Abstract
Considering the importance of characterizing germplasm banks for their conservation and use in breeding programs and the commercial and nutritional potential of snap beans, the present study aimed to characterize the genetic diversity of snap bean accessions from the germplasm bank of IDR-Paraná through morpho-agronomic descriptors and molecular markers. A total of 102 snap bean accessions were evaluated, including 60 and 42 accessions of indeterminate and determinate growth habits, respectively. Sixty descriptors were used for morpho-agronomic characterization, which were evaluated in the wet season harvests of 2017 and 2018 using a randomized block design with three repetitions. Molecular characterization was performed using amplified fragment length polymorphism markers (AFLP) with four primer combinations. There was a wide variability among the snap bean accessions and a positive correlation between morpho-agronomic and molecular descriptors. The formation of two large groups of snap bean was observed and growth habit was strongly associated with gene pool, with evidence of high diversity among the accessions of the indeterminate growth habit. The descriptors that allowed high discrimination of the accessions were those that related to crop yield and cycle. The genetically divergent accessions with desirable agronomic characteristics may be used in crosses to initiate a breeding program.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The common bean (Phaseolus vulgaris L.) is one of the major legume crops cultivated worldwide with an estimated annual production of 41.71 million ha (Rawal and Navarro 2019). This species can be grown to meet the market demand for dried beans or pods, which are consumed as a vegetable (Fukuji et al. 2019; Myers et al. 2019). Snap beans have high nutritional quality; they are an important source of fiber, minerals, and vitamins, in addition to being rich in bioactive compounds that have a positive impact on the health and well-being of consumers because of their antioxidant, hypolipidemic, and anticarcinogenic activities (Chaurasia 2020).
In Brazil, snap bean is grown mainly by smallholder farmers and is considered an important source of income and an alternative for crop diversification (Andrade et al. 2016). The main cultivars used by farmers are of the indeterminate growth habit and most of the production is intended for fresh consumption (Vaz et al. 2017). However, the market for processed pods (canned and/or frozen) has been growing in recent years and is considered promising for both domestic and international markets. In addition, the bush habit allows for mechanized harvesting. In the U.S., the snap bean occupied an area of approximately 80,000 hectares in the 2019 crop season, with a production value of $303 million, and 40% of this revenue was associated with the processing market (USDA 2020).
Bush snap bean cultivars are more suitable for the industry than those with the indeterminate growth habit, because they adapt to intensive mechanized cultivation, from sowing to harvesting, which reduces the production cost and facilitates the scheduling of production (Krause et al. 2009; Kleintop et al. 2016; Vaz et al. 2017). In this context, the development of more productive cultivars with favorable commercial pod characteristics, adequate plant size for mechanization, resistance/tolerance to biotic stresses, and adaptation to the Brazilian soil and climate conditions are considered main challenges for the improvement of this crop.
Genetic improvement involves several steps for the development of new cultivars. The first step consists of exploring the existing variability in germplasm banks to understand the genetic potential of some accessions for future use in the breeding program (Delfini et al. 2021). The Paraná Rural Development Institute—IAPAR—EMATER (IDR—Paraná) has a collection of 14,234 accessions of P. vulgaris L. introduced by several research institutions and universities as well as breeding lines improved by the institute itself (Moda-Cirino et al. 2012). These accessions have been widely characterized and used, and the IDR-Paraná stands out as one of the main institutions for common bean improvement, having released 39 cultivars. However, limited research has been conducted on the variability of the germplasm bank for the improvement of snap bean.
The characterization and evaluation of germplasms are performed using a set of descriptors, such as phenotypic and molecular descriptors, which are essential for the conservation of the germplasm bank and its use in breeding programs (Arunga et al. 2015; Moreira et al. 2018). Phenotypic characterization is of great relevance because it allows the direct measurement of descriptors of agronomic interest. However, some of these descriptors are influenced by the environment, which may limit their use in the differentiation of the accessions (Nadeem et al. 2020). On the other hand, molecular markers allow the characterization and realization of genetic divergence studies with a high degree of precision because they are not affected by environmental factors and the data obtained are stable and reproducible (Adhikari et al. 2017).
Considering the importance of characterizing and evaluating germplasm banks for the initiation of a breeding program and the nutritional and commercial potential of beans for pod consumption, the aim of the present study was to determine the genetic diversity in a panel of snap bean accessions from the germplasm bank of IDR-Paraná using phenotypic and molecular descriptors.
