fail spectacularly to describe the MBH reaction, rendering the computations basically useless.
Next, we address the question if microsolvation combined with either COSMO or COSMO-RS (semi-) continuum solvation can provide a better description of the MBH reaction. The first key questions when using microsolvation is how many explicit solvent molecules are needed and where they should be placed. This can be identified via an analysis of the screening charge density on the COSMO surface of all species including transition states and the Gibbs free energy of association (ΔGa) of methanol as described in the previous section. If the surface charge density is negative, the explicit MeOH acts as hydrogen bond donor and stabilizes the negative charge on the atom/functional group. If it is positive, mostly the case for hydrogen atoms, the explicit methanol molecule acts as hydrogen bond acceptor. The results for all reactants, products, intermediates, and transition states are compiled in Table 6. According to our analysis, the methoxide anion, intermediate species 5 and 6 (6 only for B3LYP), and most transition states except the first and last for M06-2x (only except the first for B3LYP) benefit from an explicit methanol molecule due to additional stabilization (negative ΔGa) when COSMO-RS is used as solvent model. If the simpler COSMO model is employed, the stabilization effect due to an explicit MeOH is even more pronounced, and beneficial for methoxide and all transition states (see supporting information, Table S.16). The highest stabilization is observed for the transition state of the aldol step for both, M06-2x/QZ + COSMO-RS (ΔGa = − 30 kJ/mol) and M06-2x/QZ + COSMO (ΔGa = − 58 kJ/mol).
The association of a second methanol molecule is not favored, except for zwitterionic intermediate 5 in case of COSMO-RS (with both functionals, see supporting information, Tables S.17 and S.18) and the TS of the aldol step for B3LYP/QZ + COSMO. According to our experience, and also shown in the section before, in many cases one explicit solvent molecule is enough. The advantage, i.e. slightly better stabilization, gained with more explicit solvent molecules is easily lost due to other problems, e.g. small (imaginary) vibrational frequencies that occur with increasing system size and flexibility. Thus, we included only one explicit solvent molecule when necessary.
The second question that arises when employing microsolvation is how to actually incorporate the explicitly solvated species into the reaction mechanism, i.e. how to compute the activation and reaction free energies. There is no convergent workflow for this, and we will discuss two different approaches. The first one, called approach 1 in the following, is the simpler and more straightforward one. Here, one corrects the free energies of the reactants, products, or transition states with the respective Gibbs free energy of association for one methanol molecule (ΔGa in Table 6) if stabilization is beneficial, i.e. ΔGa is negative. The corrected free energies are then used to obtain the reaction and activation free energies for a given step. As described above, methoxide anion, species 5, species 6, and all transition states need this correction. As an example, we look at the first two steps of the MBH reaction (on the M06-2x/QZ + COSMO-RS level). For the addition step, both reactants (1 and 2), the product (3) and the transition state do not benefit from explicit solvation. Thus, the reaction and activation free energies are the same as before (ΔG = 56 kJ/mol and ΔG‡ = 65 kJ/mol). For the aldol step both reactants (3 and 4) do not need explicit MeOH, but the product (5) as well as the transition state do. Thus, the Gibbs free energy of 6 is corrected with the association Gibbs free energy ΔGa of − 11 kJ/mol and the free energy of the transition state corrected by − 30 kJ/mol. This way the reaction Gibbs free energy is lowered from − 3 to − 14 kJ/mol and the activation Gibbs free energy decreases from 60 to 30 kJ/mol. The resulting thermodynamic cycle for this example is shown in Fig. 5a. Table 7 summarizes all ΔG and ΔG‡ using approach 1 to include microsolvation.
Exemplary thermodynamic cycles if microsolvation is included (for the second step of the MBH reaction with methanol as solvent) according to approach 1 (a) or approach 2 (b). ΔGsolv is computed with the COSMO-RS solvation model and ΔG‡(g)/ΔG(g) with quantum chemical methods (includes statistical thermodynamics for ideal gas)
Approach 2 is slightly less straightforward and aims at keeping the number of explicit methanol molecules constant for the computation of ΔG/ΔG‡ of a given reaction step. The reasoning behind this is the following: As discussed before, a perfect solvation model would include all interactions with the solvent molecules and correctly describe the hydrogen bond network in solvent like methanol, i.e., the Gibbs free energy of association of a methanol molecule in methanol as solvent would always be zero. Unfortunately, we do not have such a model, and therefore we assume that an explicitly solvated reactant, intermediate, or product molecule is a valid species even if ΔGa is slightly positive.
