Abstract
Efforts to elucidate the relationships between microorganisms and metal corrosion were mainly directed to understanding the formation of biofilm structures grown on corroded surfaces. The emergence of high throughput DNA sequencing techniques has helped in the description of microbial species involved directly and indirectly in the corrosion processes of alloys. Coupled with sequencing from environmental samples, other methodologies such as metatranscriptome, metaproteomics and metabolomics have allowed a new horizon to be opened on the understanding of the role of corrosive microbial biofilm. Several groups of bacteria and archaea were identified, showing the dominance of Proteobacteria in several samples analyzed and members of groups that previously received less attention, such as Firmicutes and Bacteroidetes. Our research also shows that metagenomic studies describe the presence of various Archaea domain thermophilic and methanogenic groups associated with metal corrosion. Thus, opening the prospect of describing new microbial groups as possible participants in this current global concern.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first relationship between corrosion and microorganisms was originally demonstrated in 1910 in an experiment that reported the corrosive action of bacteria on iron and steel (Gaines 1910). However, this relationship was only substantially established in the mid-1960s, when new studies presented the role of Sulfate-Reducing Bacteria (SRB) species in mild steel corrosion under anaerobic conditions and the work on widespread failures in iron pipes that described the direct association between SRB and corrosive processes (Booth and Tiller 1962; Iverson 1966; von Wolzogen Kuhr and van der Vlugt 1964). This slow progress in the knowledge of microbial corrosion in this period was due to the limited exchange of experiences between the different areas of knowledge of metallurgy, materials, electrochemistry, and microbiology. It was only in the 1980s, with the integration between these distinct fields, that concern for corrosion, especially metals, was given due importance to the impact of corrosion in productive areas (Videla and Herrera 2005).
Nowadays, metal corrosion is a worldwide concern that affects diverse productive industries. Corrosion is a process that occurs naturally and is primarily bound to the material being exposed to the surrounding water, which acts as an electrolyte, being very important in the corrosion process, as it allows ionic conduction. In addition, other compounds and corrosive environmental conditions, such as soil resistivity, moisture levels, physical stress, exposure to seawater or exposure to chemicals, especially of industrial origin, also exert an essential induction or acceleration factor in corrosion, especially in metals (Hamilton 2003). In the United States, the National Association of Corrosion Engineers (NACE) estimated costs related to the corrosion of metal structures to be $2.5 trillion, which would represent about 3.4% of global GDP in 2013. Furthermore, in this study, the annual per capita impact of corrosion was also shown to be $970 (Koch et al. 2002). These values do not include indirect costs caused by the impact of corrosion, such as loss of productivity, investment in prevention and mitigation methods and training of technicians.
Numerous methods are employed to prevent or mitigate the impact of corrosion on various types of structures. The most trivial technique consists of a passive approach, such as coating (e.g. organic and inorganic coatings, plastics and paints) structures to prevent direct contact between the material to be protected and the corrosive environment (Price and Figueira 2017). Another technique is cathodic protection, which is primarily used in offshore structures, pipelines and oil tankers and consists of creating a galvanic cell, thus acting with a "sacrifice metal" (Baeckmann 1997). Along this line, the cost of corrosion control is also significant, reaching about $121 billion worldwide, which is mainly for corrosion control, research, and development, and education and training (Koch et al. 2002).
The mechanisms involved in corrosion are complex, and go beyond the factors described above, more recently the role of microorganisms in corrosion processes has been frequently described. Microorganisms can initiate, facilitate or accelerate corrosion by altering pH and oxidation–reduction potential (Eh) in the secretion of corrosive metabolites or even directly acting in oxidation–reduction reactions through extracellular enzymes (Little and Lee 2007). This biological action is known as Microbiologically Influenced Corrosion (MIC) and plays a central role in corrosion. For example, the action of microorganisms can significantly affect these reactions, accelerating corrosion rates from 100 to 1000 times (Rajala et al. 2015; Videla 1996). Although fungi, such as Aspergillus niger, Chrysosporium merdarium, and Arthrinium phaeospermum are described as having a meaningful role in the corrosion of metals, different species of bacteria and the archaea domains are most frequently related to the degradation of metals (Li et al. 2018a, b; Lugauskas et al. 2009). Among the species commonly described as being associated with metal alloy corrosion SRB, Sulfate Oxidizing Bacteria (SOB), Iron Reducing Bacteria (IRB), Iron Oxidizing Bacteria (IOB) and Sulfate Reducing Archaea (SRA) are identified (Li et al. 2017, 2018a, b). Specimens of these bacterial groups interact with metals by adhering strongly to their surfaces by means of structures called biofilms. In fact, such complex exopolysaccharide structures create microenvironments conducive to the action of these mechanisms (Beech et al. 2014; Geesey et al. 2000). The metabolic activities that occur within structures of biofilms adhered to metal surfaces can alter conditions such as availability of dissolved oxygen, pH change, and the presence of organic acids and complexing agents. These variations, in a way, can influence the states of current electrochemical potential in a way that influences the states of, as well as the kinetics of, corrosion (Beech and Sunner 2004; Videla and Herrera 2005).
Efforts since the 1990s have deepened knowledge about the influence of microorganisms on corrosion. In recent years, molecular biology techniques such as DNA and RNA sequencing have leveraged knowledge about the dominant bacterial groups in the corrosive biofilm, the proportion of these groups in the total population, the understanding of the bacterial sequence that occurs over time and the effects of biocides on these biofilms (Beech et al. 2005; Moura et al. 2018). The use of large-scale sequencing has been widespread among MIC studies. The metagenomic analysis enables the description of the microbial groups present in the environmental sample analyzed, independent of the possibility of being cultivable, in a short space of time and at less prohibitive costs. Nowadays, the subject on MIC has received more attention, and consequently, new work has emerged as well as revisions on the subject. Moreover, due to the use of new high-through techniques, a new paradigm has arisen about microbial corrosion. Hence, this review seeks to present new approaches to the relationship between high-through techniques and microbial corrosion of metals and alloys.
Mechanisms of microbial corrosion
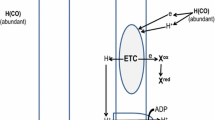
It is widely accepted that the electrochemical process is the fundamental force that drives metal corrosion (Revie 2011). Corrosion involves a transfer of electrons from zero-valent metal (Eq. 1), at an anodic site (oxidation), to an external electron acceptor, which is at the cathodic site (reduction) (Beech and Sunner 2004). The tendency of a metal to act as an electron donor, anodic site, and another component to act as acceptor, cathodic site, will depend mainly on the electrochemical potential. In this way, the flow of electrons between these two sites will be from the more negative to the more positive potential, resulting in anodic dissolution or corrosion. The kinetics between these two sites facilitate this flow of electrons between the anode and cathode sites and determine the severity of damage in anode sites as well as the rate of corrosion. In an environment where aerobic conditions prevail, the main acceptor of electrons is oxygen, and this reaction results in the formation of iron (hydro)oxides, visibly characterized by oxide deposit (Enning and Garrelfs 2014; Kaesche 2003). Over time, the speed of the reaction tends to diminish gradually, due to products that are adhered to the superficial layer of the metal. The stability of these layers will depend on their chemical and morphological nature (Beech and Sunner 2004).
In the general equation for corrosion in the oxic environment, the final acceptor of electrons is oxygen (Eq. 2) and the cathodic reaction ends with its reduction, all at a relatively high velocity. In an aqueous aerobic environment, the process has additional reactions that result in characteristic corrosion products such as iron oxide/hydroxides (Eq. 3) (Fig. 1). The reactions that occur in an aqueous aerobic environment are:
Schematic illustrations of abiotic corrosion mechanisms under oxic and anoxic conditions. The electrons resulting from the oxidation of iron, under oxic conditions are rapidly directed at reducing oxygen, whereas in anoxic conditions, the catotonic reaction is directed to the reduction of H+ in h2. In this anoxic condition the reactions occur at a slow rate
In anoxic environments, the electron acceptors for iron oxidation are protons from dissociated water, molecular hydrogen being the most common (Eq. 4). This reaction, due to the condition of electroneutrality, which is coupled to the cathodic counterpart (Eq. 5). The product of this reaction, ferrous iron, readily precipitates (Fig. 1).