Material and methods
Genetic material
A total of 102 snap bean accessions from the germplasm bank of IDR-Paraná, of which 60 were of the determinate growth habit and 42 of the indeterminate growth habit, were characterized using morpho-agronomic and molecular descriptors. These accessions were composed of improved strains from the International Center for Tropical Agriculture—CIAT (Cali, Colombia), cultivars and local varieties from Brazil, and those introduced from other countries (Table S1). All accessions were regenerated and multiplied in a protected environment at the station of the research headquarters of IDR-Paraná in Londrina, Paraná, Brazil (latitude 23° 21′ 23.81″ S, longitude 51° 9′ 41.02″ W, and altitude 589 m). The regeneration and multiplication stages occurred between September 2016 and June 2017.
Phenotyping
Two trials were established for phenotyping: one for the accessions of the determinate growth habit and another for the accessions of the indeterminate growth habit. The trials were conducted side by side and sowing was performed on the same date at the research station of the research headquarters of IDR-Paraná in the 2017 and 2018 wet season harvests (Figure S1). The two crops were grown in the same experimental area at an altitude of 585 m, latitude of 23° 21′ 43.13″ S, and longitude of 51° 9′ 52.63″ W.
The experimental design was a randomized block design with three repetitions; in the assay with the accessions of the determinate growth habit, each plot was composed of three 1-m rows with 0.5 m spaces between lines and 10 plants per linear meter. In the assay with the accessions of the indeterminate growth habit, each plot was composed of one 1.5-m row with 1.2 m spaces between lines and seven plants per linear meter.
The accessions were phenotyped using 60 morpho-agronomic descriptors (Table S2). Most of these descriptors were proposed by the International Union for the Protection of New Varieties of Plants (UPOV), the National Service for the Protection of Cultivars-SNPC/MAPA (BRAZIL 2015) for the protection of bean cultivars, and Bioversity International (IPGRI 2001) for genetic diversity studies in beans.
Sowing was performed using 350 kg ha−1 of fertilizer (formulation: 4–30–10 (N–P2O5–K2O)). Top-dressing fertilization was performed at the V4 developmental stage (fully developed third trifoliate leaf) using 40 kg ha−1 of nitrogen (source: ammonium sulfate). Disease, pest, and weed control was performed according to the technical recommendations for the crop.
Genotyping
Genomic DNA was extracted from pools of 12 plants of each access according to the protocol of Ferreira and Grattapaglia (1998) (modified) using CTAB buffer followed by isopropanol precipitation. All samples were treated with RNAse (110 ng/mL). DNA integrity was confirmed by 1% agarose gel electrophoresis.
The amplified fragment length polymorphism (AFLP) protocol was adapted from Vos et al. (1995), and DNA restriction and adapter ligation were performed simultaneously. Approximately 700 ng of DNA were digested with 5 U of EcoRI enzyme and 1 U of MseI enzyme at 37 °C for 4 h, followed by adapter ligation using 2 U of T4 DNA ligase at 22 °C for 1 h. Then, incubation was performed at 70 °C for 10 min for heat inactivation of the restriction enzymes. The preselected amplification reaction was performed using the primers EcoRI + A and MseI + C in a volume of 10 μL containing 3 μL of 1 × diluted template. The polymerase chain reaction (PCR) conditions were as follows: 72 °C for 2 min followed by 20 cycles at 94 °C for 1 s, 56 °C for 30 s, and 72 °C for 2 min, with a final extension of 30 min at 60 °C.
The selective reaction was performed using the primer pairs FAN-Eco + AAG/Mse + CTC; VIC-Eco + ACT/Mse + CTT; NED-Eco + ACA/Mse + CAC; and PET-Eco + AGC/Mse + CTGA with 2.5 μL of 6 × diluted template in a final volume of 10 μL. The amplification conditions were as follows: an initial cycle at 94 °C for 2 min followed by 65 °C for 30 s and 72 °C for 2 min; 8 cycles at 94 °C for 1 s, 64 °C for 30 s (with a decrease of 1 °C per cycle), and 72 °C for 2 min; and 23 cycles at 94 °C for 1 s, 56 °C for 30 s, and 72 °C for 2 min, with a final extension of 30 min at 60 °C. The PCR products were separated by capillary electrophoresis using an ABI 3500 XL sequencer (Applied Biosystems) with GS-600 LIZ (Applied Biosystems) as a molecular weight marker. The detection of electropherogram peaks generated by the AFLP technique and fragment analysis was performed using GeneMaPPer® v.4.1 software (Applied Biosystems). The fragments of 75 to 500 base pairs, from the four primer combinations, were combined in a single binary matrix of presence (1) and absence (0).
