Abstract
Glycosylflavonoids are a class of natural products with multiple pharmacological activities and a lot of glycosyltransferases from various plant species have been reported that they were involved in the biosynthesis of these phytochemicals. However, no corresponding glycosyltransferase has been identified from the famous horticultural and medicinal plant Iris tectorum Maxim. Here, UGT73CD1, a novel glycosyltransferase, was identified from I. tectorum. based on transcriptome analysis and functional identification. Phylogenetic analysis revealed that UGT73CD1 grouped into the clade of flavonoid 7-OH OGTs. Biochemical analysis showed that UGT73CD1 was able to glycosylate tectorigenin at 7-OH to produce tectoridin, and thus assigned as a 7-O-glycosyltransferase. In addition, it also possessed robust catalytic promiscuity toward 12 structurally diverse flavonoid scaffolds and 3, 4-dichloroaniline, resulting in forming O- and N-glycosides. This work will provide insights into efficient biosynthesis of structurally diverse flavonoid glycosides for drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Flavonoids are a versatile class of phenolic metabolites with multiple pharmacological activities in plants [1]. These natural products exhibit diverse bioactivities and confer considerable health benefits, such as antioxidant, anti-inflammatory, anti-cancer, anti-obesity, anti-diabetes, and anti-carcinogenic activities [2, 3]. Often, they are present in the form of free aglycones and glycosides [1]. Compared with free aglycones, sugar moieties attached to flavonoids are crucial for the structural diversity of plant secondary metabolites, and remarkably affect the bioactivities of these phytochemicals [4]. Glycosylflavonoids can be categorized into the O-glycosidic derivatives and C-glycosidic derivatives [5], while the former is more abundant in plants. Previous researches revealed that the stability of natural products benefit from chemical modifications in plants, such as methylation, acylation and glycosylation, eventually leading up to the formation of secondary metabolites [6, 7]. In addition, glycosylation can effectively enhanced water solubility of natural products as well as bioavailability, pharmacological properties [8].
The vast majority of glycosylation reactions are mediated by glycosyltransferase (GT; EC 2.4.x.y.) [9]. These enzymes are grouped into 114 families based on sequence similarity and collected in the Carbohydrate-Active Enzymes (CAZy) database (http://www.cazy.org//GlycosylTransferases.html). In plants, flavonoid 7-O-glycosyltransferases (7GTs) belong to the GT superfamily 1 (UGT) that is the largest multigene GT family, using uridine 5’-diphosphate (UDP) sugar as glycosyl donors. UGTs are characterized by C-terminus signature motif that is composed of a highly conserved 44-amino-acid residues, and the motif used to be called Plant Secondary Product Glycosyltransferase (PSPG) box. Plenty of 7GTs have been reported in a wide range of plant species, the overwhelming majority of which were from dicotyledon. For example, UGT73C6 and UGT89C1 were identified from Arabidopsis thaliana, a famous model plant, and led up to the formation of flavonoid glycosides and flavonoid rhamnosides respectively [10, 11]. Moreover, the canonical WsGT and UGT76F1 were sporadically distributed in other plant families, including Solanaceae and Rutaceae, displaying robust catalytic activity to structurally diverse substrates, such as flavones, flavonols, isoflavones [12, 13]. In contrast, with isoflavones as major glycosylated substrates, a considerable number of 7GTs have been reported in Leguminosae family plants, such as Pueraria lobata, Glycine max [14, 15]. It should be particularly noted that the identified 7GTs were rare in monocotyledon, and they were mainly found in Oryza sativa. For instance, UGT706D1 could catalyze flavones at their 7-OH group and thus was characterized as a 7GT in Oryza sativa [16], while UGT706C2 could catalyze flavones and flavanones [17]. However, few 7GTs have been identified in I. tectorum, a monocot Iridaceae family plant. Moreover, the substrate promiscuity of most known flavonoids glycosyltransferases are not as broad as that of UGT73AE1 [18], which would hinder the availability of bioactive natural product glycosides. Hence, it is of great practical significance to further discover 7GTs with catalytic promiscuity from I. tectorum.
I. tectorum, known as a horticultural plant, is also a famous traditional medicinal plant in China. As a traditional Chinese medicine, whose dried rhizome referred to as “Chuan She Gan”, have been widely used to treat inflammation, pharyngitis and cough for thousands of years [19]. An abundant of bioactive ingredients have been reported in I. tectorum, including flavonoids, terpenoids, and phenolics [20, 21]. Among these components, flavonoids, particularly isoflavones, are deemed to be the main compound. Previous studies have indicated that isoflavone glycosides, the most important bioactive component, determines the quality of medicinal materials [22]. As the main bioactive isoflavone 7-O-glycoside, abundant tectoridins have been found in I. tectorum, indicating that 7GTs should be involved in the biosynthesis of tectoridin in this plant. However, 7GTs in I. tectorum have not yet been identified. With the aim to character the target 7GT enzyme, de novo transcriptome of different organs of I. tectorum was investigated, and ItUGT700, a candidate gene, had been screened based on a BLAST search. By heterologous expression and enzymatic assays, a 7GT was identified as a multifunctional glycosyltransferase that could catalyze O-, N-glycosylation reactions. This finding paves the way for the biosynthesis of structurally diverse flavonoid glycosides in monocot Iridaceae family plants.