Notwithstanding these reactions do not occur rapidly in abiotic environments, microorganisms appear to play a major role in the acceleration of corrosion. Howbeit, the role of microorganisms in the redox reactions described above must also follow the laws of thermodynamics. The participation of microorganisms in the process has the characteristic of modifying the metal-solution interface through the formation of biofilm structures on the metal surface. In the MIC, the mechanism employed to capture electrons from metallic sources involves the flow from the negative to the positive potential. Corrosion influenced by microorganisms will play a role as a cathodic site, and corrosion efficiency will depend on the kinetics of the electron flux between the surface and the microbial substrate as an electron acceptor (Hamilton 2003).
Nonetheless, for corrosion to occur, some basic factors must be present, such as the source of energy, nutrients, especially the source of carbon, nitrogen, hydrogen, oxygen, phosphate, and sulfur, as well as electron donors and acceptors. The metabolic mechanism of corrosion adopted by bacterial groups will depend on which electron acceptor is employed. For example, the SRB group, one of the first to be related to metal corrosion, employs the sulfate ion (SO42−) as the final electron acceptor (Little and Lee 2007). Other associated groups described as corrosion players show different metabolic mechanisms, as iron-reducing/oxidizing-bacteria (IOB/IRB), nitrate-reducing bacteria (NRB), manganese-oxidizing bacteria (MnOB), acid-producing bacteria (APB) and methanogens (Beech and Sunner 2004; Videla and Herrera 2005).
The two metals most likely to lose electrons in the corrosion processes are iron [Fe(II)/Fe(III)] and manganese [Mn(II)/Mn(IV)] due to their high availability in the most diverse natural environments and their redox potential. In corrosion, ferrous iron is easily oxidized by iron-oxidizing bacteria because of the minimum energy required. This group of bacteria, described as acidophilic, is able to corrode in environments with low concentrations of oxygen and neutral pH (Ray et al. 2010; Andrews et al. 2013). Despite the fact manganese is chemically more stable, it is also subject to microbial oxidation. Both chemical elements are oxidized by bacteria, which are ordinarily found together in the environment. The major generators related to the oxidation of iron and manganese are Leptothrix and Crenothrix genera (Ghiorse 1984; Lovley 1991; Tebo et al. 1997). These microorganisms are also known as metal deposit bacteria due to the evident deposition of rust on corroded surfaces (Emerson 2018). A very representative bacterium of this group is Gallionella, belonging to the class Zetaproteobacteria (Kappler and Newman 2004). The reduction of iron by microorganisms occurs in a restricted group of microorganisms that changes from aerobic respiration to anaerobic, when in the absence of O2. The Fe3+ reduction process facilitates the corrosion of iron and its alloys by removing previously protected corrosion products formed in iron oxidation reactions. Among the representatives are the genera Pseudomonas and Shewanella (Beech et al. 2014). In fact, in biocorrosion studies, these members are generally found in rust present on metal surfaces (Moura et al. 2018; Procópio 2019).
Under anoxic conditions, corrosion is usually slow and therefore would not be a serious concern and, the involvement of microorganisms is frequently reported in corrosion sites on metal surfaces under anaerobic conditions (Beech and Sunner 2004; Hamilton 2003; Videla and Herrera 2005). MIC is generally described in damages to oil and gas pipelines, reservoir tanks and underground structures. MIC occurs as two unrelated types, as proposed by Li et al. (2018a, b). Type I corrosion employs sulfur or nitrate as a final electron acceptor during respiration, usually coupled to the process by extracellular oxidation of the metal. In anaerobic environments there are several acceptors, such as nitrate (Iino et al. 2015), sulfate (Enning et al. 2012, 2014) acetate (Suflita et al. 2008), carbon dioxide (CO2) and hydrogen (H2) (Dinh et al. 2004; Uchiyama et al. 2010). Species of SRBs are not the only ones to cause degradation damage in metal alloys, this group has attracted more attention in MIC studies due to the economic losses caused by its more meticulous mechanisms (Little et al. 1992). The association between SRB and anaerobic corrosion is widely described, mainly through the production of the corrosive chemical H2S (Cord-Ruwisch and Widdel 1986; Costello 1974; Enning et al. 2012) and organic acids (Iverson 1987; Pak et al. 2003). More recently strains of the species Desulfolpia corrodens and Desulfovibrio ferrophilus have been described as being able to reduce sulfate with concomitant iron oxidation, in a kinetically faster process than H2S production (Dinh et al. 2004) (Fig. 2). It seems that this ability is more restricted to the phylum Deltaproteobacteria, in the families Desulfovibrionaceae and Desulfobulbaceae (Enning and Garrelfs 2014).
Corrosion reactions associated with acetate involves the acetyl-CoA reductive biochemical pathway, with the reduction of CO2. The engagement of acetogenic microorganisms in the corrosion of metals in an anoxic environment is related to the low level of environmental sulfur (Suflita et al. 2008). Further evidence of the association between acetogenic bacteria and metal corrosion processes has recently been reported by the bacterium Acetobacterium sp., which was described as using iron as the sole source of electrons (Mand et al. 2014). Large production of ferrous iron in the process of iron oxidation and the concomitant production of acetate was described by Kato et al. (2015), demonstrating the role of Sporomusa sp. in the direct role of metallic iron corrosion.
The participation of methanogenic microorganisms in the corrosion of metals under anaerobic conditions was proposed initially in the 1980s (Boopathy and Daniels 1991; Daniels et al. 1987). The reaction involves the production of methane with the dependence of iron employing CO2 or H2 as the final electron acceptor, during the anaerobic respiration. Methanogenic archaea related to metal corrosion were described later, for example, Methanobacterium sp. IM1 (Dinh et al. 2004), Methanococcus maripaludis KA1 and Mic1c10 (Mori et al. 2010; Uchiyama et al. 2010). These methanogens have been related as a participant in MIC, leading to corrosion pitting of steel pipelines in anoxic environments (Larsen et al. 2010; Usher et al. 2014).
The relationship between iron oxidation and MIC and nitrate-reduction mediated corrosion is a well-known and widely occurring phenomenon in MIC (Iino et al. 2015). During iron corrosion, the microorganisms utilize NO−3 as an oxidizing agent and the acceptor final in an anaerobic environment (Eq. 6) (Kato et al. 2015). Interestingly, in gas and oil reservoirs, nitrate injection used to be applied in the mitigation of corrosion by SRBs. The purposeful addition of NO−3 stimulates the growth of NRBs, which compete with SRB for hydrocarbons. Subsequently, new findings point to the participation of NRB in MICs, with the phylum Bacteroidetes members as active agents, with the isolate Prolixibacter sp. strain MIC-1 described as capable of using metal iron electrons as a donor of electrons (Iino et al. 2015).
Type II is characterized by the secretion of metabolites of the microorganisms involved in the corrosion of the metal, this process is also called Metabolites-MIC (M-MIC), or APB in the case of acid producers, specifically. The APB includes heterotrophic fermenters able to produced organic acids, such as acetic acid, lactic acid, and formic acid. The presence of these acidic compounds occurs in the biofilm, between the biofilm-metal interface (Dong et al. 2018; Xu et al. 2016). Members described as acidophilic bacteria, e.g. the Thiobacillus genus, which can survive in pH near 1 (Peccia et al. 2000). APB members can grow in different conditions, as in the presence and absence of oxygen (Bond et al. 2002), which makes this group relevant concern for corrosion mitigation procedures.