As an example, we again look at the aldol addition step of the MBH reaction (M06-2x/QZ + COSMO-RS). Both reactants do not need explicit solvation but the product and the transition state benefit from it. Therefore, reactant 3 is also explicitly solvated with one methanol molecule to keep the number of hydrogen bonds in the calculation of ΔG‡ constant. With this, the reaction Gibbs free energy results in − 16 kJ/mol and the activation Gibbs free energy in 28 kJ/mol. The resulting thermodynamic cycle is shown in Fig. 5b. Table 8 summarizes all ΔG and ΔG‡ using approach 2 to include microsolvation. A disadvantage of approach 2 is, that summing over all reaction free energies of all steps of the MBH reaction does not result in the exact same overall thermodynamics that was discussed before (computed without any explicit MeOH) and the values differ by 1 kJ/mol for both, M06-2x/QZ + MS + COSMO-RS and M06-2x/QZ + MS + COSMO, respectively. The advantage is, that a general solvent model and solvent dependent offset (see e.g. COSMO values for solvent–solvent interaction in Table 1) for the addition of the explicit solvent is probably better compensated.
The comparison of approaches 1 and 2 shows that for all activation and reaction free energies the values differ by maximally 8 kJ/mol for both, M06-2x/QZ + MS + COSMO-RS and M06-2x/QZ + MS + COSMO. The analogous results for B3LYP-D3/QZ with both approaches to include microsolvation can be found in the supporting information, Tables S.10 and S.11.
We will now discuss the results that can be obtained using microsolvation (approach 1) with respect to the four points as before, i.e., (1) The overall error of activation and reaction free energies compared to experiment, (2) the difference in barrier heights for the aldol-step and the C-protonation of 8, (3) the difference in barrier heights for the proton shuttle and acid/base mechanisms for conversion from 5 to 6, and (4) the overall rate-limiting step. Figure 6 shows the reaction profile for the M06-2x/QZ computations with microsolvation (approach 1) combined with COSMO or COSMO-RS continuum solvation.

taken from Ref. [58]
Experimental and computed free energies for the MBH reaction on the M06-2x/QZ level with microsolvation approach 1 (MS, one MeOH molecule) combined with the COSMO and COSMO-RS continuum solvation models. Experimental values are
In general, the errors are much lower when microsolvation is included and the agreement with experiment improves significantly. As before, M06-2x/QZ is more accurate than B3LYP-D3/QZ and we will focus on the M06-2x/QZ results. The mean absolute error for ΔG drops from 20 to 10 kJ/mol when microsolvation is used together with COSMO, and from 18 to 10 kJ/mol when it is used in combination with COSMO-RS. The maximal absolute error of ΔG (conversion of 9 to 6) decreases from 43 to 22 kJ/mol for MS + COSMO and from 46 to 24 kJ/mol for MS + COSMO-RS. The improvement for the reaction barriers is less evident. The maximal error is still found for the barrier of the aldol step, which was overestimated without microsolvation and is underestimated when including microsolvation. The resulting error is 30 kJ/mol and 18 kJ/mol when using microsolvation in combination with COSMO and COSMO-RS, respectively. The errors for the individual steps are similar for COSMO and COSMO-RS but when looking at the full reaction profile (see Fig. 5), the overall agreement with experiment is slightly better when COSMO-RS is used as continuum solvation model.
Next, we look at the barriers of the aldol step 3 + 4 and the C-protonation of 3. When microsolvation is employed, the correct order of the two pathways is obtained and the aldol step has the lower barrier. For M06-2x/QZ + MS + COSMO the aldol step has a barrier of 67 kJ/mol and the C-protonation a barrier of 84 kJ/mol. In case of M06-2x/QZ + MS + COSMO-RS the barriers are 86 and 106 kJ/mol for the aldol step and C-protonation, respectively. The differences of 17–20 kJ/mol in both cases are larger than in experiment (6 kJ/mol).