Data analysis
The quantitative phenotypic data were subjected to the mixed-model analysis of variance. The statistical model used was \(y=Xr+Zg+Wi+e,\) where \(y\) is the vector of observations, \(r\) is the vector of the repetition effect (considered as fixed) added to the general mean, \(g\) is the vector of the genotype effects (considered as variable), \(i\) the vector of the interaction between the genotype and environment effects (random), and \(e\) is the vector of the error (random). \(X\), \(Z, \mathrm{and}\) \(W\) represent the incidence matrices for the abovementioned effects. Based on the analysis of the models, the genetic parameters were estimated by restricted maximum likelihood (REML) and the genetic values were predicted by the best linear unbiased prediction (BLUP). All analyses were performed using software Selegen REML/BLUP (Resende 2016). The BLUP values were used for correlation and box plot analysis, and the accessions were grouped according to their origin (cultivar, breeding line and landrace) for analysis. To analyze the morpho-agronomic genetic divergence among the accessions, the qualitative and quantitative descriptors (BLUPs) were used. The Ward-MLM method was used, with the ideal number of groups determined by likelihood analysis. These analyses were performed using SAS (SAS 2000) and R (R CORE TEAM 2020) software with corrplot (Wei and Simko 2017) and cluster (Mächler et al. 2019) packages.
For the molecular analysis, the data were transformed into a binary matrix of presence (1) and absence (0) and subsequently into a Jaccard dissimilarity matrix and principal component analysis (PCA). Binary matrix and AFLPsurv 1.0 software were used to calculate the genetic diversity index (Fst). The dendrogram was obtained using the neighbor-joining method with the cluster package (Machler et al. 2019), and PCA analysis was conducted using Tidyverse (Wickham et al. 2019) and Ggrepel (Slowikowski 2020) packages. These analyses were performed using R software (R CORE TEAM 2020).
The genetic structure of the accessions was calculated using molecular data and a clustering algorithm, namely, sparse non-negative matrix factorization algorithm, implemented with the LEA package (Frichot and François 2015). The number of clusters (K) was determined using the cross-entropy criterion (CEC), in which the lowest value indicates the most likely number of clusters (Frichot et al. 2014). K was estimated to range from 1 to 10 clusters with 100 runs per K. Finally, the best run of the 100 repetitions (lowest CEC) corresponding to the selected K was used for the graphical representation of the population structure.
Correlation analysis between the matrices was performed based on the dissimilarity matrices of the morpho-agronomic data (Gower’s matrix) and molecular data (Jaccard’s matrix). The significance of the correlation was assessed using the t-test and the Mantel test based on 10,000 simulations. The analysis was performed using Genes software (Cruz 2013).
Results
Phenotyping
Most of the qualitative descriptors showed variations among the snap bean accessions from the germplasm bank of IDR-Paraná, except for the descriptor pod surface texture, and all accessions were classified as smooth pods. Three descriptors showed variations in only three accessions, namely, the presence of secondary color in the pod, the secondary color of the pod, and the density of spots on the secondary color of the pod, and it was demonstrated that most accessions had pods with only one color.
Considering the predominance of multicategory descriptors, the accessions from the snap bean germplasm bank of IDR-Paraná can be described as having the following characteristics: absence of rough leaf (80.4% of the accessions), circular to quadrangular terminal shape of the central leaflet (80.4%), medium bracts (44.1%), white wing color (63.7%), elliptical cross section of the pod (36.3%), pod with median green color (48%), slightly curved pod profile (92.2%), presence of string at the ventral suture of the pod (63.7%), pod with a low degree of curvature (61.8%), apical tooth of pod with a small curvature (55.9%), moderate constriction in dry pod (54.9%), reniform longitudinal section of seed (65.7%), seed with only one color (84.3%), white colored seed (49%), seed venation absent or very weak (43.1%), opaque seed (49%), and absence of halo in seed (87.3%).
The analysis of the quantitative morpho-agronomic descriptors showed that all traits had a significant effect on the analysis of variance for the source of genotypic variation, which indicates a wide variability among the accessions. The values of heritability (h2g) ranged from 0.03 to 0.94, and most values were > 0.60 (Table 1). The traits beak length (ALP) and seed length had low h2g (0.03 and 0.07, respectively) and accuracy (Ac) (0.27 and 0.42, respectively) values, which indicates a strong environmental influence on these descriptors. Most of the other traits had accuracy values > 90%.