High-through sequencing techniques
With the genome of the Haemophilus influenza bacterium sequenced in 1995 by the Sanger method, the modern bacterial genomic era was inaugurated (Fleischmann et al. 1995). Thus, rapid advances in sequencing techniques allowed the increase of information on genomes of particular bacterial species. After about 10 years later, the advent of techniques known as next-generation technologies or high-through enabled results to be obtained in days or even hours, unlike months or even years of work, when compared with the technique of Sanger. Another significant barrier was the required sequencing cost inputs, and currently, most of the available platforms offer to sequence for about US$ 1.000 per running (Loman et al. 2012).
The large-scale sequencing technologies provided particular techniques for solving large-scale sequencing problems. In spite of the numerous sequencing platforms, e.g. Roche's Platform 454 GS Junior GS FLX+, Ion PGM and Proton from Life Technologies, PacBio RS from Pacific Biosciences and MiSeq and HiSeq from Illumina, present different methodological approaches, it is possible to divide them into two broad groups. The first group that uses the production of libraries of clonally amplified templates, while a second, more recent group, which performs the sequencing from a single molecule (Metzker 2005). Describing in a general way, the first groups of sequencing platforms use three workflow stages. In the first step, the library is assembled or prepared. Here, a limiting step is the amount of biological material required, which may range from a few nanograms to a 10 microg scale, and this 10 µm value may be a step to consider when the sample to be analyzed comes directly from the environment (Hennig et al. 2018). The second stage consists of template amplification, which helps to develop a strong detectable signal in the next stage of sequencing. Such clonal amplification may occur, depending on the sequencing platform, under emulsification PCR (emPCR) conditions, where the beads are encased in aqueous phase microreactors or in bridge PCR when the models are immobilized on a solid surface (Kojima et al. 2005; Shao et al. 2011). Finally, in the last stage, although all platforms described here use a synthesis sequencing project, they show a difference between the chemistry details and the strategy employed to read the sequence. Illumina sequencing employs the reversible termination (RT) approach (Reuter et al. 2015). Differently, the hydrogen ion sequencing performed by 454 and Ion Torrent presents an approach to measure the pH change induced by hydrogen ion release during DNA strand amplification (Eid et al. 2009; Rothberg et al. 2011). On all these available platforms, the most routine sequencing-related errors are base deletion and insertion, and errors can also be produced at 6 bp distant repetitions due to the correlation between the number of incorporated bases and the voltage change (Liu et al. 2012; Rothberg et al. 2011).
The third generation of sequencing known as NGS technology or single molecule real-time (SMRT) has the important advantage of eliminating PCR bias introduced by clonal amplification, employed in the technologies described above. NGS technologies employ the sequencing of a single DNA strand, rather than the sequencing of clonally amplified template, thereby reducing the use of biochemical steps and reagents throughout the process, as well as leading the sequencing process at the nanoscale level. The main platforms are Helicos' Genetics Analysis System, Pacific Biosciences Single Molecular Real-Time and Oxford Nanopore's nanopore sequencing. The first technology developed was Helicos, which presents the facility of not requiring the prior preparation of DNA libraries, consists of sheared and tailed DNA with poly-A (Thompson and Steinmann 2010). The Helicos platform has the advantage of allowing the sequencing of RNA molecules without the previous cDNA conversion since it allows the direct hybridization of the RNA in the flow cell. Nonetheless, this technology has the disadvantage of producing small read lengths, with only 24–70 bases, as well as low data output, about 20 Gb, which entails subsequent time and cost in the production of results (Harris et al. 2008). The Pacific Biosciences sequencing employs DNA polymerase along with a single strand DNA template, which is attached at the bottom of a nanophotonic confinement structure called Zero Mode Waveguides (ZMW) (Eid et al. 2009). The last platform described here is the DNA Sequencing of Nanopore from Oxford Technologies, which employs the approach of a nanopore device with a diameter of 1 ηm, analogous to an electrophoresis method. Similar to the Helicos system, this platform does not use library preparation and is capable of sequencing DNA and RNA molecules directly, without the retrotranscription step (Greninger et al. 2015).
These techniques described above allowed the study of genomes and genes of non-cultivable microorganisms directly from the environment, in the technique called metagenomics. From the shotgun sequencing methods of environmental samples, it is possible to describe the microbial composition present in a taxonomic approach from marker genes identification. For example, microbial amplicons of the 16S rRNA genes enabled the description of the taxonomic profile and functional profiling of complex systems of microbial populations present in unlike samples from natural environmental sites, buildings and the human body (Rondon et al. 2000; Tyson et al. 2004). However, the use of metagenomic sequencing techniques also presents limitations. The sequencing of a sample will present a cross-section of that community at a given time only, regardless of the dynamics that occur over time. Another concern in the use of metagenomics is in the fact that the results on phylogeny are at the species level, while much of the biogeochemical or physiological processes occur at the level of strains. Finally, the number of sequenced samples may not be a faithful representation of the populations present in the environment in question (Gutleben et al. 2018).
Other 'omics' approaches
Other techniques for studying the microbial communities involved in corrosion include the description of messenger RNAs (mRNAs), proteins and metabolites involved in the corrosive biofilm. The survey involving the sequencing of all the RNA produced by the microorganisms present in the environmental sample is called Metatranscriptome. It seems obvious to establish the transcriptional profile with the community physiology participating in corrosive processes. However, it should be considered that not all mRNAs will really be translated into proteins, nor that these proteins are involved in corrosion reactions. Despite these limitations, the use of microbial mRNA has been shown to be useful in describing the microbial activity profile, especially in the biogeochemical cycle, although no studies have been directed to biocorrosion processes (Li et al. 2018a, b; Vigneron 2018). The first challenge of the metatranscriptome technique lies in the fact that most of the RNA is ribosomal RNA (rRNA), generally above 95% of the total RNA. Thus, the mRNA enrichment procedure is required through the depletion of rRNA prior to sequencing (Giannoukos et al. 2012). In addition, it is also routine to convert mRNA into cDNA, the binding of barcoding, which can make the total cost of the processes an impediment to the choice of this strategy. Finally, whereas a combination of metagenome and metatranscriptome allows a prediction of the taxonomic profile, a limitation lies in the presumption of the under-representation of the genes present in the environmental sample, as well as the presence, when not necessary, of the presence of viral genes (Brum et al. 2013).
Measuring the expression of proteins in an environmental sample, called metaproteomics, can bring more real data about the microbial activity. The abundance of proteins present in a given sample can be quantified by means of mass spectrometry methods based on shotgun quantification. From the mass and abundance of peptides, the patterns of these fragments allow for identifying the full-length proteins by homology of sequences deposited in databases. For example, metaproteomic methodologies were applied to describe the influence of temperature gradient over the biofilm physiology in an environmental site (Mosier et al. 2015). This technique has also been in describing marine bacteria adaptation to oligotrophic environments, where there is a nutrient depletion scenario. In relation to biocorrosion there are few studies employing this technique. In a study combining the two techniques of metagenomics and metaproteomics, simulating incoming sulfur compounds deep underground waters, the results from metaproteomic analysis show that even with low microbial diversity there is a possibility of biocorrosion occurring (Bell et al. 2018).