Materials and Methods
Chemicals and Materials
Substrates including tectorigenin (1), formononetin (2), genistein (3), daidzein (4), apigenin (5), luteolin (6), naringenin (7), eriodictyol (8), hesperetin (9), kaempferol (10), isorhamnetin (11), 3,4-dichloroaniline (13), 3,4-dichlorobenzenethiol (14), emodin (15), tectoridin (16), puerarin (17), vitexin (18), orientin (19), and mangiferin (20) were purchased from Sichuan Weikeqi Bio-Technology CO., LTD. (Chengdu, China) and Chengdu Must Bio-Technology Co., Ltd (Chengdu, China) while the compound phloretin (12) as a gift was obtained from Dr. Peng Wu, Guangzhou University of Chinese Medicine. UDP-glucose was purchased from Tianjin Heowns Bio-Technology Co., Ltd. (Tianjin, China). Analytical or High-Performance Liquid Chromatography (HPLC) grade solvents were obtained from Thermo Fisher Scientific Co., Ltd. (Shanghai, China). Unless otherwise stated, all other chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO, USA) and Shanghai Sangon Bio-Technology CO., LTD. (Shanghai, China).
Plant Materials, RNA Isolation, Transcriptome Sequencing and Transcriptome Assembly
I. tectorum, plants were collected from the Institute of Botany, Chinese Academy of Sciences in Beijing and they were cultivated in the Guangzhou University of Chinese Medicine. Total RNA was extracted from the rhizomes, leaves and flowers of I. tectorum, using the Quick RNA Isolation Kit (Waryong Biotech., Beijing, China). All samples were performed in triplicate. The transcriptome sequencing was performed by Illumina HiSeq™ 2000 (Biomarker Technologies, Beijing, China). Transcriptome assembly was conducted using Trinity 2 with min_kmer_cov set to 2 by default and all other parameters were set by default.
Phylogenetic Analysis
The diverse UGTs protein sequences from plants and microorganism were aligned by Clustal W. A Neighbor-Joining phylogenetic tree was then constructed by MEGA 7.0 software.
Molecular Cloning of UGT73CD1
The total RNA of the rhizomes of I. tectorum was extracted using the Quick RNA Isolation Kit (Huayueyang Biotech, China) and reverse-transcribed to cDNA with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa Biotech, Dalian, China) according to the manufacturer’s instructions. ItUGT700, as a candidate unigene with the full-length coding sequences, was screened out in the transcriptome databases. It was amplified by PCR using Phanta MAX Super-Fidelity DNA Polymerase (Vazyme Biotech, Nanjing, China) with gene-specific primers. The PCR product was then cloned between the EcoRI and XhoI restriction sites of the expression vector pET-32a (+) (Novagen, USA) using the ClonExpressII One Step Cloning Kit (Vazyme Biotech, Nanjing, China) and was confirmed by sequencing.
Heterologous Expression and Protein Purification of UGT73CD1
The recombinant plasmid was then introduced into E.coli BL21 (DE3) (Transgen Biotech, Beijing, China) for heterologous expression. E.coli cells were cultured (2 mL) overnight in Luria–Bertani (LB) medium containing 100 μg/mL ampicillin at 37 °C with shaking (200 rpm). These cultures were then added into 500 mL of LB medium containing 100 μg/mL ampicillin and grown at 37 °C with shaking (200 rpm). After the OD600 value reached 0.4–0.6, the cells were induced with 0.1 mM isopropyl β-D-thiogalactoside (IPTG) at 18 °C for 18 h with shaking (150 rpm). The cells were harvested by centrifugation at 4000 rpm for 10 min at 4 °C, and then resuspended in 20 mL lysis buffer (50 mM Tris–HCl pH 8.0) and ruptured by sonication on ice. Then the cell lysate was removed by centrifugation at 12,000 rpm for 30 min at 4 °C. The supernatant was mixed with 3 mL of Ni–NTA resin (TaKaRa Biotech, Dalian, China) pre-equilibrated with 15 mL binding buffer (pH 8.0, 50 mM Na3PO4, 300 mM NaCl, 20 mM imidazole) and incubated for 1 h at 4 °C. The target protein was eluted by the elution buffer (pH 8.0, 50 mM Na3PO4, 300 mM NaCl) with 50, 80, 150, 250 and 300 mM imidazole, respectively. After sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) analysis, the purified target protein was concentrated by Amicon Ultra-30 K (Millipore, USA) and the protein concentration was determined by Bradford Protein Assay Kit (Sangon Biotech, Shanghai, China).