Considering the 102 accessions from the germplasm bank of IDR-Paraná, the following results were obtained: a mean mass of 250.6 g for 1000 seeds, a cycle of 39.7 days until the beginning of flowering and 58 days until the beginning of harvest, a mean of 5.3 seeds per pod, commercial pod mass (COM) of 274 g per plant, 55 commercial pods per plant, and mean pod mass of 5.43 g. The accessions of the determinate growth habit had higher values of trifoliate leaf length (LL), leaf width (LW), pod thickness (PT), and seed thickness (ST). The accessions of the indeterminate growth habit had higher values of cycle until the beginning of flowering (FC), cycle until the beginning of harvest (HAC), COM, and commercial pod number (MPN) (Fig. 1).
Boxplots for the characterization of the snap bean germplasm bank of IDR-Paraná considering the main quantitative morpho-agronomic descriptors. LL central leaflet length (cm), LW central leaflet width (cm), PL pod length (cm), PT pod thickness (mm), W1000 thousand seed mass (g), ST seed thickness (mm), FC cycle until the beginning of flowering (days), HAC cycle until the beginning of harvest (days), CPM commercial pod mass (g), CPN commercial pod number
The accessions of the determinate growth habit were divided into three groups (breeding line, cultivar and landrace). Breeding lines showed the highest values for LL, LW, and pod length (PL) amongst all the groups (Figure S3). Cultivars showed higher HAC and MPN, whereas landraces varieties showed higher thousand seed mass (W1000) and ST (Fig. 1). The accessions of the indeterminate growth habit were divided into two groups (cultivars and landraces). Cultivars showed higher values of PL, PT, FC, HAC, and COM, whereas landraces had higher values of W1000 and ST.
Pearson's correlation analysis showed that LL and LW were highly correlated with each other and negatively correlated with total and commercial number of pods (TPN and MPN, respectively) and with total and commercial pod mass (TPM and COM, respectively) (Fig. 2). Cycle length (FC and HAC) and descriptors related to production components (TPM, TPN, MPM, and MPN) were positively correlated. A negative correlation was observed between PT and FC, HAC, TPM, TPN, COM, and MPN.
Pearson's correlation (p ≤ 0.05) among the 22 quantitative morpho-agronomic descriptors used in the characterization of snap bean accessions from the germplasm bank of IDR-Paraná. LL central leaflet length (cm), LW central leaflet width (cm), PL pod length (cm), PW pod width (mm), PT pod thickness (mm), PT/PW pod thickness to pod width ratio, ALP apical tooth length (cm), W1000 thousand seed mass (g), ST seed thickness (mm), SW seed width (cm), SL seed length (cm), SL/SW seed length to width ratio, ST/SW seed thickness to width ratio, FC cycle until the beginning of flowering (days), HAC cycle until the beginning of harvest (days), LNP number of loci per pod, SNP number of seeds per pod, TPM total pod mass (g), TPN total number of pods, CPM commercial pod mass (g), CPN commercial number of pods, PMA mass per pod unit
Multivariate Ward-MLM analysis showed that the ideal number of groups was two, according to the log-likelihood function (Table S3). Considering the first canonical variable, the quantitative descriptors that most contributed to the discrimination of the accessions were PT/PW, SL/SW, ST/SW, FC, HAC, TPM, TPN, MPM, and MPN (Table 2). Considering the second canonical variable, the descriptors that contributed the most were FC, HAC, and MPM. The quantitative descriptors that showed less ability to differentiate the accessions were PL, ALP, W1000, ST, SL, LNP, SNP, and PMA.
The graphical representation of the first two canonical variables for the two groups created by the Ward-MLM cluster analysis showed that group 1 was mostly formed by the accessions of the determinate growth habit and group 2 was mostly formed by the accessions of the indeterminate growth habit (Fig. 3). The accessions of group 1 were placed adjacent, whereas the accessions of group 2 were dispersed. Separation between cultivars, breeding lines, and landraces was not observed in any group.
Genotyping
In the molecular characterization using AFLP markers, the four primer combinations produced 968 polymorphic loci. Of these, 130 (13.43%) were generated by the primer pair FAM-Eco + AAG/Mse + CTC, 222 (22.93%) by the primer pair NED-Eco + ACA/Mse + CAC, 424 (43.8%) by the primer pair PET-Eco + AGC/Mse + CTGA, and 192 (19.84%) by the primer pair VIC-Eco + ACT/Mse + CTT. The coefficient of genetic differentiation (Fst) between the accessions of the determinate and indeterminate growth habits was 0.13 (Table 3). A total of 747 polymorphic loci were generated within the accessions of the determinate growth habit, with an Fst of 0.084 among the cultivar, breeding line, and landrace subpopulations. Within the accessions of the indeterminate growth habit, there were 737 polymorphic loci and an Fst of 0.103 existed between cultivars and landraces.