Another technique that makes it possible to study the physiology of the biofilm involved in corrosion processes is the metabolomics. The metabolomic describes the presence of metabolites in the studied microbial community. This technique employs the detection and quantification of metabolites and other small molecules by chromatographic techniques. For this, the high-performance liquid chromatography allows separating the compounds present in the sample, whereas mass spectrometry quantifies and identifies them (Turnbaugh and Gordon 2008). Similar to other techniques described above, the metabolomics also faces several challenges. The metabolite compounds present a wide catalog of characteristics, which may make the analysis more complicated than other classes of molecules, such as proteins and DNA/RNA. Metabolites, besides being made up of several small molecules, may have structures composed of sugars and lipids, as well as complex structures, such as those with antibiotics. Despite this, the metabolomics brings extensive subsidies for the understanding of what occurs during corrosion induced by microorganisms.
A good example of the contribution of metabolomic techniques in understanding biocorrosion can be seen in a survey between two high and low corrosion oil pipeline systems under similar physicochemical conditions (Bonifay et al. 2017). Initially, the anaerobic degradation of hydrocarbons expected in this condition cannot be confirmed by the detection of related genes. However, metabolomic analyses confirmed the presence of expected metabolites for this condition was detected in abundance. The detection of corrosive products produced by Halanaerobium congolense WG strain was enabled by metabolomic techniques (Booker et al. 2019). In this study, conducted in a system simulating under high-pressure subsurface conditions, showed that metabolic shifts were associated with low carbon presence, resulting in the generation of organic acids. Metabolic redirection was also detected in Desulfovibrio vulgaris during biofilm formation and maintenance of carbon steel (Zhang et al. 2016). Compared metabolites between planktonic cells and adhered cells over the carbon steel surface, indicated that D. vulgaris exchanged the central carbon metabolism for 58 different metabolites, especially related to the production of fatty acids.
The metagenomic studies in microbial corrosion
There are still few works on the application of high-performance sequencing techniques in the study of biocorrosion, but the rapid growth in the use of this methodology is notable. The most relevant studies still give a descriptive and punctual look at microbial communities that are influencing the corrosion process on metal surfaces. Nevertheless, when analyzing such data, similarities between the results are evident when environmental conditions are closely comparable, indicating that the metagenomic technique translates the real microbial composition present. Bacteria groups and members of the archaea domain are among the most present in the metagenomic analyzes. Among the bacteria, the phylum Proteobacteria stands out as being described as dominant in several communities present in corrosive biofilms, while among the Archaea representatives the thermophilic and methanogenic strains are prevalent.
Despite all classes of Proteobacteria phylum are related to metal corrosion, there are particular patterns of the presence of the classes according to environmental conditions and the type of metal analyzed. For example, when analyzing Gammaproteobacteria and Deltaproteobacteria classes, the two classes most present in metagenomic analyzes, there is a clear dominance of one group over the other when different environmental samples are analyzed. While species of Gammaproteobacteria class are described mainly in aerobic environments and in the initial processes of formation of biofilms, the members of the class Deltaproteobacteria are detected in more extreme environments, of anoxia and higher temperatures. Species of the Gammaproteobacteria class are described at diverse levels of biofilm formation, even so, they are associated with the beginning of the colonization of the bacteria on the surface of the metal having a role in the formation of the initial biofilm (Table 1). Metagenomic studies always show data that drive the presence of several species in environmental samples associated with metal corrosion. In fact, the analyses of the 16S rRNAs sequenced point to the Gammaproteobacteria as the dominant class in most samples. A comparative study between two particular environmental samples, described as low and high corrosion levels, in oil production pipelines, showed the dominant presence of species of the class Gammaproteobacteria (Bonifay et al. 2017). Studies on bacterial succession over a period of up to 90 days describe high numbers of Operational Taxonomic Unit (OTU) of Gammaproteobacteria in the early stages of biofilm formation on carbon steel surfaces (Moura et al. 2018). Deltaproteobacteria is a class always described in the corrosion of metals under anaerobic conditions. In fact, the preeminent representatives of Deltaproteobateria are the species of the SRB group, such as the genera Desulfovibrio, Desulfobacter, Desulfotignum and Desulforomonas (Li et al. 2017; McBeth and Emerson 2016; Park et al. 2011; Ramírez et al. 2016; Vigneron et al. 2016, 2018) (Table 1). The relative abundance of species of this class involved in corrosion in the marine environment is to be expected since these genera present physiology related to Fe-reduction. The detection techniques of SRBs groups involve, in addition to primes for the 16S rRNA gene, the use of specific primers for dsrAB genes, which encode enzymes involved in the initial iron- and sulfur-reduction reactions. Both approaches show the increase of transcripts of dsrAB and OTUs with the concomitant maturation of the biofilm on metallic surfaces, where there are formations of anoxic niches inside, which allow the growth of SRBs species and the support for the conditions for sulfate and iron reduction (McBeth and Emerson 2016; Vigneron et al. 2016).
Other classes of the phylum Proteobacteria are also identified in biofilms involved in biocorrosion. Alpha-, Beta-, Epsilon- and Zetaproteobacteria were described in several metagenomic studies (Table 1). Species of the class Alphaproteobacteria are more detected in the early stages of the formation of biofilms, along with representatives of the class Gammaproteobacteria. The representatives of species detected in metagenomic analysis are of the orders Rhodobacterales, Rhodopirillales, and Sphigomonadales (Dang et al. 2011; McBeth and Emerson 2016; Moura et al. 2018). Another class, Betaproteobacteria were also described in some corrosion work. One of the explanations of the participation of Betaprobacteria strains lies in the fact that the metabolism of the Burkholderiales and Hydrogenophilales families use hydrogen as an energy source, thus acting in metabolic processes coupled with other species in redox processes (Stöhr et al. 2001). Species of Betaproteobacteria were described as the most abundant bacterial class in all carbon steel samples incubated in groundwater, with emphasis on the oxidative iron species Leptothrix and Sideroxydans (Rajala et al. 2015). The Epsilon- and Zetaproteobacteria classes are usually associated with hydrothermal vents or volcanic environments (Emerson and Moyer 1997; Ruehland and Dubilier 2010). The presence of strains related to corrosion of metallic surfaces has always been underestimated. On the other hand, more recent studies have shown specimens of this class as settlers of metal alloy surfaces (Emerson et al. 2007). Sequences of 16S rRNA genes from bacterial communities influencing the corrosion of carbon steel during a period of up to 7 days allowed the identification of the presence of lithotrophic species Epsilonproteobacteria and Zetaproteobacteria (Dang et al. 2011). The non-identification of these species previously was probably limited by the difficulty of cultivation in laboratories, and this obstacle is overcome by the use of sequencing techniques directly from environmental samples.
Firmicutes are described in metagenomic analyzes in the usual way. High-through approaches indicate the presence of Clostridium and Bacillus genera together with representatives of biofilm-associated Proteobacteria grown on metal alloys (Table 1). In a metagenomic analysis associating the sequencing of 16S rRNA and culture of bacterial isolates from samples grown in oil pipelines, different strains of the Bacillus and Clostridium species, especially Bacillus cereus, were widely identified (Rajasekar et al. 2010). Furthermore, the unusual presence of representatives of the Bacilli class was noted in a metagenomic study of corroded oil pipelines surfaces, of which the total sequence of eubacteria, the Bacilli 16S genes accounted for about 29% (Lenhart et al. 2014). Although these techniques suggest these species play a role in the formation and maintenance of these structures, studies of isolates of Bacillus species in the laboratory show their corrosive action on metallic surfaces (Bonifay et al. 2017; Moura et al. 2018).
The Bacteroidetes phylum, in spite of not being described as having a direct role in the corrosion of metal alloy surfaces, is present in diverse metagenomic studies, especially in marine environments. Its role seems to be mainly related to the formation and maintenance of the structures that make up the biofilm (Dang et al. 2011). The most representative groups of this class described in corrosive biofilms include the orders Flavobacteriales, Bacteroidales and Sphingobacteriales (Table 1). The Bacteroidetes phylum was described in a temporal study of biocorrosion in metal coupons in marine water, which showed the participation of members of the families Flavobacteriacea and Bacteroidales in the initial days of corrosion (Moura et al. 2018). Other recent metagenomic studies have detected the diversity of Bacteroidetes in metal bio-corroding microbiota in seawater environments (Dang et al. 2011; Li et al. 2017; Rajala et al. 2015).
