Abstract
Objectives
To characterize a glycosyltransferase (UGT74AN3) from Catharanthus roseus and investigate its specificity toward cardiotonic steroids and phenolic compounds.
Results
UGT74AN3, a novel permissive GT from C. roseus, displayed average high conversion rate (> 90%) toward eight structurally different cardiotonic steroids. Among them, resibufogenin, digitoxigenin, and uzarigenin gave 100% yield. Based on LC–MS, 1H-NMR and 13C-NMR analysis, structure elucidation of eight glycosides was consistent with 3-O-β-d-glucosides. We further confirmed UGT74AN3 was permissive enough to glycosylate curcumin, resveratrol, and phloretin. The cDNA sequence of UGT74AN3 contained an ORF of 1,425 nucleotides encoding 474 amino acids. UGT74AN3 performed the maximum catalytic activity at 40 °C, pH 8.0, and was divalent cation-independent. Km values of UGT74AN3 toward resibufogenin, digitoxigenin, and uzarigenin were 7.0 µM, 12.3 µM, and 17.4 µM, respectively.
Conclusions
UGT74AN3, a glycosyltransferase from a noncardenolide-producing plant, displayed catalytic efficiency toward cardiotonic steroids and phenolic compounds, which would make it feasible for glycosylation of bioactive molecules.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
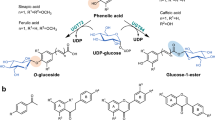
Cardiotonic steroids (CTS) have long been used as cardiac agents for centuries and served as promising agents for the treatment of cancer (Menger et al. 2013). However, serious cardiovascular toxicity, low bioavailability, and poor water solubility limit their wider application for the treatment of other diseases (Gantt et al. 2011). C3 glycosylation of the steroidal core has been recognized as a practical way to improve their therapeutic index, which can improve the solubility, pharmacokinetics, and pharmacodynamics of drug candidates (Iyer et al. 2010).
Chemical synthesis of cardiotonic glycosides is faced with poor regio- and stereoselectivities, the requirement of hazardous and expensive chemical derivatives for protection and deprotection of functional groups (Beale and Taylor 2013). Instead, glycosyltransferases (GTs) are powerful synthetic tools which can alleviate these disadvantages (Gantt et al. 2011). However, the application of microbial GTs for C3 glycosylation of CTS was generally hindered by low efficiency and limited regiospecificity (Li et al. 2017; Zhou et al. 2012).
In comparison, plant GTs have advantages in glycosylating botanical natural products (Xie et al. 2014). In previous work, we reported the identification of UGT74AN1 for CTS glycosylation from Asclepias curassavica, a cardenolide-producing plant from Asclepiadaceae family (Wen et al. 2018). Interestingly, the plant cell suspension cultures of several noncardenolide-producing species such as Catharanthus roseus, have been used for biotransformation of cinobufagin to yield cinobufagin 3-O-β-d-glucoside, which imply the existence of permissive but regiospecific GTs (Ye et al. 2002). As several multi-functional GTs have been purified or cloned from C. roseus (Kaminaga et al. 2004; Masada et al. 2009; Oguchi et al. 2007; Piovan et al. 2010), it is of great interest in further mining of permissive GTs from C. roseus for natural product glycodiversification. The aim of this study was to identify the specific GTs and to evaluate their catalytic efficiency and synthetic utility in vitro.
Herein, a novel permissive GT from C. roseus, named UGT74AN3, was cloned and characterized. UGT74AN3 displayed catalytic efficiency and regiospecificity toward cardenolide and bufadienolide aglycons to form 3-O-β-d-glucosides, which makes it to be the first identified GT from a noncardenolide-producing plant for CTS 3-O-glycosylation. These results suggest that UGT74AN3 could serve as a powerful enzymatic tool for glycosylation of CTS and may have a good potential for application in glycosylation of drug-like scaffolds.
Materials and methods
Plant material and chemicals
The wildly grown C. roseus (L.) was collected from Guangzhou city, Guangdong province and cultivated in college of pharmacy, Jinan University. All chemicals and reagents were purchased from Sigma-Aldrich, J & K Scientific Ltd. and Baoji Chenguang Biotech Co., Ltd., unless otherwise stated.