Glycosyltransferase Activity Assays
The whole reactions were individually conducted in a final volume of 100 µL containing 50 mM Tris–HCl buffer (pH 8.0), 0.1 mM aglycone, 0.5 mM sugar donor, and 50 µg of purified ItUGT700. Reactions were incubated at 37 °C for 2 h and were terminated as previously described. After centrifugation at 12,000 rpm for 30 min, the supernatants were collected to be analyzed by HPLC on an Agilent 1200 instrument and Agilent 1200 ultrahigh performance liquid chromatography couple to Agilent 6540 Q-TOF MS (LC–MS) system fitted with an ESI source (Agilent Technologies, USA). All samples were separated on a Eclipse plus C18 column (3.0 × 100 mm, 1.8 μm, Agilent, USA). For quantitative analysis, the system was operated in positive ion mode at a flow rate of 0.4 mL/min using solvent A (water containing 0.1% formic acid) and solvent B (acetonitrile), accompanied by the column temperature set at 50 °C. The gradient elution programs were described in Table S3. An optimal data was collected with UV spectra scanned from 190 to 400 nm and full scan mode set from 100 to 1500 m/z at a scan rate of 3 scans per second. The other scan ion source parameters was as follows: gas temperature, 320 °C; dry gas flow rate, 11 L/min; gas flow of the atomizer, 12 L/min; skimmer voltage, 65 V; capillary voltage, 3,500 V; fragmentor, 130 V; nebulizer, 35 psi. The reaction products were identified based on a comparison of their retention times, UV spectra and MS spectra with that of standards or the published literature. The conversion rates were obtained through calculating peak areas of products and substrates from HPLC–UV/ESIMS.
Biochemical Properties of UGT73CD1
To test the optional pH value for ItUGT700 activity, the reactions were performed in different reaction buffers ranged in pH values from 4.0 to 5.0 (citric acid-sodium citrate buffer), 6.0–8.0 (Na2HPO4-NaH2PO4 buffer), and 9.0–11.0 (Na2CO3-NaHCO3 buffer). To investigate the optimal reaction temperature for ItUGT700, the reactions were performed at different temperatures (4, 25, 37, 42, 50, 60 °C). To test the dependence of divalent metal ions for ItUGT700, CaCl2, BaCl2, MgCl2, ZnCl2, CoCl2 and EDTA were added into the reactions individually at 5 mM. After 30 min, all of the reactions were terminated with an equal volume of ethyl acetate and extracted for three times. The extracts were dissolved in HPLC-grade methanol after being dried in vacuum, and centrifuged at 12,000 rpm for 30 min, then the supernatants were analyzed by HPLC. The above reactions were conducted in triplicate.
Determination of Kinetic Parameters of UGT73CD1
For kinetic studies of ItUGT700, enzymatic assays were performed in a final volume of 100 μL containing 50 mM Na2HPO4-NaH2PO4 buffer (pH 7.0), 20 μg of purified ItUGT700, 500 μM of saturated uridine 5'-diphosphate glucose (UDP-Glc), and varying concentrations(10, 20, 30, 50, 80, 100, 200 μM)of 1. All of the reactions were conducted at 37 °C for 30 min. Then the reactions were extracted with ethyl acetate and centrifuged for HPLC analysis as previously described. The value of Km was calculated using Michaelis–Menten plot method.
Results
Candidate Gene Screening and Phylogenetic Analysis
In this work, we analyzed the transcriptome data of I. tectorum and a BLAST search was used for candidate gene screening. A few of (iso)flavonoid 7GTs nucleotide sequences were used as query sequences (Table S2), including the flavonoid 7GT gene UGT73C6 (Arabidopsis thaliana), UGT73C8 (Medicago truncatula), and isoflavonoid 7GT gene PlUGT4 (Pueraria lobata), etc. Through homology analysis and gene annotation based on different databases, ItUGT700, a (iso)flavonoid 7GT with full-length coding sequences, was preliminarily identified. ItUGT700 was observed with characteristic PSPG Box (Fig. 1), in keeping with other flavonoid UGTs. The open reading frame (ORF) of ItUGT700 encodes 494 amino acids, and the molecular weight of ItUGT700 was predicted to be 54.8 kDa.