It is curious that despite the fact that species of Archaea are habitual participants in MIC biofilm, especially in conditions of anoxia marine environments, and as co-participants alongside the Deltaproteobacteria class, not many specific studies bring data of archaeas populations participating in metal corrosion. Despite that, when analyzing the results of OTUs of Archaea participating in metal corrosion, the dominance of the Methanobacteria class among the community analyzed is clear. This is in accordance with what has been described above, namely that a corrosion process with concomitant methane production seems to prevail among the metabolism of corrosive Archaea species. Metagenomic techniques, using the MiSeq Illumina and 454 platforms, for analysis of 16S rRNA of the microbial community involved in the corrosion of pipelines confirmed the presence of Archaea representatives, with almost 30 percent of the composition of OTUs, followed by the Deltaproteobacteria group (An et al. 2016). Studies on the composition of the microbial community in a temperature gradient involved in the corrosion of carbon steel used in oil pipelines, also showed the dominance of the methanogenic group (Li et al. 2017). More than half of the species described in the composition of the pipe wall community belong to the family Methanobacteriaceae (Park et al. 2011). In a metagenomic approach to evaluating the microbial succession in offshore oil production facilities, five orders, Methanobacteriales, Methanococcales, Methanomicrobiales, Methanosarcinales and Methanomassiliicoccales, have driven the prevalence of methanogenic strains in corrosion processes (Vigneton et al. 2016). Indeed, such surveys show that methanogenic lineage specimens are regularly described as dominant participants, alongside species of the Deltaproteobacteria class, in the microbiologically influenced corrosion of oil facilities. These conditions of anoxia, high temperatures and the presence of hydrocarbons in these samplings indicate a unique metabolism when we analyze the composition of the microbial community, favoring the colonization and formation of corrosive biofilms on metal surfaces.
Other important considerations
In the face of all the difficulties inherent in micro-organism cultivation in laboratories, at first glance high-throughput amplicon-sequencing proves to be an elegant solution for the analysis of communities from environmental samples. All these technologies bring various limitations and problems together with the promised solutions. The first barrier to be considered lies in procedures well before the analysis of sequences by the above platforms. The collection of environmental samples is a crucial step to be considered. Microbial communities present in soil environments may show heterogeneity in terms of a few meters distance between them, and at a higher level concerning to the depth collected (Certini et al. 2004). The same problem can be described in aquatic environments, where environmental factors such as water flow, depth, luminosity have an enormous impact on the microbial community present. The temporal dimension is also a crucial factor in experimental designed. The frequency and how long the analysis should occur has considerable impacts on results obtained, especially in situations of microbial corrosion, where the change in microbial community profile is closely correlated with the all process.
As the environmental sample is stored it can influence the obtaining of biological material, especially DNA, which can suffer damages or even degradation, and microbial cells that can modify its metabolism or its population profile. Despite that the procedure to freeze specimens in laboratories seems to be the most advisable, continuous thawing/freezing can cause damage to cells and DNA, especially long fragments (Thomson et al. 2010; Todorova et al. 2012). The process of DNA extraction is related to bias and artifacts in sampling. Environmental physical–chemical characteristics can influence the biological material during the extraction method, which can result in inefficient disruption of cells, walls, and membranes, and underestimated the real community profile present in the sample environmental. Another concern is the satisfactory removal of contaminants, such as organic acids and rust, commonly present in biocorrosion studies and that can inhibit the following DNA amplification or cDNA synthesis from environmental RNA. In relation to the amplification of DNA by Polymerase Chain Reaction (PCR) an additional consideration is how much the amplified fragment represents a certain taxonomic species. PCR depends strictly on the region of interest of the DNA that was flanked by the primers. Further analysis of these amplified DNA fragments, by sequencing, for example, may not indicate the correct species or microbial strain. A possible solution would be the use of a mixture of degenerate primers, which could increase the percentage of targeted taxa. But even the solution of using degenerate primers encounters problems, since the increased odds of the amplified DNA fragments, can result in a diversity not corresponding to the real found in the environmental sample. Besides that, frequent errors in DNA amplification, such as nucleotide substitutions, insertions, and deletions, should be considered, especially in a sequencing platform employing initial enrichment steps of the templates.
The final product of the sequencing technologies described above digital data in the form of 16S rRNA gene sequences. These hundreds of thousands of sequences flood the databases, which entails how much they represent the reality of microbial structures analyzed in natural environments. It is assumed that the 16S or 18S gene represents a single species. This brings us to the problematic of the real concept of species. The initial concept proposed by Ernst Mayr in 1994 conceptualizes species as "a group of organisms that maintains similar phenotype characteristics throughout the generations, in spite of the recombination between them" (Dobzhansky and Mayr 1944). However, even if this initial concept evolved from this one, as the molecular concept of > 97% identity, the task of conceptualizing species in groups of bacteria and archaeas still remains arduous. It should be for this reason the choice of many researchers to use the terms OTU rather species, inclusive in diversity measures.
Added to the techniques described above, other methodologies to aid in the description of microbial structures are measures of diversities, especially measures species-based. In the most recent studies, quantitative and qualitative data are prevalent to aggregate information on the ecological foundations of the microbial communities under study (Hugerth and Andersson 2017). Diversity measures can aid understanding in several ways, but some seem to be more preferred among scientists. In particular quantitative and qualitative measures within a single community (alpha diversity) and the partitioning of diversity into two or more communities (beta diversity). The alpha diversity concept was first proposed by Robert Whittaker, together with the terms gamma diversity and beta diversity (Whittaker 1972). In studies on microbial diversity from environmental samples, alpha-diversity generally corresponds to the measure of diversity within a sample or set of replicates at the same site and analysis. The alpha diversity index is a logarithmic serial parameter, where its calculation is the first necessary step for adjusting the number distribution of the "species", or OTUs, in relation to the total number of individuals. Although there is considerable availability of measures available to determine the alpha diversity index, most studies on microbial ecology employ quantitative species-based measures, such as Simpson and Shannon (Shannon and Weaver 1963) and qualitative species-based measures, as Chao 1 or ACE (Chao 1983; Chazdon 1998).
The second metric broadly employed to estimates diversity in microbial communities is beta diversity. This measure describes the degree of dissimilarities or how distinct both are. Thus, in essence, while the alpha diversity describes the characteristic of a defined specific unit, the beta diversity reflects the biotic alteration or the substitution of species in two or more different units. As they occur in measures of alpha diversity, there are many methods to describe beta diversity. But, they are all within three categories. In the first category, the indices describe the differences between two or more units of alpha diversity in relation to total diversity, which is applied as a measure of total species richness. This group generally uses the Whittaker Bw measure (Whittaker 1972). The second category examines differences in species composition between alpha diversity areas, considering complementary or similarity/dissimilarity measures. Jaccard and Bray–Curtis metrics are adopted, which are the most present in environmental microbial diversity studies. Finally, the latter group explores the relationship between species and areas and evaluates the totality related to the accumulation of individuals present in the area.