Cloning of candidate CrGTs
Total RNA was extracted from the leaf tissues of C. roseus and reverse transcribed into cDNA using PrimeScript™ RT reagent Kit (TaKaRa). The obtained cDNA was used as template for PCR of candidate CrGTs. The primers were designed based on the candidate genes that generated by BLAST search of Transcriptome Shotgun Assembly (TSA) database of C. roseus (Genbank No. AYC35244) (Supplementary Table 1). The PCR product was subcloned into pET28a vector using In-fusion cloning Kit (TaKaRa).
Expression and purification of candidate CrGTs
Each recombinant CrGT-pET28a plasmid was transferred into Transetta (DE3) Escherichia coli for expression. As OD600 reached 0.4–0.6, IPTG was subsequently added to a final concentration of 0.25 mM and the cells were grown for 16 h at 18 °C and 180 rpm. After harvesting, the cells were resuspended in 10 mL chilled lysis buffer (20 mM phosphate buffer, 50 mM NaCl, 10 mM imidazole, pH 7.5) and disrupted by sonication. The recombinant proteins were purified using 5 mL high affinity Ni–NTA resin. Protein purity was analyzed by 12% SDS-PAGE and protein concentration was determined by the Bradford method.
Characterization of the recombinant enzymes
Enzyme activity assays were conducted in a final reaction volume of 200 µL consisting of 50 mM Tris/HCl (pH 7.5), 500 μM UDP-glucose, 200 μM sugar acceptor, 5 mM MgCl2, and 500 μg of purified CrGTs. Two separate control reactions that withheld either enzyme or UDP-glucose were conducted in parallel. After incubation at 37 °C for 12 h, all the reactions were quenched with 200 µL methanol and analyzed by analytical reverse-phase HPLC equipped with an Polar RP C18 Column (Welch). The analytes were eluted by the mobile phase consisting of eluent A (1% formic acid) and eluent B (100% acetonitrile) using a gradient program: 0–20 min, 10–75% B; 20–22 min, 75–100% B; 22–27 min, 100% B. The reactions which displayed potential new products via HPLC were further confirmed by LC–MS. Percent conversions were determined by HPLC and calculated by dividing the integrated area of glycosylated product by the total peak area of glycosylated product and remaining substrate.
Phylogenetic analysis
For the phylogenetic analysis, the full-length amino acid sequences of UGT74AN3 were aligned with glycosyltransferases from other species using ClustalW. The resulting alignment was used to construct an unrooted phylogenetic tree using the neighbor-joining method in the MEGA 7.0 package. One thousand bootstrapped datasets were used to estimate the confidence of each tree clade.
Effects of pH, temperature, divalent metal ions and reaction time
To examine the optimal pH value for enzyme activity, the reactions were performed in different reaction buffers with pH values ranged from 4.0 to 6.0 (Citric acid/sodium citrate buffer) and 7.0 to 11.0 (Tris/HCl buffer). To test the optimal reaction temperature for enzyme activity, the reactions were incubated at various temperatures ranging from 25 to 60 °C. To study the effects of divalent metal ions on enzyme activities, CaCl2, CuCl2, MgCl2, MnCl2, NiCl2, PbCl2, ZnCl2 and EDTA were dissolved in reaction mixtures to a final concentration of 5 mM. To investigate the reaction time, typically 12 time points between 5 and 720 min were studied. All assays were performed in triplicate using 50 μg purified UGT74AN3 as the catalyst, UDP-glucose as the donor and resibufogenin as the acceptor, and each reaction was terminated with methanol and centrifuged for HPLC analysis. All experiments were performed in triplicate. One unit of the enzyme was defined as the quantity that produced 1 μmol product per minute under the optimum conditions (40 °C, pH 8.0, and 5 mM Mg2+).
Determination of kinetic parameters
For kinetic studies, assays were performed in a final volume of 100 µL, contained 50 mM Tris–HCl (pH 8.0), 5 mM MgCl2, 1 mM of saturating UDP-glucose and varying concentration (5–100 µM) of resibufogenin, digitoxigenin, and uzarigenin. The reactions were quenched with an equal volume of methanol and centrifuged at 13,500 g for 30 min. Supernatants were collected and analyzed by reverse-phase HPLC. Three parallel assays were performed routinely. The value of Km was calculated by using Lineweaver–Burk plot method.
Preparative-scale reactions
The reactions mixture consisted of 15 µmol substrate, 50 µmol UDP-glucose, 10 mg purified UGT74AN3 in 10 mL Tris–HCl (50 mM, pH 8.0). The reactions were gently agitated at the optimum conditions. The glycosylated products were separated by reversed-phase semi-preparative HPLC and characterized by HR-ESI–MS and NMR.