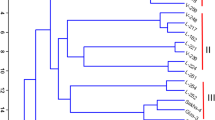
PCA and hierarchical clustering identified four groups (Fig. 4). Groups 1 and 4 were formed by the accessions of the determinate and indeterminate growth habits, respectively. Groups 2 and 3 exhibited a transition profile and were composed of the accessions of different growth habits. Mean dissimilarity was 0.57, and the smallest distance (0.27) occurred between the accessions of the determinate growth habit, Macarrão Rasteiro and Zegane, whereas the largest distance (0.75) was observed between the landrace of the determinate growth habit, Montcalm 23, and the cultivar of the indeterminate growth habit, Tempesta. Considering only the groups formed according to the growth habit, the determinate and indeterminate growth habit groups had a mean genetic distance of 0.55 and 0.60, respectively. When the accessions were analyzed separately according to cultivars, breeding line and landraces, the mean genetic distances in cultivars, breeding lines, and landraces were 0.58, 0.55, and 0.58, respectively.
In the population structure analysis, based on the minimum cross-entropy value criterion, the optimum number of groups was five (k = 5) (Fig. 3C). The first group (G1) was formed by cultivars and breeding lines of the determinate growth habit. In G2, the only accessions of the indeterminate growth habit was the cultivar Strada; the remaining accessions in the group were cultivars and breeding lines of the determinate growth habit. G3 was composed mainly of the landraces of both determinate and indeterminate growth habits. G4 was formed by the landraces of the indeterminate growth habit, except for cultivar, Macarrão Baixo, which is of the determinate growth habit. G5 was composed of five of the seven cultivars of the indeterminate growth habit and one landrace. The comparison between hierarchical grouping and the population structure analysis showed 93% agreement. The correlation analysis between Gower's dissimilarity matrices obtained through characterization with 60 morpho-agronomic descriptors and Jaccard's matrix generated with molecular characterization data using AFLP markers (968 polymorphic loci) showed a significant correlation of 0.43.
Discussion
The narrow genetic base of cultivars and the low utilization of genetic resources are some of the main factors limiting the increase in productivity of non-commodity legumes. The characterization of genetic variability in germplasm banks through pre-breeding enables the introgression of favorable genes for the development of cultivars that are more productive, more resistant/tolerant to biotic and abiotic stresses, and have better nutritional characteristics (Sharma et al. 2013). The wide variability observed in the accessions from the snap bean germplasm bank of IDR-Paraná shows the potential of using these accessions in breeding programs for the development of productive cultivars with adequate pod characteristics.
Because snap bean is a vegetable, the choice of cultivar should consider aspects beyond productivity, such as elliptical (PT/PW: approximately 1) and non-curved shape, longer length, reduced fiber content, absence of strings at the ventral suture, and white and cylindrical seeds (Hagerty et al. 2016). It was observed that the accessions of the germplasm bank have preferred qualitative traits that meet the requirements of the consumer market.
The high heritability observed for most of the traits indicates that they are minimally influenced by the environment and that selection based on the phenotype can result in great success in the selection of genotypes (Katuuramu et al. 2020). Furthermore, a high accuracy was observed for most of the traits, which indicates a high precision in the inference of the genotype means. This parameter informs the correct classification of accessions for selection purposes and the efficacy of the conclusions regarding the genotypic value of the accession, which is a correlation between the predicted and true genotypic values (Resende 2009). Moreover, a low accuracy value for the traits ALP and SL may hinder the selection of accessions because these variables are highly influenced by the environment.
A descriptor’s ability to discriminate the accessions should be the main factor to consider when defining the descriptors to be used in germplasm characterization (Laura et al. 2018). Broadly considering the estimates of heritability, selective accuracy, and contribution to accession differentiation through the canonical variables, the following quantitative descriptors should be prioritized for the characterization of snap bean germplasm: production components (TPM, MPN and MPM), cycle (FC and HC), PT/PW, SL/SW, and ST/SW. Descriptors with low ability to differentiate the accessions of a germplasm bank should not be utilized because the use of ineffective descriptors increases the costs and time of the work (Delfini et al. 2017). Under the conditions of low availability of human and financial resources, the descriptors ALP, LNP, SNP, and PMA should not be prioritized.