Conclusions
Currently, the concern with corrosion, especially corrosion influenced by microorganisms, has driven the search for the understanding of the dynamics of the corrosive process, how it is initiated, and which are the effective microorganisms that participate in the corrosion. The limitations of laboratory studies, because they do not faithfully replicate the actual conditions encountered in the environment or simply because of the difficulty of cultivating more corrosive strains, have been promoting for efforts to find technologies that describe the behavior of the microbial community involved in the corrosion of metal surfaces. Large-scale sequencing today does not present the prohibitive costs of before and brings a wealth of biological data in a short period of time. These data allow us to infer the action of representatives of microbial groups in the corrosion processes. Regardless of, all the techniques described above represent the state of the art in the taxonomic and physiological description of microorganisms present in the environmental sample, the improvement of the experimental design still presents a challenge in most of the studied studies, especially in laboratory conditions. Many times, conditions in laboratories, such as microcosms or mesocosms, do not simulate all the factors that occur in real situations in the environment. Throughout the life of metallic structures, these are under conditions of physical–chemical stresses inherent to their use in the industry. Another aspect to be considered would be the in situ approaches, where the collection of environmental samples could express a more realistic scenario of the action of microorganisms in the corrosion, but even this possibility faces challenges. One should carefully consider whether sample collection sites represent the situation that may occur at another location or in a future period. The time scale should also be seriously considered since many times mitigating the corrosion process in the initial stages can reduce costs, downtime and personnel displacement for repairs.
References
An D, Dong X, An A, Park HS, Strous M, Voordouw G (2016) Metagenomic analysis indicates Epsilonproteobacteria as a potential cause of microbial corrosion in pipelines injected with bisulfite. Front Microbiol 28:7–28
Andrews S, Norton I, Salunkhe AS, Goodluck H, Aly WSM, Mourad-Agha H, Cornelis P (2013) Control of iron metabolism in bacteria. In: Banci L (ed) Metallomics and the cell. Springer, Switzerland, pp 203–239
Baeckmann WV (1997) Handbook of cathodic corrosion protection. Elsevier, Amsterdam
Beech IB, Sunner J (2004) Biocorrosion: towards understanding interactions between biofilms and metals. Curr Opin Biotechnol 15:181–186
Beech IB, Sunner JA, Hiraoka K (2005) Microbe-surface interactions in biofouling and biocorrosion processes. Int Microbiol 8:157–168
Beech IB, Sztyler M, Gaylarde CC, Smith WL, Sunner J (2014) Biofilms and biocorrosion. In: Liengen T, Féron D, Basséguy R, Beech IB (eds) Understanding biocorrosion: fundamentals and applications. Elsevier, Amsterdam, pp 33–56
Bell E, Lamminmäki T, Alneberg J, Andersson AF, Qian C, Xiong W, Hettich RL, Balmer L, Frutschi M, Sommer G, Bernier-Latmani R (2018) Biogeochemical cycling by a low-diversity microbial community in deep groundwater. Front Microbiol 9:2129. https://doi.org/10.3389/fmicb.2018.02129
Bond DR, Holmes DE, Tender LM, Lovley DR (2002) Electrode-reducing microorganisms that harvest energy from marine sediments. Science 295:483–485
Bonifay V, Wawrik B, Sunner J, Snodgrass EC, Aydin E, Duncan KE, Callaghan AV, Oldham A, Liengen T, Beech I (2017) Metabolomic and metagenomic analysis of two crude oil production pipelines experiencing differential rates of corrosion. Front Microbiol 8:99. https://doi.org/10.3389/fmicb.2017.00099
Booker AE, Hoyt DW, Meulia T, Eder E, Nicora CD, Purvine SO, Daly RA, Moore JD, Wunch K, Pfiffner SM, Lipton MS, Mouser PJ, Wrighton KC, Wilkins MJ (2019) Deep-subsurface pressure stimulates metabolic plasticity in shale-colonizing Halanaerobium spp. Appl Environ Microbiol. https://doi.org/10.1128/AEM.00018-19
Boopathy R, Daniels L (1991) Effect of pH on anaerobic mild steel corrosion by methanogenic bacteria. Appl Environ Microbiol 57:2104–2108
Booth GH, Tiller AK (1962) Polarization studies of mild steel in cultures of sulphate-reducing bacteria. Part 2. Thermophilic organisms. Transact Faraday Soc 58:110–115
Brum JR, Culley AI, Steward GF (2013) Assembly of a marine viral metagenome after physical fractionation. PLoS ONE 8(4):e60604. https://doi.org/10.1371/journal.pone.0060604
Certini G, Campbell CCD, Edwards AC (2004) Rock fragments in soil support a different microbial community from the fine earth. Soil Biol Biochem 36:1119–1128
Chao A (1983) Non-parametric estimation of the classes in a population. Scand J Stat 11:265–270. https://doi.org/10.2307/4615964
Chazdon RL (1998) A tropical rain forest feast. Trends Ecol Evol 13:421–422
Cord-Ruwisch R, Widdel F (1986) Corroding iron as a hydrogen source for sulfate reduction in growing cultures of sulfate-reducing bacteria. Appl Microbiol Biotechnol 25:169–174
Costello JA (1974) Cathodic depolarization by sulphate-reducing bacteria. S Afr J Sci 70:202–204
Dang H, Chen R, Wang L, Shao S, Dai L, Ye Y, Guo L, Huang G, Klotz M (2011) Molecular characterization of putative biocorroding microbiota with a novel niche detection of Epsilon- and Zetaproteobacteria in Pacific Ocean coastal seawaters. Environ Microbiol 13:3059–3074
Daniels L, Belay N, Rajagopal BS, Weimer PJ (1987) bacterial methanogenesis and growth from CO2 with elemental iron as the sole source of electrons. Science 237:509–511
Dinh HT, Kuever J, Mussmann M, Hassel AW, Stratmann M, Widdel F (2004) Iron corrosion by novel anaerobic microorganisms. Nature 427:829–832
Dobzhansky T, Mayr E (1944) Experiments on sexual isolation in Drosophila: I. Geographic strains of Drosophila willistoni. Proc Natl Acad Sci USA 30:238–344
Dong Y, Jiang B, Xu D, Jiang C, Li Q, Gu T (2018) Severe microbiologically influenced corrosion of S32654 super austenitic stainless steel by acid producing bacterium Acidithiobacillus caldus SM-1. Bioelectrochemistry 123:34–44
Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D et al (2009) Real-time DNA sequencing from single polymerase molecules. Science 323:133–138. https://doi.org/10.1126/science.1162986
Emerson D (2018) The role of iron-oxidizing bacteria in biocorrosion: a review. Biofouling 34:989–1000. https://doi.org/10.1080/08927014.2018.1526281
Emerson D, Moyer C (1997) Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl Environ Microbiol 63:4784–4792
Emerson D, Rentz JA, Lilburn TG, Davis RE, Aldrich H, Chan C, Moyer CL, Reysenbach A-L (2007) A novel lineage of proteobacteria involved in formation of marine Fe-oxidizing microbial mat communities. PLoS ONE 2(8):e667
Enning D, Garrelfs J (2014) Corrosion of iron by sulfate-reducing bacteria: new views of an old problem. Appl Environ Microbiol 80:1226–1236
Enning D, Venzlaff H, Garrelfs J, Dinh HT, Meyer V, Mayrhofer K et al (2012) Marine sulfate-reducing bacteria cause serious corrosion of iron under electroconductive biogenic mineral crust. Environ Microbiol 14:1772–1787
Fleischmann RD, Adams MD, White O, Clayton RA, Kirkness EF, Kerlavage AR, Bult CJ, Tomb JF, Dougherty BA, Merrick JM et al (1995) Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 269:496–512
Gaines HA (1910) Bacterial activity as a corrosion induced in the soil. J Eng Ind Chem 2:128–130
Geesey GG, Beech I, Bremmer PJ, Webster BJ, Wells D (2000) Biocorrosion. In: Bryers JD (ed) Biofilms II: process analysis and applications, Wiley-Liss, Hoboken pp 281–326
Ghiorse WC (1984) Biology of iron- and manganese-depositing bacteria. Annu Rev Microbiol 38:515–550
Giannoukos G, Ciulla DM, Huang K, Haas BJ, Izard J, Levin JZ, Livny J, Earl AM, Gevers D, Ward DV, Nusbaum C, Birren BW, Gnirke A (2012) Efficient and robust RNA-seq process for cultured bacteria and complex community transcriptomes. Genome Biol 13:R23. https://doi.org/10.1186/gb-2012-13-3-r23