Results
Functional identification of UGT74AN3
A search of the transcriptome shotgun assembly (TSA) database of C. roseus (Genbank No. PRJNA358259) was performed by using the amino acid sequences of UGT74AN1 as a query and produced several putative candidate genes. These candidate genes shared greater than 40% sequence identity with sequences of UGT74AN1 and were tentatively named as CrGT1, CrGT2, CrGT3, CrGT4, and CrGT5, respectively (Supplementary Fig. 1). The full length cDNAs of five CrGTs were cloned and expressed in E. coli. To isolate the specific GT for CTS glycosylation, the detecting reactions were performed using resibufogenin as the sugar acceptor and the recombinant CrGTs as the catalyst. Of the five recombinant CrGTs, only CrGT1 showed glycosylation activity toward resibufogenin with high yields (> 90%) (Supplementary Fig. 2). Based on LC–MS, 1H NMR, and 13C NMR spectroscopic analysis, the structure of this glycoside was consistent with the resibufogenin-3-O-β-d-glucoside.
The cDNA sequence of CrGT1 contained an ORF of 1,425 nucleotides (GenBank Accession No. MF942418) encoding a protein of 474 amino acids. CrGT1 used to be named as CrUGT9 by other researchers, but its catalytic activity remains unknown (Miettinen et al. 2014). In this paper, CrGT1 was further named as UGT74AN3 by the UGT Naming Committee (https://prime.vetmed.wsu.edu/resources/udp-glucuronsyltransferase-homepage) and showed the 59.92% amino acid sequence identity to UGT74AN1. A phylogenetic tree was constructed and revealed UGT74AN3 was clustered into a separate clade with UGT74AN1, UGT74AN2, UGT74AC1, and UGT74AE2 (Fig. 1). Among them, UGT74AN1 is a cardiotonic 3-O-GT from A. curassavica. UGT74AN2 is a predicted GT from Calotropis gigantea; UGT74AC1 is a mogrol 3-O-GT from Siraitia grosvenorii (Dai et al. 2015); UGT74AE2 is a protopanaxadiol 3-O-GT from Panax ginseng (Jung et al. 2014).
Exploring the catalytic specificity of UGT74AN3
To study the catalytic promiscuity, regio- and stereospecificity of UGT74AN3 for CTS glycosylation, eight representative bufadienolide and cardenolide aglycons were selected as substrates, including resibufogenin (1), cinobufagenin (2), bufarenogin (3), gamabufotalin (4), 14β,16β-dihydroxy-5α-bufa-20,22-dienolide (5), digitoxigenin (6), uzarigenin (7), and digoxigenin (8) (Fig. 2). The glycosylated products were initially analyzed by HPLC and LC–MS. Comparing with the control reactions, the HPLC chromatograms of each compounds displayed only one new peak at different retention time. UGT74AN3 showed average high catalytic activity (> 90%) toward each tested CTS (Table 1, Supplementary Figs. 2 and 3). Among them, Compound 1, 6, 7 gave 100% yield. To elucidate the structures of glucosylated products, a total of eight products were isolated from the preparative-scale reactions. HPLC, LC–MS, and NMR spectroscopic data analysis confirmed that the sugar side chain of each of the compounds was linked at the C3 position of steroid core (Supplementary Sect. 1; Supplementary Figs. 2–4). The monoglucosylated product of compounds 1–8a were resibufogenin 3-O-β-d-glucoside, cinobufotalin 3-O-β-d-glucoside, bufarenogin 3-O-β-d-glucoside, gamabufotalin 3-O-β-d-glucoside,14β,16β-dihydroxy-3β-(β-d-glucopyranosyloxy)-5α-bufa-20,22-dienolide, digitoxigenin 3-O-β-d-glucoside, uzarigenin 3-O-β-d-glucoside, digoxigenin 3-O-β-d-glucoside, respectively.