The 60 morpho-agronomic descriptors and the AFLP molecular markers provided a broad characterization of the snap bean accessions from the germplasm bank of IDR-Paraná, with the separation of the accessions of the determinate and indeterminate growth habits. Similarly, Andrade et al. (2016) reported the separation of accessions according to the determinate and indeterminate growth habits using AFLP markers.
The group formed by the accessions of the indeterminate growth habit in the morpho-agronomic characterization exhibited higher genetic variability than the group formed by the accessions of the determinate growth habit. Studies suggest that the snap bean accessions of the determinate growth habit are closer to the Andean center of origin, whereas the accessions of the indeterminate growth habit are from the Mesoamerican center of origin (Blair et al. 2010; Andrade et al. 2016). As high genetic variability occurs in the Mesoamerican center of origin than in the Andean center (Kwak and Gepts 2009; Bitocchi et al. 2012), the higher genetic divergence among the accessions of the indeterminate growth habit may be associated with the center of origin and evolutionary characteristics. Wallace et al. (2018) reported that snap bean has a narrower genetic basis than common bean. The authors also found evidence of the possibility of unique combinations in crosses between snap bean and common bean. This information demonstrates the potential of using snap bean accessions in bean breeding programs for dry bean consumption and the need for maintaining or expanding the genetic base in snap bean breeding programs.
In characterization studies using AFLP molecular markers, the estimated Fst indicates that most of the variation exists within the accession groups (both determinate and indeterminate) and that only 13% of the genetic variation occurs between these groups. The Fst is directly linked to the variation in allele frequency between populations; therefore, the higher the Fst, the higher the genetic differentiation between the analyzed populations and the lower the differentiation within each population (Bashir et al. 2020). In addition, the separate analysis of the accessions of the determinate and indeterminate growth habits showed a lower diversity among cultivars, breeding lines and landraces. In populations of autogamous species, such as beans, the coefficient of genetic differentiation is usually low (≤ 0.05) or moderate (> 0.05 to ≤ 0.15), because there is less migration of genes and populations tend to be homozygous (Bradshaw 2016; Pipan and Meglič 2019; Bashir et al. 2020; Delfini et al. 2021).
The correlation between morpho-agronomic and molecular data can be linked to the number of descriptors used. In the present study, 60 morpho-agronomic descriptors were used. The results of the present study indicate the need to use a high number of morpho-agronomic descriptors with the ability to discriminate the accessions because germplasm characterization studies that used molecular markers and few morpho-agronomic descriptors did not show a significant correlation between the phenotype and the genotype (Roldán-Ruiz et al. 2001; Ghalmi et al. 2010; Arunga et al. 2015; Baba et al. 2016; Maras et al. 2016; Delfini et al. 2017).
Despite the increasing use of molecular markers to characterize and estimate genetic diversity in germplasm bank accessions, phenotypic evaluation is still crucial to determine the local adaptation and agronomic potential of plant genetic resources (Chávez-servia et al. 2016). The combination of morpho-agronomic and molecular characterization proves to be the best strategy for performing complete genetic diversity studies in germplasm banks (Guidoti et al. 2018).
Conclusions
Based on the molecular and morpho-agronomic descriptors, a wide genetic variability was observed among the snap bean accessions from the germplasm bank of IDR-Paraná, which indicates the potential of these accessions for breeding programs. A higher variability was observed within the accessions of the indeterminate growth habit than within those of the determinate growth habit. There was a positive correlation between morpho-agronomic and molecular descriptors.
Data availability
All data generated or analyzed during this study are included in this article and its supplementary information files.