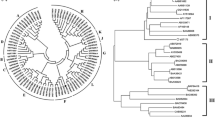
For the better understanding of the relationship of ItUGT700 with functionally characterized (iso) flavonoid UGTs, a phylogenetic tree was constructed by aligning the protein sequences of these UGTs. The phylogenetic tree divided into six clusters, including Cluster I (Microbial CGTs), Cluster II (Flavonoid 5-OH OGTs), Cluster III (Flavonoid 3-OH OGTs), Cluster IV (Flavonoid 7-OH OGTs), Cluster V ( (Iso)flavone 7-OH OGTs), Cluster VI (plant CGTs) (Fig. 2). As expected, ItUGT700 was grouped into the Cluster IV, and it was close to UGT73AE1, UGT73C6 and UGT73C8, indicating that ItUGT700 may have similar function with them. Both UGT73C6 from Arabidopsis thaliana and UGT73C8 from Medicago truncatula were functionally characterized as canonical flavonoid 7GTs, with flavone units as substrates [10, 23]. ItUGT700 shared a 45.06% similarity with UGT73C8, slightly more than UGT73C6 (44.93%). In contrast, ItUGT700 shared the highest similarity (45.51%) with UGT73AE1 [18], implying that it was able to glycosylate structurally diverse flavonoid compounds.
Phylogenetic analysis of UGT73CD1 together with functionally characterized flavonoid UGTs. Cluster I: microbial CGTs; Cluster II: flavonoid 5-OH OGTs; Cluster III: flavonoid 3-OH OGTs; Cluster IV: flavonoid 7-OH OGTs; Cluster V: (Iso) flavone 7-OH OGTs; Cluster VI: plant CGTs. “Red star” indicates UGT73CD1. Accession numbers and species are shown in Table S3
Molecular Cloning and Functional Characterization of UGT73CD1
The full-length cDNA of ItUGT700 were obtained from reverse transcription with total RNA of I. tectorum, and they were subsequently cloned into the pET-32a (+) vector so as to be transformed into E.coli BL21 (DE3). The recombinant protein contained fused Trx·tag, S·tag and 6 × His tag, leading up to a molecular weight of 75.2 kDa when analyzed by SDS-PAGE, and it was purified by Ni–NTA affinity Chromatography (Fig. S1).
I. tectorum is highly rich in 1, so it was used as sugar acceptor to conduct enzymatic activity assay, while UDP-Glc was used as the only sugar donor. As shown in Fig. 3, new products 1a was observed, while control assay showed no product, displaying that ItUGT700 exhibited significantly glycosylation activity toward 1. The MS/MS spectrum of 1a showed fragment ions at m/z 301.0738, which was the characteristic fragment ions [M + H-162] +, indicating the presence of 1 and the catalytic products was O-glycoside [24]. The retention time and mass fragmentation pattern of 1a was essentially identical to an authentic reference standard 16, thus the glycosylated product 1a was identified as tectoridin. The above result unequivocally established ItUGT700 as a first permissive plant UGT identified from I. tectorum, able to glycosylate isoflavone at 7-OH. ItUGT700 was further sent to be named as UGT73CD1 by the UGT Nomenclature Naming Committee [25].
Substrate Promiscuity of UGT73CD1
To further explore the promiscuity and regiospecificity of UGT73CD1 for flavonoid substrates, structurally diverse flavonoid compounds were tested by enzymatic assay, with UDP-Glc as the sugar donor. Unexpectedly, UGT73CD1 showed robust promiscuity towards diverse flavonoid substrates. It could catalyze the whole 11 structurally diverse scaffolds to form mono products, including isoflavones (1–4), flavones (5, 6), flavanones (7–9), dihydrochalcone (12) and simple aromatic compound (13). For some representative obtained glycosides, their structures were established by the standard references. For example, compared with the Daidzin standard, the product catalyzed by UGT73CD1 using the compound 4 as the substrate was identified as Daidzin, a 7-O-isoflavone glycoside (Fig. S2). Besides, the other glycosylated products were tentatively characterized as O-glycosides based on a comparison of their retention times, MS spectrum and MS/MS spectrum with the published literature. Remarkably, UGT73CD1 could accept compound 7–9 at relative high conversion rates (Fig. S7-S9). Moreover, apparent glycosylation conversion rates (> 20%) were observed with compound 1, 3 and 4, with the one exception of 2 (Fig. 4, Fig. S2–S4). Notably, the methoxy group in the B ring of compound 2 hindered conversion rates. Overall, the above results proved that UGT73CD1 as an efficient and regiospecific UGT, was capable of glycosylating isoflavones, flavones and flavanones at 7-OH. It is important to point out that there were three different-sites glycosylated products when flavonols (10, 11) was used as substrates, indicating that UGT73CD1 had the potential to make full use of flavonols, as an efficient instrument for glycosylation (Fig. S10–S11).