Greninger AL, Naccache SN, Federman S, Yu G, Mbala P, Bres V, Stryke D, Bouquet J, Somasekar S, Linnen JM, Dodd R, Mulembakani P, Schneider BS, Muyembe-Tamfum JJ, Stramer SL, Chiu CY (2015) Rapid metagenomic identification of viral pathogens in clinical samples by real-time nanopore sequencing analysis. Genome Med 7:99. https://doi.org/10.1186/s13073-015-0220-9
Gutleben J, Chaib De Mares M, van Elsas JD, Smidt H, Overmann J, Sipkema D (2018) The multi-omics promise in context: from sequence to microbial isolate. Crit Rev Microbiol 44:212–229. https://doi.org/10.1080/1040841X.2017.1332003
Hamilton WA (2003) Microbially influenced corrosion as a model system for the study of metal microbe interactions: a unifying electron transfer hypothesis. Biofouling 19:65–76
Harris TD, Buzby PR, Babcock H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, Dimeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z (2008) Single-molecule DNA sequencing of a viral genome. Science 320:106–109. https://doi.org/10.1126/science.1150427
Hennig BP, Velten L, Racke I, Tu CS, Thoms M, Rybin V, Besir H, Remans K, Steinmetz LM (2018) Scale low-cost NGS library preparation using a robust Tn5 purification and tagmentation protocol. G3 (Bethesda) 8:79–89. https://doi.org/10.1534/g3.117.300257
Hugerth LW, Andersson AF (2017) Analysing microbial community composition through amplicon sequencing: from sampling to hypothesis testing. Front Microbiol 8:1561. https://doi.org/10.3389/fmicb.2017.01561
Iino T, Ito K, Wakai S, Tsurumaru H, Ohkuma M, Harayama S (2015) Iron corrosion induced by nonhydrogenotrophic nitrate-reducing Prolixibacter sp. strain MIC1-1. Appl Environ Microbiol 81:1839–1846. https://doi.org/10.1128/AEM.03741-14
Iverson WP (1966) Direct evidence for the cathodic depolarization theory of bacterial corrosion. Science 151:986–988
Iverson WP (1987) Microbial corrosion of metals. Adv Appl Microbiol 32:1–36
Kaesche H (2003) Corrosion of metals: physicochemical principles and current problems, 1st edn. Springer, Berlin, GM
Kappler A, Newman D (2004) Formation of Fe(III)-minerals by Fe(II)-oxidizing photoautotrophic bacteria. Geochim Cosmochim Acta 68:1217–1226. https://doi.org/10.1016/j.gca.2003.09.006
Kato S, Yumoto I, Kamagata Y (2015) Isolation of acetogenic bacteria that induce biocorrosion by utilizing metallic iron as the sole electron donor. Appl Environ Microbiol 81:67–73. https://doi.org/10.1128/AEM.02767-14
Kip N, Jansen S, Leite MFA, de Hollander M, Afanasyev M, Kuramae EE, Van Veen JA (2017) Methanogens predominate in natural corrosion protective layers on metal sheet piles. Sci Rep 7(1)
Koch GH, Brongers MPH, Thompson NG, Virmani YP, Payer JH (2002) Corrosion cost and preventive strategies in the United States. Federal Highway Administration, Washington, D.C., pp 1–12
Kojima T, Takei Y, Ohtsuka M, Kawarasaki Y, Yamane T, Nakano H (2005) PCR amplification from single DNA molecules on magnetic beads in emulsion: application for high-throughput screening of transcription factor targets. Nucleic Acids Res 33:e150
Larsen J, Kim J R, Rasmussen, Pedersen HS, Sørensen KB, Lundgaard T, Skovhus TL (2010) Consortia of mic bacteria and archaea causing pitting corrosion in top side oil production facilities. NACE—International Corrosion Conference Series, pp 14–18.
Lenhart TR, Duncan KE, Beech IB, Sunner JA, Smith W, Bonifay V, Biri B, Suflita JM (2014) Identification and characterization of microbial biofilm communities associated with corroded oil pipeline surfaces. Biofouling 30:823–835
Li X, Duan J, Xiao H, Li Y, Liu H, Guan F, Zhai X (2017) Analysis of bacterial community composition of corroded steel immersed in Sanya and Xiamen seawaters in China via method of illumina MiSeq sequencing. Front Microbiol 8:1737. https://doi.org/10.3389/fmicb.2017.01737
Li Y, Tang K, Zhang L, Zhao Z, Xie X, Chen CA, Wang D, Jiao N, Zhang Y (2018a) Coupled carbon, sulfur, and nitrogen cycles mediated by microorganisms in the water column of a shallow-water hydrothermal ecosystem. Front Microbiol 9:2718. https://doi.org/10.3389/fmicb.2018.02718
Li Y, Xu D, Chena C, Li X, Jia R, Zhang D, Sand W, Wang F, Gu T (2018b) Anaerobic microbiologically influenced corrosion mechanisms interpreted using bioenergetics and bioelectrochemistry: a review. J Mater Sci Technol 34:1713–1718. https://doi.org/10.1016/j.jmst.2018.02.023
Little B, Lee JS (2007) Microbiologically influenced corrosion, 1st edn. Wiley-Interscienc, New York
Little B, Wagner P, Mansfeld F (1992) An overview of microbiologically influenced corrosion. Electrochim Acta 37:2185–2194
Liu L, Li Y, Li S, Hu N, He Y, Pong R, Lin D, Lu L, Law M (2012) Comparison of next-generation sequencing systems. J Biomed Biotechnol 2012:251364. https://doi.org/10.1155/2012/251364
Loman NJ, Constantinidou C, Chan JZ, Halachev M, Sergeant M, Penn CW, Robinson ER, Pallen MJ (2012) High-throughput bacterial genome sequencing: an embarrassment of choice, a world of opportunity. Nat Rev Microbiol 10:599–606
Lovley DR (1991) Dissimilatory Fe(III) and Mn(IV) reduction. Microbiol Rev 55:259–287
Lugauskas A, Prosyčevas I, Ramanauskas R, Grigucevičienė A, Selskienė A, Pakštas V (2009) The influence of micromycetes on the corrosion behaviour of metals (steel, al) under conditions of the environment polluted with organic substances. Mat Sci 15:224–235
Mand J, Park HS, Jack TR, Voordouw G (2014) The role of acetogens in microbially influenced corrosion of steel. Front Microbiol 5:268
McBeth JM, Emerson D (2016) In situ microbial community succession on mild steel in estuarine and marine environments: exploring the role of iron-oxidizing bacteria. Front Microbiol 7:767. https://doi.org/10.3389/fmicb.2016.00767
Metzker ML (2005) Emerging technologies in DNA sequencing. Genome Res 15:1767–1776
Mori K, Tsurumaru H, Harayama S (2010) Iron corrosion activity of anaerobic hydrogen-consuming microorganisms isolated from oil facilities. J Biosci Bioeng 110:426–430
Mosier AC, Li Z, Thomas BC, Hettich RL, Pan C, Banfield JF (2015) Elevated temperature alters proteomic responses of individual organisms within a biofilm community. ISME J 9:180–194. https://doi.org/10.1038/ismej.2014.113
Moura V, Ribeiro I, Moriggi P, Capão A, Salles C, Bitati S, Procópio L (2018) The influence of surface microbial diversity and succession on microbiologically influenced corrosion of steel in a simulated marine environment. Arch Microbiol 10:1447–1456. https://doi.org/10.1007/s00203-018-1559-2
Pak KR, Lee HJ, Lee HK, Kim YK, Oh YS, Choi SC (2003) Involvement of organic acid during corrosion of iron coupon by Desulfovibrio desulfuricans. J Microbiol Biotechnol 13:937–941
Park HS, Chatterjee I, Dong X, Wang SH, Sensen CW, Caffrey SM, Jack TR, Boivin J, Voordouw G (2011) Effect of sodium bisulfite injection on the microbial community composition in a brackish-water-transporting pipeline. Appl Environ Microbiol 77:6908–6917. https://doi.org/10.1128/AEM.05891-11
Peccia J, Marchand EA, Silverstein J, Hernandez M (2000) Development and application of small-subunit rRNA probes for assessment of selected Thiobacillus species and members of the genus Acidiphilium. Appl Environ Microbiol 66:3065–3072
Price SJ, Figueira RB (2017) Corrosion protection systems and fatigue corrosion in offshore wind structures: current status and future perspectives. Coatings 7:1–51. https://doi.org/10.3390/coatings7020025
Procópio L (2019) The role of biofilms in the corrosion of steel in marine environments. World J Microbiol Biotechnol 35:73. https://doi.org/10.1007/s11274-019-2647-4
Rajala P, Carpén L, Vepsäläinen M, Raulio M, Sohlberg E, Bomberg M (2015) Microbially induced corrosion of carbon steel in deep groundwater environment. Front Microbiol 6:647. https://doi.org/10.3389/fmicb.2015.00647
Rajasekar A, Anandkumar B, Maruthamuthu S, Ting YP, Rahman PK (2010) Characterization of corrosive bacterial consortia isolated from petroleum-product-transporting pipelines. Appl Microbiol Biotechnol 85:1175–1188. https://doi.org/10.1007/s00253-009-2289-9
Ramírez GA, Hoffman CL, Lee MD, Lesniewski RA, Barco RA, Garber A, Toner BM, Wheat CG, Edwards KJ, Orcutt BN (2016) Assessing marine microbial induced corrosion at Santa Catalina Island, California. Front Microbiol 7:1679
Ray RI, Lee JS, Little BJ (2010) Iron-oxidizing bacteria: a review of corrosion mechanisms in fresh water and marine environments. NACE Corrosion 2010 Conference, p 1–19.