References
Adhikari S, Saha S, Biswas A et al (2017) Application of molecular markers in plant genome analysis: a review. Nucleus 60:283–297. https://doi.org/10.1007/s13237-017-0214-7
Arunga E, Kinyua M, Ochuodho J et al (2015) Genetic diversity of determinate French beans grown in Kenya based on morpho-agronomic and simple sequence repeat variation. J Plant Breed Crop Sci 7:240–250. https://doi.org/10.5897/JPBCS2015.0503
Baba VY, Rocha KR, Gomes GP et al (2016) Genetic diversity of Capsicum chinense accessions based on fruit morphological characterization and AFLP markers. Genet Resour Crop Evol 63:1371–1381. https://doi.org/10.1007/s10722-015-0325-4
Bashir H, Bashir Z, Mahajan R et al (2020) Molecular characterization and insights into the origin of common bean (Phaseolus vulgaris L.) landraces of north western Himalayas. Nucleus 63:271–279. https://doi.org/10.1007/s13237-020-00323-2
Bitocchi E, Nanni L, Bellucci E et al (2012) Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.1108973109
Blair MW, Chaves A, Tofiño A et al (2010) Extensive diversity and inter-genepool introgression in a world-wide collection of indeterminate snap bean accessions. Theor Appl Genet 120:1381–1391. https://doi.org/10.1007/s00122-010-1262-4
Bradshaw JE (2016) Genetic structure of landraces. In: Bradshaw JE (ed) Plant breeding: past, present and future. Springer, p 693
Chaurasia S (2020) Green beans. In: Jaiswal AK (ed) Nutritional composition and antioxidant properties of fruits and vegetables. Academic Press, p 766
Chávez-servia JL, Heredia-garcía E, Chávez-servia JL et al (2016) Diversity of common bean (Phaseolus vulgaris L.) landraces and the nutritional value of their grains. In: de Ron AM (ed) Grain legumes. IntechOpen, p 438
Cruz CD (2013) GENES: a software package for analysis in experimental statistics and quantitative genetics. Acta Sci Agron 35:271–276
da Vaz DC, de Morais Júnior OP, Peixoto N (2017) Agro-morphological characterization and genetic divergence assessment in bush snap bean genotypes1. Pesquisa Agropecuária Tropical 47:134–144. https://doi.org/10.1590/1983-40632016v4743525
de Andrade FA, Gonçalves LSA, Miglioranza É et al (2016) Análise de AFLP na diversidade genética em acessos de feijão-de-vagem de hábito determinado e indeterminado. Acta Sci Agron 38:29–34. https://doi.org/10.4025/actasciagron.v38i1.25577
Delfini J, Moda-Cirino V, Ruas CDF et al (2017) Distinctness of Brazilian common bean cultivars with carioca and black grain by means of morphoagronomic and molecular descriptors. PLoS ONE. https://doi.org/10.1371/journal.pone.0188798
Delfini J, Moda-Cirino V, dos Santos NJ et al (2021) Population structure, genetic diversity and genomic selection signatures among a Brazilian common bean germplasm. Sci Rep. https://doi.org/10.1038/s41598-021-82437-4
Ferreira ME, Grattapaglia D (1998) Introdução ao uso de marcadores moleculares em análise genética. Introdução ao uso de marcadores moleculares em análise genética
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929. https://doi.org/10.1111/2041-210X.12382
Frichot E, Mathieu F, Trouillon T et al (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983. https://doi.org/10.1534/genetics.113.160572
Fukuji AYS, Constantino LV, Zeffa DM et al (2019) Amino acid concentration, total phenolic compound content and antioxidant activity of snap bean genotypes. Braz J Food Technol 22:2–7. https://doi.org/10.1590/1981-6723.06918
Ghalmi N, Malice M, Jacquemin JM et al (2010) Morphological and molecular diversity within Algerian cowpea (Vigna unguiculata (L.) Walp.) landraces. Genet Resour Crop Evol 57:371–386. https://doi.org/10.1007/s10722-009-9476-5
Guidoti DT, Gonela A, Vidigal MCG et al (2018) Interrelação entre características morfológicas, agronômicas e moleculares na análise de diversidade genética em feijão comum. Acta Sci Agron 40:1–9. https://doi.org/10.4025/actasciagron.v40i1.33032
Hagerty CH, Cuesta-Marcos A, Cregan P et al (2016) Mapping snap bean pod and color traits, in a dry bean × snap bean recombinant inbred population. J Am Soc Horticult Sci 141:131–138. https://doi.org/10.21273/jashs.141.2.131
IPGRI (2001) Descritores para Phaseolus vulgaris. International Plant Genetic Resources Institute, Roma
Katuuramu DN, Luyima GB, Nkalubo ST et al (2020) On-farm multi-location evaluation of genotype by environment interactions for seed yield and cooking time in common bean. Sci Rep 10:1–12. https://doi.org/10.1038/s41598-020-60087-2
Kleintop AE, Myers JR, Echeverria D et al (2016) Total phenolic content and associated phenotypic traits in a diverse collection of snap bean cultivars. J Am Soc Horticult Sci 141:3–11. https://doi.org/10.21273/jashs.141.1.3