Structures of cardiotonic steroids (1–11) and corresponding glucosylated products (1–8a). Compound identification: 1, resibufogenin; 2, cinobufagenin; 3, bufarenogin; 4, gamabufotalin; 5, 14β,16β-dihydroxy-5α-bufa-20,22-dienolide; 6, digitoxigenin; 7, uzarigenin; 8, digoxigenin; 9, curcumin; 10, resveratrol; 11, phloretin
To access the catalytic ability of UGT74AN3 toward other scaffolds, several polyhydroxy compounds such as curcumin (9), resveratrol (10), and phloretin (11), were tested as sugar acceptors. Based on HPLC and LC–MS analysis, high conversion rates and multi-products were observed for three compounds (Table 1, Supplementary Fig. 4). Among them, phloretin gave as many as five products.
To test the sugar donor specificity of UGT74AN3, UDP-glucose, UDP-galactose, UDP-glucuronic acid, UDP-rhamnose, UDP-xylose, were used sugar donors and resibufogenin was used as the sugar acceptor. The results showed UGT74AN3 exclusively selected UDP-glucose as the sugar donor. No product was observed when other activated sugars were tested (data not shown).
Biochemical characterization of UGT74AN3
To study the biochemical properties and dynamic parameters, the recombinant His6-UGT74AN3 was expressed in a large-scale culture and purified to near homogeneity by His-tag affinity chromatography (Fig. 3). By using resibufogenin as a sugar acceptor and UDP-glucose as a donor, the purified UGT74AN3 was found to perform the maximum conversion rates at 40 °C, pH 8.0, and was divalent cation-independent (Supplementary Fig. 5). Specific activities of UGT74AN3 for glucosylation of compound 1, 6, and 7 were 11.2 mU mg−1, 32.6 mU mg−1, and 21.3 mU mg−1, respectively. Km values of UGT74AN3 toward compound 1, 6, and 7 were found to be 7.0 µM, 12.3 µM, and 17.4 µM, respectively (Supplementary Fig. 6), and the corresponding kcat/Km values were found to be 641.4 M−1 s−1, 752.3 M−1 s−1, and 446.0 M−1 s−1, respectively. For preparative synthesis of cardiotonic glycosides, each reaction achieved the maximum yield within 2 h. The productivities for compound 1, 6, and 7 were 5.88 g L−1 d−1, 6.36 g L−1 d−1, and 7.8 g L−1 d−1, respectively.
Discussion
To date, glycosyltransferases (GTs) have been classified into 105 families (GT1-GT105, https://www.cazy.org/GlycosylTransferases). The family 1 GTs constitute the maximum number of GT candidates in plants, which catalyze the glycosylation of secondary metabolites such as terpenoids, alkaloids, flavonoids, phenylpropanoids, and lignans (Tiwari et al. 2016). Plant GTs have a promising role in glycodiversification of drug-like compounds for both in vitro and in vivo use (Xie et al. 2014). However, there are still few reports about plant GTs which are capable of glycosylating CTS. Herein, we report the identification of a novel permissive GT, named UGT74AN3, which displayed average high conversion rates (> 90%) toward 8 structurally different CTS, including 5α and 5β cardenolide or bufadienolide aglycons (Table 1). Like UGT74AN1, UGT74AN3 also exhibited identical catalytic activity toward bufadienolide or cardenolide aglycons with cis or trans fused A/B ring juncture. Besides, UGT74AN3 specifically generated 3-O-glucosides, when polyhydroxy steroids were used as substrates, such as bufarenogin, gamabufotalin, and digoxigenin. These results confirmed UGT74AN3 can be used as a powerful synthetic tool for the regiospecific CTS 3-O-glycosylation. In fact, the characterized plant sterol glycosyltransferases (SGTs) are also regiospecific enzymes. For example, four tomato SGTs, SlSGT1-4, catalyze the glycosylation of the free hydroxyl group at C3 position of sterols (Ramirez-Estrada et al. 2017). DzS3GT is a SGT catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis (Ye et al. 2017). The majority of identified SGTs are insoluble membrane bound enzymes, while UGT74AN3 is a soluble protein, which makes it suitable for industrial applications.