Reuter JA, Spacek DV, Snyder MP (2015) High-throughput sequencing technologies. Mol Cell 58:586–597. https://doi.org/10.1016/j.molcel.2015.05.004
Revie RW (2011) Uhlig's corrosion handbook, 3rd edn. Wiley-Interscienc, New Jersey
Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA, MacNeil IA, Minor C, Tiong CL, Gilman M, Osburne MS, Clardy J, Handelsman J, Goodman RM (2000) Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 66:2541–2547
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M, Leamon JH, Johnson K, Milgrew MJ, Edwards M, Hoon J, Simons JF, Marran D, Myers JW, Davidson JF, Branting A, Nobile JR, Puc BP, Light D, Clark TA, Huber M, Branciforte JT, Stoner IB, Cawley SE, Lyons M, Fu Y, Homer N, Sedova M, Miao X, Reed B, Sabina J, Feierstein E, Schorn M, Alanjary M, Dimalanta E, Dressman D, Kasinskas R, Sokolsky T, Fidanza JA, Namsaraev E, McKernan KJ, Williams A, Roth GT, Bustillo J (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. https://doi.org/10.1038/nature10242
Ruehland C, Dubilier N (2010) Gamma- and epsilonproteobacterial ectosymbionts of a shallow-water marine worm are related to deep-sea hydrothermal vent ectosymbionts. Environ Microbiol 2:2312–2326
Shannon CE, Weaver W (1963) The mathematical theory of commuication. University of Illinois Press
Shao K, Ding W, Wang F, Li H, Ma D, Wang H (2011) Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS ONE 6:e24910. https://doi.org/10.1371/journal.pone.0024910
Stöhr R, Waberski A, Liesack W, Völker H, Wehmeyer U, Thomm M (2001) Hydrogenophilus hirschii sp. nov., a novel thermophilic hydrogen-oxidizing beta-proteobacterium isolated from Yellowstone National Park. Int J Syst Evol Microbiol 51:481–488
Suflita JM, Phelps TJ, Little B (2008) Carbon dioxide corrosion and acetate: a hypothesis on the influence of microorganisms. Corros Sci 64:854–859. https://doi.org/10.5006/1.3279919
Tebo BM, Ghiorse WC, Van Waasbergen LG, Siering PL, Caspi R (1997) Bacterially mediated mineral formation: insights into manganese(h) oxidation from molecular genetic and biochemical studies. Rev Miner 35:259–266
Thompson JF, Steinmann KE (2010) Single molecule sequencing with a HeliScope genetic analysis system. Curr Protoc Mol Biol 92(7):10
Thomson LK, Fleming SD, Barone K, Zieschang JA, Clark AM (2010) The effect of repeated freezing and thawing on human sperm DNA fragmentation. Fertil Steril 93:1147–1156. https://doi.org/10.1016/j.fertnstert.2008.11.023
Todorova T, Pesheva M, Stamenova R, Dimitrov M, Venkov P (2012) Mutagenic effect of freezing on nuclear DNA of Saccharomyces cerevisiae. Yeast 29:191–199. https://doi.org/10.1002/yea.2901
Turnbaugh PJ, Gordon JI (2008) An invitation to the marriage of metagenomics and metabolomics. Cell 134:708–713. https://doi.org/10.1016/j.cell.2008.08.025
Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF (2004) Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 428:37–43
Uchiyama T, Ito K, Mori K, Tsurumaru H, Harayama S (2010) Iron-corroding methanogen isolated from a crude-oil storage tank. Appl Environ Microbiol 76(6):1783–1788. https://doi.org/10.1128/AEM.00668-09
Usher KM, Kaksonena AH, MacLeod ID (2014) Marine rust tubercles harbour iron corroding archaea and sulphate reducing bacteria. Corros Sci 83:189–197
Videla HA (1996) Manual of biocorrosion, 1st edn. Lewis Publishers, Florida
Videla HA, Herrera LK (2005) Microbiologically influenced corrosion: looking to the future. Int Microbiol 3:169–180
Vigneron A, Alsop EB, Chambers B, Lomans BP, Head IM, Tsesmetzis N (2016) Complementary microorganisms in highly corrosive biofilms from an offshore oil production facility. Appl Environ Microbiol 82:2545–2554. https://doi.org/10.1128/AEM.03842-15
Vigneron A, Head IM, Tsesmetzis N (2018) Damage to offshore production facilities by corrosive microbial biofilms. Appl Microbiol Biotechnol 102:2525–2533. https://doi.org/10.1007/s00253-018-8808-9
von Wolzogen Kuhr CAH, van der Vlugt LS (1964) The graphitization of cast iron as an electrobiochemical process in anaerobic soils. Water 18:147–165
Whittaker RH (1972) Evolution and measurement of species diversity. Taxon 21:213–225
Xu D, Li Y, Gu T (2016) Mechanistic modeling of biocorrosion caused by biofilms of sulfate reducing bacteria and acid producing bacteria. Bioelectrochemistry 110:52–58. https://doi.org/10.1016/j.bioelechem.2016.03.003
Zhang Y, Pei G, Chen L, Zhang W (2016) Metabolic dynamics of Desulfovibrio vulgaris biofilm grown on a steel surface. Biofouling 32:725–736. https://doi.org/10.1080/08927014.2016.1193166
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Procópio, L. The era of ‘omics’ technologies in the study of microbiologically influenced corrosion. Biotechnol Lett 42, 341–356 (2020). https://doi.org/10.1007/s10529-019-02789-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02789-w