Krause W, Rodrigues R, Gonçalves LSA et al (2009) Genetic divergence in snap bean based on agronomic traits and resistance to bacterial wilt. Cropp Breed Appl Biotechnol 9:246–252. https://doi.org/10.12702/1984-7033.v09n03a06
Kwak M, Gepts P (2009) Structure of genetic diversity in the two major gene pools of common bean (Phaseolus vulgaris L., Fabaceae). Theor Appl Genet 118:979–992. https://doi.org/10.1007/s00122-008-0955-4
Laura EYL, Azize O, Arlette A et al (2018) Morphological characterization of common bean (Phaseolus vulgaris L.) landraces of Central region of Benin Republic. J Plant Breed Crop Sci 10:304–318. https://doi.org/10.5897/jpbcs2018.0766
Mächler M, Rousseeuw P, Struyf A, et al (2019) Cluster: cluster analysis basics and extensions. R package version 2.1.0
Maras M, Ibusoska A, Kratovalieva S et al (2016) Genetic diversity of common bean accessions from Former Yugoslav Republic of Macedonia as revealed by molecular and morphological markers. Genetika 48:729–742. https://doi.org/10.2298/GENSR1602729M
Moda-Cirino V, Gerage AC, Riede CR et al (2012) Plant breeding at Instituto Agronômico do Paraná: IAPAR. Crop Breed Appl Biotechnol 12:25–30. https://doi.org/10.1590/s1984-70332012000500004
Moreira AFP, Ruas PM, de Ruas CF et al (2018) Genetic diversity, population structure and genetic parameters of fruit traits in Capsicum chinense. Sci Hortic 236:1–9. https://doi.org/10.1016/j.scienta.2018.03.012
Myers JR, Wallace LT, Moghaddam SM et al (2019) Improving the health benefits of snap bean: genome-wide association studies of total phenolic content. Nutrients 11:1–17. https://doi.org/10.3390/nu11102509
Nadeem MA, Karaköy T, Yeken MZ et al (2020) Phenotypic characterization of 183 Turkish common bean accessions for agronomic, trading, and consumer-preferred plant characteristics for breeding purposes. Agronomy. https://doi.org/10.3390/agronomy10020272
Pipan B, Meglič V (2019) Diversification and genetic structure of the western-to-eastern progression of European Phaseolus vulgaris L. germplasm. BMC Plant Biol 19:1–16. https://doi.org/10.1186/s12870-019-2051-0
R CORE TEAM (2020) R: a language and environment for statistical computing.
Rawal V, Navarro DK (2019) The Global economy of pulses. FAO, Rome
Resende MDV (2009) Genética biométrica e estatística no melhoramento de plantas perenes. Embrapa Informação Tecnológica, Brasília
Resende MDV (2016) Software Selegen-REML/BLUP: a useful tool for plant breeding. Crop Breed Appl Biotechnol 16:330–339
Roldán-Ruiz I, van Euwijk FA, Gilliland TJ et al (2001) A comparative study of molecular and morphological methods of describing relationships between perennial ryegrass (Lolium perenne L.) varieties. Theor Appl Genet 103:1138–1150. https://doi.org/10.1007/s001220100571
SAS (2000) SAS software
Sharma S, Upadhyaya HD, Varshney RK, Gowda CLL (2013) Pre-breeding for diversification of primary gene pool and genetic enhancement of grain legumes. Front Plant Sci. https://doi.org/10.3389/fpls.2013.00309
Slowikowski K (2020) ggrepel: Automatically Position Non-Overlapping Text Labels with ’ggplot2
USDA (2020) Vegetables 2019 Summary
Vos P, Hogers R, Bleeker M et al (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414. https://doi.org/10.1093/nar/23.21.4407
Wallace L, Arkwazee H, Vining K, Myers JR (2018) Genetic diversity within snap beans and their relation to dry beans. Genes 9
Wei T, Simko V (2017) R package “corrplot”: Visualization of a Correlation Matrix (Version 0.84)
Wickham H, Averick M, Bryan J et al (2019) Welcome to the Tidyverse. J Open Sour Softw 4:1686. https://doi.org/10.21105/joss.01686
Acknowledgements
The authors thank the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for supporting this study.
Funding
This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001.
Author information
Authors and Affiliations
Contributions
JSN, VMC and LSAG conceived and designed the research. JSN, JD, DMZ, AFN and LSAG conducted the experiments and analyzed the data. JSN, JD, DMZ and AFN participated in parts of the experiments and data analysis. JSN drafted the manuscript, and VMC and LSAG revised the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that no competing or conflicts of interest exist.
Ethical approval
The experiments were performed in compliance with the current laws of Brazil.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
dos Santos Neto, J., Moda-Cirino, V., Delfini, J. et al. Morpho-agronomic and molecular characterization of snap bean diversity panel. Genet Resour Crop Evol 70, 399–411 (2023). https://doi.org/10.1007/s10722-022-01434-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-022-01434-2