Inspired by the robust substrate promiscuity for flavonoid compounds, several structurally diverse compounds were used as substrates to further tested. The substrates include simple aromatic compounds with various nucleophilic groups of -SH, -NH2 group (13, 14), anthraquinone (15) and flavonoid glycosides (16–20). Interestingly, a new glycosylated product come out when compound 13 used as a substrate, different from the glycosylating pattern for above identified flavonoid substrates (Fig. S13). In contrast, UGT73CD1 showed no obvious activity to compound 14, a representative compound containing S-based nucleophile. Moreover, no product was observed when compound 16–20 as glycosides were incubated with the enzyme, indicating that UGT73CD1 could catalyze aglycones instead of glycosides (Fig. 4). In general, these results showed that UGT73CD1 was a promising enzyme for the synthesis of glycosides with diverse structures and bioactivity.
The Kinetic Parameters of UGT73CD1
To examine kinetic parameters of UGT73CD1, several vital parameters was screened by enzymatic assay using tectorigenin as an acceptor and UDP-Glc as sugar donor. The result demonstrated that the optimum pH value was determined at 7.0 (Na2HPO4-NaH2PO4 buffer) (Fig. 5a) and the optimum temperature was 37 °C (Fig. 5b), similar to previously reported plant flavonoid UGTs [26, 33]. UGT73CD1 was independent of divalent cations, supporting that was a GT-B type enzyme (Fig. 5c). Based on the above optimum parameters, kinetic parameters of UGT73CD1 reaction were calculated and Km value was 60 µM (Fig. 5d).
Biochemical properties of UGT73CD1. a Different pH conditions (pH 4.0–11.0); b Temperature (°C); c Divalent metal ions. 1 was used as the substrate and UDP-Glc was used as the sugar donor. An optimized reaction time of 30 min was used for a, b and c. UGT73CD1 exhibited its highest activity at pH 7.0 (50 mM Na2HPO4-NaH2PO4 buffer) and 37 °C, and this enzyme was independent of divalent cations; d Determination of kinetic parameter for UGT73CD1. Using 1 as the acceptor and UDP-Glc as the sugar donor at 37 °C and pH 7.0 for 30 min. The apparent Km values and Vmax were 60 μM and 2.22 × 10−5 mmol/min/μg, respectively
Discussion
Glycosides from medicinal plants play a critical role in drug discovery. For instance, tectoridin exhibits anti-tumor, anti-inflammatory, anti-oxidant activities [27,28,29] and inhibits osteoclastogenesis and bone loss [30]. Recently, the lead compounds being synthesized by UGTs from different plant species in drug discovery has attracted considerable interest, especially for discovery of plant UGTs possessing robust promiscuity and regiospecificity. Typical examples include UGT73AE1 and UGT74AN1, which are from Carthamus tinctorius and Asclepias curassavica respectively [18, 31], and as multifunctional O-, S-, and N-glycosyltransferases, they are capable of glycosylating structurally diverse substrates. MiCGT from Mangifera indica exhibited C-, O-, and N-glycosylation activity, and 35 structurally diverse druglike scaffolds could be glycoslated [32]. Moreover, TcCGT1 from Trollius chinensis and GgCGT from Glycyrrhiza glabra could afford a more S-glycosylation than MiCGT [26, 33]. However, no corresponding UGTs have been discovered in I. tectorum. (Iso)flavonoids, especially tectoridin, were major bioactive components in I. tectorum, having a great influence on the pharmacological actions. A previous study has indicated that the bioactivity and potency of a compound are positively correlated with the position of the glycosidic bond [34]. Compared to the corresponding substrates, glycosides were reported to be better in druggabilities [6]. Synthesis of glycosides are usually divided into chemical synthesis and biosynthesis [35, 36]. Glycosyltransferase, known as a tool enzyme with high efficiency and regiospecificity, were further studied to promoting synthesis of glycosides.
In this study, we identified a new plant UGT named as UGT73CD1 from monocot I. tectorum, which accepted various flavonoid substrates and displayed regiospecificity toward isoflavones and flavones aglycons. Specifically, the diverse substrates of UGT73CD1 was resulting from an acception of simple aromatic compound with-NH2 group. Among these tested substrates, tectorigenin (1) as an endogenous compound was accepted by UGT73CD1 at a relative high conversion, leading to production of tectoridin. Hence, we tentatively put forward that UGT73CD1 might be involved in tectoridin biosynthesis in I. tectorum. Compared with tectorigenin (1), formononetin (2) was converted into a formononetin 7-O-glycoside at a lower conversion rate, indicating that it was badly affected by the position of the methoxy group on the B ring. In addition, UGT73CD1 also converted apigenin (5) and luteolin (6) into corresponding glycosides at a relatively low level. In contrast, UGT73CD1 possessed the highest conversions when flavanones (7–9) used as substrates. Considering that C2-C3 double bond exists at the C ring of flavones while flavanones are just a single bond at the same position, we speculate that subtle changes of substrate configurations may disturb sugar acceptors deeply entering the binding pocket of the enzemy, resulting in poor catalytic activity. It should be noted that UGT73CD1 showed no catalytic activity with flavonoid glycosides (16–20). It indicated that a structural alteration would interfere with the secondary glycosylation of UGT73CD1, contrary to the catalytic characteristic of PlUGT2 [37]. Intriguingly, multiple products were observed when flavonols (10, 11) were catalyzed by UGT73CD1. Previous studies also indicated that a UGT was capable of transferring a glycosyl moiety to different hydroxyl groups of flavonoids [26, 33], while no exactly catalytic mechanism has been reported for that. Moreover, 3, 4-dichloroaniline (13) as a proper N-glycosylation substrate could be catalyzed to form an N-glycoside. Hence, we were further convinced that UGT73CD1 was a multifunctional glycosyltransferase with robust promiscuity towards structurally diverse substrates.