Next, we discuss the most critical step in the whole MBH reaction when considering the controversy between computed and experimental results, the acid–base step for the conversion of 5 to 6. Even when using microsolvation combined with the COSMO-RS solvation model for both density functionals and combined with COSMO for B3LYP, the shuttle mechanism is still favored. For M06-2x/QZ the proton shuttle and the acid–base step have the same barrier (110 kJ/mol) when COSMO is used together with microsolvation. Nevertheless, for COSMO-RS the difference is significantly decreased, and the shuttle is favored by only 12 kJ/mol compared to 23 kJ/mol without MS. This clearly shows that even a large effort to include solvation effects might not be enough and better solvation models are still needed. The dissipation of a proton into the methanol solvent that somewhere also contains solvated methoxide anions is the worst-case scenario for all available static quantum chemistry protocols. The methoxide does not necessarily have to be very close to the species that is deprotonated as the hydrogen bond network can transfer the proton trough the methanol. Evidently, these dynamics of the environment cannot be captured with static approaches as used in this work. One can also argue that due to the hydrogen bond network of the solvent the acid/base mechanism is basically like a proton shuttle, concerted but not synchronous. The more explicit methanol molecules are added to our static calculation, the more freedom the system has, or the other way around, if fewer explicit MeOH are used, the system is forced to a more specific transition state. This effect can already be seen when one compares the transition state geometries for the deprotonation of 9 and the shuttle from 5 to 6 with different amounts of explicit methanol molecules (see Fig. 7). Already with one explicit MeOH the bond length equalizes dramatically and with two explicit MeOH the transition states for proton shuttle and deprotonation are indistinguishable after the conformational sampling and picking the energetically lowest conformer.
Finally, the rate limiting step of the MBH reaction is correctly identified when microsolvation is included. The experimental barrier for the deprotonation of cationic intermediate 9 by MeO− to yield 6 is 89 kJ/mol. M06-2x/QZ + MS + COSMO yields a barrier of 110 kJ/mol for the proton shuttle and the acid base mechanism and M06-2x/QZ + MS + COSMO-RS provides a barrier of 92 kJ/mol for the shuttle and 104 kJ/mol for the acid–base mechanism. Without microsolvation the aldol step was identified as rate limiting step. Here, the barriers of the conversion of 5 to 6 are 43 and 6–18 kJ/mol higher than the aldol step, for M06-2x/QZ + MS + COSMO and M06-2x/QZ + MS + COSMO-RS, respectively. Again, the differences are larger than in experiment (4 kJ/mol) for COSMO but similar for COMO-RS.
Compared to previous computational studies, our approach has a similar accuracy with respect to the experimental results. With their approach based on DLPNO-CCSD(T) energies and molecular dynamics to include microsolvation Harvey, Sunoj and coworkers obtained absolute errors in the relative Gibbs free energy (relative to the initial reactants) in the range of 3–20 kJ/mol and an MAD of 6 kJ/mol [81]. The paramedic approach of Basdogan and Keith to include microsolvation in a static way in combination with DLPNO-CCSD(T)/TZ energies yields absolute relative errors in the range of 4–16 kJ/mol and an MAD of 10 kJ/mol [38]. With our approach based on M06-2x/QZ energies and a much simpler static approach to include microsolvation in combination with the COSMO-RS solvation model absolute relative errors in the range of 1–20 kJ/mol and an MAD of 13 kJ/mol were obtained. Overall, our approach tends to overestimate the relative free energies and the approach of Basdogan and Keith tends to underestimate them. A comparison of the reaction profiles is provided in the supporting information in Figure S.4.