Phylogenetic analysis showed UGT74AN3 was clustered into a separate clade with UGT74AN1, UGT74AC1 and UGT74AE2 (Fig. 1). The activity of UGT74AN3 was consistent with its phylogenetic grouping, as UGT74AN1, UGT74AC1 and UGT74AE2 were regiospecific 3-O-GTs that recognized steroid-like compounds with 6/6/6/5 fused-ring skeleton. Km values of UGT74AN3 toward resibufogenin, digitoxigenin, and uzarigenin were found to be 7.0 µM, 12.3 µM, and 17.4 µM, respectively, which were comparable with that of UGT74AN1 (Supplementary Fig. 6). In comparison, the Km value of UGT74AC1 for mogrol and UGT74AE2 for protopanaxadiol is 41 µM and 25 µM, respectively (Dai et al. 2015; Jung et al. 2014). Besides, kinetic data also suggest that the catalytic efficiency of UGT74AN3 was higher than the other two GTs. For example, the kcat/Km value of UGT74AN3 for resibufogenin was 641.4 M−1 s−1, which was much higher than that of UGT74AE2 for protopanaxadiol (2.24 M−1 s−1).
However, it is important to know that C. roseus is a noncardenolide-producing plant, which means CTS are endogenous substrates for UGT74AN3. Further experiments revealed UGT74AN3 was flexible enough to glycosylate other scaffolds, including curcumin, resveratrol, and phloretin. Unlike CTS, each polyhydroxy compound led to multiple products (Supplementary Fig. 4). This indicates that UGT74AN3 is a multi-functional GT with broad substrate spectra. In previous reports, several multi-functional GTs have been characterized from C. roseus. For example, CaUGT2 can be used for in vitro production of curcumin glucoside (Kaminaga et al. 2004). CaUGT3 is a permissive flavonoid glucoside 1,6-glucosyltransferase with the ability to catalyze various monoglucosides (Masada et al. 2009). One possible reason is that the broad substrate specificity of GTs is a proposed mechanism that plants use to detoxify endogenous and exogenous substances (Brazier-Hicks et al. 2007; Tiwari et al. 2016). Moreover, given the established substrate promiscuity, UGT74AN3 can be applied to glycodiversification of bioactive molecules in drug discovery.
Conclusion
UGT74AN3, a novel permissive glycosyltransferase from C. roseus, displayed robust capabilities for the regiospecific C3 glycosylation of cardenolide and bufadienolide aglycons to form 3-O-β-D-glucosides. To the best of our knowledge, it is the first identified plant GT for cardiotonic steroid 3-O-glycosylation from a noncardenolide-producing plant. Notablely, UGT74AN3 was permissive enough to catalyze 5α and 5β cardenolide or bufadienolide aglycons, and even accept structurally different drug-like compounds. The space–time yields for resibufogenin, digitoxigenin, and uzarigenin were 5.88 g L−1 d−1, 6.36 g L−1 d−1, and 7.8 g L−1 d−1, respectively. Therefore, UGT74AN3 could serve as a powerful regiospecific catalyst for glycosylation of cardiotonic steroids and may be considered as a promising tool for application in glycosylation of drug-like scaffolds.
References
Beale TM, Taylor MS (2013) Synthesis of cardiac glycoside analogs by catalyst-controlled, regioselective glycosylation of digitoxin. Org Lett 15:1358–1361
Brazier-Hicks M, Edwards LA, Edwards R (2007) Selection of plants for roles in phytoremediation: the importance of glucosylation. Plant Biotechnol J 5:627–635
Dai L, Liu C, Zhu Y, Zhang J, Men Y, Zeng Y, Sun Y (2015) Functional characterization of cucurbitadienol synthase and triterpene glycosyltransferase involved in biosynthesis of mogrosides from Siraitia grosvenorii. Plant Cell Physiol 56:1172–1182
Gantt RW, Peltier-Pain P, Thorson JS (2011) Enzymatic methods for glyco(diversification/randomization) of drugs and small molecules. Nat Prod Rep 28:1811–1853
Iyer AK, Zhou M, Azad N, Elbaz H, Wang L, Rogalsky DK, Rojanasakul Y, O'Doherty GA, Langenhan JM (2010) A direct comparison of the anticancer activities of digitoxin MeON-neoglycosides and O-glycosides: oligosaccharide chain length-dependent induction of caspase-9-mediated apoptosis. ACS Med Chem Lett 1:326–330
Jung SC, Kim W, Park SC, Jeong J, Park MK, Lim S, Lee Y, Im WT, Lee JH, Choi G, Kim SC (2014) Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol 55:2177–2188