Generally, most of the known 7GTs cluster in the same branch of the phylogenetic tree and exhibit similar function based on sequence similarity. For example, UGT73C6, UGT73C8 and UGT73AE1 clustered in the UGT73 family group, sharing similar catalytic activities toward flavonoids though UGT73AE1 was able to accept other structurally diverse substrates. Nevertheless, a recent study demonstrated that BcUGTs held a close relationship with the UGT706 family and displayed different catalytic activities toward different sugar acceptors while they shared high sequence similarity of more than 80% [38], hinting the activities of UGTs are subject to substrate structures and enzyme-binding sites. The hypothesis was validated through the crystal structure analysis and molecular docking assay of TcCGT1 [33], together with the site-directed mutagenesis studies of GgCGT [26]. Likewise, the activities of UGT73CD1 depend on the key active sites around the substrate binding pocket of itself. We note that AbCGT was able to improve the catalytic activity and enhance the yield of glycosylated products through mutating Val183, a key amino acid residue that is interacting with the 4’-hydroxyl group of 2-hydroxynaringenin [36]. However, no corresponding amino acid residue of UGT73CD1 has been explored in this work. Therefore, key active sites of UGT73CD1 still need to be screened out based on sequence alignment and molecular docking, combined with site-directed mutagenesis. In addition, further studies also need to be performed to improve the catalytic activities of UGT73CD1 and boost production of glycosylflavonoids. Equipping UGT73CD1 with such capabilities is challenging and meaningful for the synthesis of biological glycosylflavonoids in drug discovery.
Conclusion
In summary, UGT73CD1 as a novel glycosyltransferase identified from the medicinal plant I. tectorum, was highlighted by the robust catalytic promiscuity to structurally different compounds. In this study, prokaryotic expression, enzymatic assays, and LC–MS analyses revealed that UGT73CD1 displayed regiospecificity toward isoflavones and led to the formation of 7-O-β-D-glucosides. In addition, flavones, flavanones, dihydrochalcone, flavonols could also be glycosylated to form mono- or multiple products. Unexpectedly, UGT73CD1 could catalyze 3, 4-dichloroaniline to generate corresponding N-glycoside. It expanded the substrate scope of UGT73CD1 and further manifested that UGT73CD1 was a multifunctional glycosyltransferase that could catalyze O-, N-glycosylation reactions. This study provides insights into the characterization of multifunctional glycosyltransferase involving in structurally diverse glycosylflavonoids.
References
Nabavi, S. M., Šamec, D., Tomczyk, M., Milella, L., Russo, D., Habtemariam, S., & Xu, S. W. (2020). Flavonoid biosynthetic pathways in plants: versatile targets for metabolic engineering. Biotechnology Advances, 38, 107316.
Moharram, F. A., Nagy, M. M., El-Dib, R. A., El-Tantawy, M. M., El-Hossary, G. G., & El-Hosari, D. G. (2021). Pharmacological activity and flavonoids constituents of Artemisia judaica L aerial parts. Journal of Ethnopharmacology, 270, 1137.
Kawser, H. M., Abdal, D. A., Han, J., Yin, Y., Kim, K., Kumar, S. S., & Cho, S. G. (2016). Molecular mechanisms of the anti-obesity and anti-diabetic properties of flavonoids. International Journal of Molecular Sciences, 17, 569.
Dai, Y., Zhang, S. S., Liu, D. C., Li, H. M., Ma, T., Huo, Q., & Wu, C. Z. (2018). Enzymatic biosynthesis of novel bavachin glucosides via Bacillus UDP-glycosyltransferase. Phytochemistry Letters, 23, 9–14.
Xiao, J., Capanoglu, E., Jassbi, A. R., & Miron, A. (2016). Advance on the flavonoid C-glycosides and health benefits. Critical Reviews in Food Science and Nutrition, 56(sup1), S29–S45.
Cheynier, V., Comte, G., Davies, K. M., Lattanzio, V., & Martens, S. (2013). Plant phenolics: Recent advances on their biosynthesis, genetics, and ecophysiology. Plant Physiology and Biochemistry, 72, 1–20.