In the end, we want to briefly discuss a problem that can occur when using microsolvation and which concerns the entropy. Small vibrational frequencies appear more often when the system size increases and when flexible non-covalent interactions are formed due to explicit solvent molecules. Low-lying vibrational frequencies are inaccurate in the harmonic approximation and numerical noise easily enters in the computations, especially when structural optimizations are performed with the COSMO solvation model, causing further errors. Thus, the error of the resulting entropy can be severe. To remedy this problem, Grimme introduced an approach that replaces the vibrational entropy of the low-lying modes below a certain threshold by the entropy of a rigid-rotor [84]. However, it was checked that when using this approach with a threshold of 100 cm−1, the resulting reaction and activation free energies within the above mechanism change by only 1–3 kJ/mol. Even when one MeOH is explicitly included the systems are obviously still quite small and thus, the effect on the entropy is negligible. For larger systems this might not be the case and the effect of small vibrational modes on the entropy has to be investigated carefully.
Conclusion
Whenever a chemist writes down a reaction scheme, it seems that there is a clear distinction between reactants and reaction conditions. However, when the reaction has to be translated into molecular models for quantum chemical studies, often the problem emerges how to most adequately mimic molecular reality. The fact that, e.g., reagents or an acid catalyst “on top of the arrow” require explicit consideration in the molecular model is rather clear, but the solvent and/or environment as a sometimes equally crucial factor remains often neglected. Microsolvation represents one approach to tackle this problem. Our results clearly show that a systematic protocol when and how to apply it leads to improved results for cases where the limits of state-of-the-art solvation models are reached. Specifically, for the COSMO and COSMO-RS solvation models considered in this study, microsolvation is practically unnecessary for many neutral molecules but can become crucial when the solute is an ion or zwitterion with either little charge delocalization or a high overall charge (larger than ± 1). The proposed criterion to apply microsolvation whenever a (good) solvation model thermodynamically prefers the association of a solvent (∆Ga < 0 for the quasi-reaction between solute and solvent) at interactions sites with high screening charge densities appears reasonable for the two presented use-cases, aqueous acid–base chemistry and the mechanism of the DABCO and methanol catalyzed Morita–Baylis–Hillman reaction. In these examples, microsolvation was able to reduce errors for the worst described species by a factor of more than two.
It should, however, be noted that microsolvation does not represent a simple and generally applicable solution to cases wherever solvation models reach their limits. One problem is that when a solvent molecule is added to a system, association entropies of around 100 J/mol/K for the interaction between solute and solvent (resulting in Gibbs free energy contributions of around 30 kJ/mol) are encountered suddenly. This is in contrast to real systems where the interaction between a solute and its environment increases gradually. Furthermore, as mentioned above, the general calculation of molecular entropies accumulates more and more noise when computational models become larger and larger.
Thus, further advances in solvation models would be highly desirable, so that microsolvation really represents an ultimate resort but which can be avoided as much as possible. The presented criterion that solvent–solvent interactions should exhibit a ∆Ga as close to zero as possible could be one ingredient on the way towards more consistent solvation models that yield reliable results for systems where microsolvation is sometimes required and sometimes not.
Data availability
Supplementary data available.