Kaminaga Y, Sahin FP, Mizukami H (2004) Molecular cloning and characterization of a glucosyltransferase catalyzing glucosylation of curcumin in cultured Catharanthus roseus cells. FEBS Lett 567:197–202
Li K, Feng J, Kuang Y, Song W, Zhang M, Ji S, Qiao X, Ye M (2017) Enzymatic synthesis of bufadienolide O-glycosides as potent antitumor agents using a microbial glycosyltransferase. Adv Synth Catal 359:3765–3772
Masada S, Terasaka K, Oguchi Y, Okazaki S, Mizushima T, Mizukami H (2009) Functional and structural characterization of a flavonoid glucoside 1,6-glucosyltransferase from Catharanthus roseus. Plant Cell Physiol 50:1401–1415
Menger L, Vacchelli E, Kepp O, Eggermont A, Tartour E, Zitvogel L, Kroemer G, Galluzzi L (2013) Trial watch: cardiac glycosides and cancer therapy. Oncoimmunology 2:e23082
Miettinen K, Dong L, Navrot N, Schneider T et al (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5:3606
Oguchi Y, Masada S, Kondo T, Terasaka K, Mizukami H (2007) Purification and characterization of UDP-glucose: curcumin glucoside 1,6-glucosyltransferase from Catharanthus roseus cell suspension cultures. Plant Cell Physiol 48:1635–1643
Piovan A, Cozza G, Caniato R, Moro S, Filippini R (2010) A novel glucosyltransferase from Catharanthus roseus cell suspensions. Process Biochem 45:655–659
Ramirez-Estrada K, Castillo N, Lara JA, Arró M, Boronat A, Ferrer A, Altabella T (2017) Tomato UDP-glucose sterol glycosyltransferases: a family of developmental and stress regulated genes that encode cytosolic and membrane-associated forms of the enzyme. Front Plant Sci 8:984
Tiwari P, Sangwan RS, Sangwan NS (2016) Plant secondary metabolism linked glycosyltransferases: an update on expanding knowledge and scopes. Biotechnol Adv 34:714–739
Wen C, Huang W, Zhu X, Li X, Zhang F, Jiang R (2018) UGT74AN1, a permissive glycosyltransferase from Asclepias curassavica for the regiospecific steroid 3-O-glycosylation. Org lett 20:534–537
Xie K, Chen R, Li J, Wang R, Chen D, Dou X, Dai J (2014) Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Org Lett 16:4874–4877
Ye M, Dai J, Guo H, Cui Y, Guo D (2002) Glucosylation of cinobufagin by cultured suspension cells of Catharanthus roseus. Tetrahedron Lett 43:8535–8538
Ye T, Song W, Zhang J-J, An M, Feng S, Yan S, Li J (2017) Identification and functional characterization of DzS3GT, a cytoplasmic glycosyltransferase catalyzing biosynthesis of diosgenin 3-O-glucoside in Dioscorea zingiberensis. Plant Cell Tiss Org 129:399–410
Zhou M, Hou Y, Hamza A, Pain C, Zhan CG, Bugni TS, Thorson JS (2012) Probing the regiospecificity of enzyme-catalyzed steroid glycosylation. Org Lett 14:5424–5427
Acknowledgements
The study was financially supported by the Doctor Fundation of Jinggangshan University, China (No. JZB1821).
Supporting information
Section 1—MS, 1H NMR, 13C NMR data of compound 1a–8a.
Supplementary Table 1—The Genebank accession numbers and the primers of CrGT1-5.
Supplementary Table 2—Genebank accession numbers of the GTs used for phylogenetic analysis.
Supplementary Fig. 1—Alignment of five glycosyltransferases from Catharanthus roseus with UGT74AN1.
Supplementary Fig. 2—HPLC chromatogram of UGT74AN3 enzymatic products of bufadienolide aglycons.
Supplementary Fig. 3—HPLC chromatogram of UGT74AN3 enzymatic products of cardenolides aglycons.
Supplementary Fig. 4—HPLC chromatogram of UGT74AN3 enzymatic products of curcumin (9), resveratrol (10), and phloretin (11).
Supplementary Fig. 5—Effects of temperature (A), pH (B), divalent metal ions (C) and reaction time (D) on the activity of UGT74AN3.
Supplementary Fig. 6—Determination of kinetic parameters for UGT74AN3.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wen, C., Huang, W., He, MM. et al. Cloning and characterization of a glycosyltransferase from Catharanthus roseus for glycosylation of cardiotonic steroids and phenolic compounds. Biotechnol Lett 42, 135–142 (2020). https://doi.org/10.1007/s10529-019-02756-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10529-019-02756-5