Li, J., Liu, X., Gao, Y., Zong, G., Wang, D., Liu, M. Z., & Shen, Y. Q. (2019). Identification of a UDP-glucosyltransferase favouring substrate-and regio-specific biosynthesis of flavonoid glucosides in Cyclocarya paliurus. Phytochemistry, 163, 75–88.
Hostetler, G. L., Ralston, R. A., & Schwartz, S. J. (2017). Flavones: Food sources, bioavailability, metabolism, and bioactivity. Advances in Nutrition, 8, 423–435.
Liang, D. M., Liu, J. H., Wu, H., Wang, B. B., Zhu, H. J., & Qiao, J. J. (2015). Glycosyltransferases: Mechanisms and applications in natural product development. Chemical Society Reviews, 44, 8350–8374.
Jones, P., Messner, B., Nakajima, J., Schäffner, A. R., & Saito, K. (2003). UGT73C6 and UGT78D1, glycosyltransferases involved in flavonol glycoside biosynthesis in Arabidopsis thaliana. Journal of Biological Chemistry, 278, 43910–43918.
Yonekura-Sakakibara, K., Tohge, T., Niida, R., & Saito, K. (2007). Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. Journal of Biological Chemistry, 282, 14932–14941.
Kumar, R. J. S., Ruby, Singh, S., Sonawane, P. D., Vishwakarma, R. K., & Khan, B. M. (2013). Functional characterization of a glucosyltransferase specific to flavonoid 7-O-glucosides from Withania somnifera. Plant Molecular Biology Reporter, 31, 1100–1108.
Liu, X., Lin, C., Ma, X., Tan, Y., Wang, J., & Zeng, M. (2018). Functional characterization of a flavonoid glycosyltransferase in sweet orange (Citrus sinensis). Frontiers in Plant Science, 9, 166.
Funaki, A., Waki, T., Noguchi, A., Kawai, Y., Yamashita, S., Takahashi, S., & Nakayama, T. (2015). Identification of a highly specific isoflavone 7-O-glucosyltransferase in the soybean (Glycine max (L.) Merr.). Plant and Cell Physiology, 56, 1512–1520.
Wang, X., Li, C., Zhou, Z., & Zhang, Y. (2019). Identification of Three (Iso)flavonoid glucosyltransferases from Pueraria lobata. Frontiers in Plant Science, 10, 28.
Peng, M., Shahzad, R., Gul, A., Subthain, H., Shen, S., Lei, L., & Luo, J. (2017). Differentially evolved glucosyltransferases determine natural variation of rice flavone accumulation and UV-tolerance. Nature Communications, 8, 1975.
Zhang, F., Guo, H., Huang, J., Yang, C., Li, Y., Wang, X., & Luo, J. (2020). A UV-B-responsive glycosyltransferase, OsUGT706C2, modulates flavonoid metabolism in rice. Science China Life Sciences, 63, 1037–1052.
Xie, K., Chen, R., Li, J. H., Wang, R. S., Chen, D. W., Dou, X. X., & Dai, J. G. (2014). Exploring the catalytic promiscuity of a new glycosyltransferase from Carthamus tinctorius. Organic Letters, 16, 4874–4877.
Xu, H. Y., Ren, J. H., Su, Y., Ren, F., Zhou, Y. J., Jiang, H., & Chen, J. (2020). Anti-hepatitis B virus activity of swertisin isolated from Iris tectorum Maxim. Journal of Ethnopharmacology, 257, 112787.
Sun, Y., Li, W., & Wang, J. (2011). Ionic liquid based ultrasonic assisted extraction of isoflavones from Iris tectorum Maxim and subsequently separation and purification by high-speed counter-current chromatography. Journal of Chromatography B, 879, 975–980.
Koch, W. K., Sieniawska, E., Widelski, J., Urjin, O., Głowniak, P., & Woźniak, K. S. (2015). Major secondary metabolites of Iris spp. Phytochemistry Reviews, 14, 51–80.
Wu, Z., Ren, S., Chen, T., Hui, A., & Zhang, W. (2020). Separation and purification of six isoflavones from Iris tectorum Maxim by macroporous resin-based column chromatography coupled with preparative high-performance liquid chromatography. Separation Science and Technology, 55, 1686–1694.
He, X. Z., Li, W. S., Blount, J. W., & Dixon, R. A. (2008). Regioselective synthesis of plant (iso)flavone glycosides in Escherichia coli. Applied Microbiology and Biotechnology, 80, 253–260.
Chen, K., Hu, Z. M., Song, W., Wang, Z. L., He, J. B., Shi, X. M., & Ye, M. (2019). Diversity of O-glycosyltransferases contributes to the biosynthesis of flavonoid and triterpenoid glycosides in Glycyrrhiza uralensis. ACS Synthetic Biology, 8, 1858–1866.