References
Tajti A, Szalay PG, Csaszar AG, Kallay M, Gauss J, Valeev EF, Flowers BA, Vazquez J, Stanton JF (2004) J Chem Phys 121:11599
Bomble YJ, Vazquez J, Kallay M, Michauk C, Szalay PG, Csaszar AG, Gauss J, Stanton JF (2006) J Chem Phys 125:064108
Harding ME, Vazquez J, Ruzcic B, Wilson AK, Gauss J, Stanton JF (2008) J Chem Phys 128:114111
Marx D, Hutter J (2009) Ab initio molecular dynamics. Cambridge
Warshel A, Levitt M (1976) J Mol Biol 103:227–249
Chandler D, Andersen HC (1972) J Chem Phys 57:1930–1937
Ikeguchi M, Doi JJ (1995) Chem Phys 103:5011
Beglov D, Roux B (1997) J Phys Chem B 101:7821
Du Q, Beglov D, Roux B (2000) J Phys Chem B 104:796
Kovalenko A, Hirata F (1998) Chem Phys Lett 290:237
Hoffgaard F, Heil J, Kast SM (2013) J Chem Theory Comput 9:4718–4726
Tomasi J, Mennucci B, Cammi R (2005) Chem Rev 105:2999–3093
Miertus S, Scrocco E, Tomasi J (1981) Chem Phys 55:117
Tomasi J, Persico M (2027) Chem Rev 1994:94
Tomasi J, Cammi R, Menucci B (1999) Int J Quantum Chem 75:767
Cancès E, Menucci B, Tomasi J (1997) J Chem Phys 107:3032–3041
Foresman JB, Keith TA, Wiberg KB, Snoonian J, Frisch MJ (1996) J Phys Chem 100:16098–16104
Marenich AV, Cramer CJ, Truhlar DG (2009) J Phys Chem B 113:6378–6396
Cramer CJ, Truhlar DG (2008) Acc Chem Res 41:760
Cramer CJ, Truhlar DG (2009) Acc Chem Res 42:493
Marenich AV, Cramer CJ, Truhlar DG (2009) J Phys Chem B 113:4538–4543
Klamt A, Schüürmann G (1993) J Chem Soc Perkin Trans II:799
Maurizio Cossi M, CarloAdamo C, Barone V (1998) Chem Phys Lett 297:1–7
Tao DJ, Slutskyy Y, Muuronen M, Le A, Kohler P, Overman L (2018) J Am Chem Soc 140(8):3091–3102
Klamt A (1995) J Phys Chem 99:2224–2235
Eckert F, Klamt A (2002) AIChE J 48:369–385
Ashcraft RW, Raman S, Green WH (2008) J Phys Chem 112:7577
Deglmann P, Müller I, Becker F, Schäfer A, Hungenberg K-D, Weiß H (2009) Macromol React Eng 3:496
Deglmann P, Schenk S (2012) J Comput Chem 33:1304
Gadre SR, Yeole SD, Sahu N (2014) Chem Rev 114:12132–12173
Pliego Jr, JR, Riveros JM (2001) J Chem Phys A 105:7241–7247
Pliego Jr JR, Riveros JM (2002) J Chem Phys A 106:7434–7439
Pliego Jr JR, Riveros JM (2004) Chem Phys 306:273–280
Eckert F, Diedenhofen M, Klamt A (2010) Mol Phys 108:229–241
Ho J, Coote M (2010) Theor Chem Acc 125:3–21
Ho J, Ertem MZ (2016) J Phys Chem B 120:1319–1329
Kelly CP, Cramer CJ, Truhlar DG (2005) J Chem Theory Comput 1:1133–1152
Basdogan Y, Keith JA (2018) Chem Sci 9:5341–5346
Hadad C, Florez E, Acelas N, Merino G, Restreppo A (2019) Int J Quant Chem 119:e25766
Florez E, Acelas N, Ramirez F, Hadad C, Restreppo A (2018) Phys Chem Chem Phys 20:8909–8916
Ahlrichs R, Armbruster MK, Bär M, Baron H-P, Bauernschmitt R, Crawford N, Deglmann P, Ehrig M, Eichkorn K, Elliott S, Furche F, Haase F, Häser M, Hättig C, Hellweg A, Horn H, Huber C, Huniar U, Kattannek M, Kölmel C, Kollwitz M, May K, Nava P, Ochsenfeld C, Öhm H, Patzelt H, Rappoport D, Rubner O, Schäfer A, Schneider U, Sierka M, Treutler O, Unterreiner B, von Arnim M, Weigend F, Weis P, Weiss H (2018) TURBOMOLE 7.3, Universität Karlsruhe. http://www.turbomole.com.
Furche F, Ahlrichs R, Hättig C, Klopper W, Sierka M, Weigend F (2014) WIREs Comput Mol Sci 4:91–100
Tao J, Perdew J, Staroverov V, Scuseria G (2003) Phys Rev Lett 91:146401
Weigend F, Ahlrichs R (2005) Phys Chem Chem Phys 7:3297–3305
Grimme S, Antony J, Ehrlich S, Krieg H (2010) J Chem Phys 132:154104
Johnson ER, Becke AD (2005) J Chem Phys 123:24101
Becke AD, Johnson ER (2005) J Chem Phys 123:154101
Eckert F, Klamt A (2018) COSMOtherm, Version C3.0, Release 18.01; COSMOlogic GmbH & Co. KG, Leverkusen, Germany
Perdew JP (1986) Phys Rev B 33:8822–8824
Becke AD (1988) Phys Rev A 38:3098–3100
Schäfer A, Huber C, Ahlrichs R (1994) J Chem Phys 100:5829–5235
Staroverov VN, Scuseria GE, Tao J, Perdew JP (2003) J Chem Phys 119:12129
Lee C, Yang W, Parr RG (1988) Phys Rev B 37:785
Vosko SH, Wilk L, Nusair M (1980) Can J Phys 58:1200
Stephens PJ, Devlin FJ, Chabalowski CF, Frisch MJ (1994) J Phys Chem 98:11623
Zhao Y, Truhlar DG (2008) Theor Chem Acc 120:215
Eichkorn K, Treutler O, Öm H, Häser M, Ahlrichs R (1995) Chem Phys Lett 242:652–660
Weigend F (2006) Phys Chem Chem Phys 8:1057–1065
Treutler O, Ahlrichs R (1995) J Chem Phys 102:346
Plata RE, Singleton DA (2015) J Am Chem Soc 137:3811
Jensen J (2015) PhysChemChemPhys 17:12441
Zimmerman PM (2013) J Chem Theory Comput 9:3043–3050
Zimmerman PM (2013) J Chem Phys 138:184102
Zimmerman PM (2015) Comput Chem 36:601
Legault CY (2009) CYLview, b.; Université de Sherbrooke. http://www.cylview.org
Kildgaard JV, Mikkelsen KV, Bilde M, Elm J (2018) J Phys Chem A 122:5026
Kildgaard JV, Mikkelsen KV, Bilde M, Elm J (2018) Phys Chem A 122:8549
Simm GN, Türtscher PL, Reiher M (2020) J Comput Chem 41:1144–1155
Bruice TC, Schmir GL (1958) J Am Chem Soc 80:148
Robiette R, Aggarwal VK, Harvey JN (2007) J Am Chem Soc 129:15513
Cantillo D, Kappe CO (2010) J Org Chem 75:8615
Xu J (2006) J Mol Struct Theochem 767:61
Fan J-F, Yang C-H, He L-J (2009) Int J Quantum Chem 74:3031
Li J, Jiang W-Y (2010) J Theor Comput Chem 9:65
Dong L, Qin S, Su Z, Yang H, Hu C (2010) Org Biomol Chem 8:3985
Roy D, Sunoj RB (2007) Org Lett 9:4873
Harvey JN (2010) Faraday Discuss 145:487
Martelli G, Orena M, Rinaldi S (2012) Eur J Org Chem 2012:4140
Roy D, Sunoj RB (2008) Chem Eur J 14:10530
Roy D, Patel C, Sunoj RB (2009) J Org Chem 74:6936
Liu Z, Patel C, Harvey JN, Sunoj RB (2017) Phys Chem Chem Phys 19:30647–30657
Goerigk L, Hansen A, Bauer CA, Ehrlich S, Najibi A, Grimme S (2017) Phys Chem Chem Phys 19:32184
Check CE, Gilbert TM (2005) J Org Chem 70:9828–9834
Grimme S (2012) Chem Eur J 18:9955–9964
Acknowledgements
The authors thank Stefan Grimme for providing the code to compute the vibrational entropy with the quasi-rigid-rotor-harmonic-oscillator approach, Oliver Welz and Mikko Muuronen for helpful discussions, and the reviewers for their thorough work.
Author information
Authors and Affiliations
Contributions
MM and AP carried out preliminary studies. PD performed the computations for the introduction and the section “general strategy” and RS performed those for the acid–base reactions and the MBH mechanism. RS and PD wrote the manuscript.
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sure, R., el Mahdali, M., Plajer, A. et al. Towards a converged strategy for including microsolvation in reaction mechanism calculations. J Comput Aided Mol Des 35, 473–492 (2021). https://doi.org/10.1007/s10822-020-00366-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-020-00366-2