Homepage of the UGT Nomenclature Committee, Retrieved June 2, 2021 from http://prime.vetmed.wsu.edu/resources/udp-glucuronsyltransferase-homepage.
Zhang, M., Li, F. D., Li, K., Wang, Z. L., Wang, Y. X., He, J. B., & Ye, M. (2020). Functional characterization and structural basis of an efficient Di-C-glycosyltransferase from Glycyrrhiza glabra. Journal of the American Chemical Society, 142, 3506–3512.
Ha, D. T., Binh, B. T., Thu, N. T., Bich Thu, N. T., Thanh Tung, P. H., & Oh, W. K. (2019). Four new compounds isolated from the aerial part of Belamcanda chinensis (L.) and their effect on vascular smooth muscle cell (VSMC) Proliferation. Chemical & Pharmaceutical Bulletin, 67, 41–46.
Monthakantirat, O., De-Eknamkul, W., Umehara, K., Yoshinaga, Y., Miyase, T., Warashina, T., & Noguchi, H. (2005). Phenolic constituents of the rhizomes of the Thai medicinal plant Belamcanda chinensis with proliferative activity for two breast cancer cell lines. Journal of Natural Products, 68, 361–364.
Wang, C. L., Li, D., Wang, C. D., Xiao, F., Zhu, J. F., Shen, C., & Chen, X. D. (2017). Anti-inflammatory and anti-osteoarthritis effects of tectorigenin. Biology Open, 6, 1130–1136.
Wang, J., Tang, Y., Lv, X., Zhang, J., Ma, B., Wen, X., & Wang, G. F. (2020). Tectoridin inhibits osteoclastogenesis and bone loss in a murine model of ovariectomy-induced osteoporosis. Experimental Gerontology, 140, 111057.
Wen, C., Huang, W., Zhu, X. L., Li, X. S., Zhang, F., & Jiang, R. W. (2018). UGT74AN1, a permissive glycosyltransferase from Asclepias curassavica for the regiospecific steroid 3-O-glycosylation. Organic Letters, 20, 534–537.
Chen, D. W., Chen, R., Wang, R., Li, J., Xie, K., Bian, C., & Dai, J. G. (2015). Probing the Catalytic promiscuity of a regio-and stereospecific C-glycosyltransferase from Mangifera indica. Angewandte Chemie International Edition, 54, 12678–12682.
He, J. B., Zhao, P., Hu, Z. M., Liu, S., Kuang, Y., Zhang, M., & Ye, M. (2019). Molecular and structural characterization of a promiscuous C-glycosyltransferase from Trollius chinensis. Angewandte Chemie International Edition, 58, 11513–11520.
Liu, X., Zhang, L., Feng, X., Lv, B., & Li, C. (2017). Biosynthesis of glycyrrhetinic acid-3-O-monoglucose using glycosyltransferase UGT73C11 from Barbarea vulgaris. Industrial & Engineering Chemistry Research, 56, 14949–14958.
Chen, D. W., Fan, S., Chen, R., Xie, K., Yin, S., Sun, L., & Dai, J. G. (2018). Probing and engineering key residues for bis-C-glycosylation and promiscuity of a C-glycosyltransferase. ACS Catalysis, 8, 4917–4927.
Xie, K., Zhang, X., Sui, S., Ye, F., & Dai, J. G. (2020). Exploring and applying the substrate promiscuity of a C-glycosyltransferase in the chemo-enzymatic synthesis of bioactive C-glycosides. Nature Communications, 11, 5162.
Wang, X., Fan, R., Li, J., Li, C., & Zhang, Y. S. (2016). Molecular cloning and functional characterization of a novel (Iso)flavone 4′,7-O-diglucoside glucosyltransferase from Pueraria lobata. Frontiers in Plant Science, 7, 387.
Zhang, X., Zhu, Y., Ye, J., Ye, Z., Zhu, R., Xie, G., & Qin, M. J. (2021). Iris domestica (iso)flavone 7- and 3’-O-glycosyltransferases can be induced by CuCl2. Frontiers in Plant Science, 12, 632.
Funding
This study was supported by Guangdong Key Laboratory for translational Cancer research of Chinese Medicine (No. 2018B030322011), the National Natural Science Foundation of China (CN) (No.81874333 and 82003895) and the Guangdong Basic and Applied Basic Research Foundation (No.2020A1515010926).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors in this article indicated no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, J., Li, J., Yue, J. et al. Functional Characterization of a Novel Glycosyltransferase (UGT73CD1) from Iris tectorum Maxim. for the Substrate promiscuity. Mol Biotechnol 63, 1030–1039 (2021). https://doi.org/10.1007/s12033-021-00364-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-021-00364-1