Abstract
Vascularized organoid-on-a-chip (VOoC) models achieve substance exchange in deep layers of organoids and provide a more physiologically relevant system in vitro. Common designs for VOoC primarily involve two categories: self-assembly of endothelial cells (ECs) to form microvessels and pre-patterned vessel lumens, both of which include the hydrogel region for EC growth and allow for controlled fluid perfusion on the chip. Characterizing the vasculature of VOoC often relies on high-resolution microscopic imaging. However, the high scattering of turbid tissues can limit optical imaging depth. To overcome this limitation, tissue optical clearing (TOC) techniques have emerged, allowing for 3D visualization of VOoC in conjunction with optical imaging techniques. The acquisition of large-scale imaging data, coupled with high-resolution imaging in whole-mount preparations, necessitates the development of highly efficient analysis methods. In this review, we provide an overview of the chip designs and culturing strategies employed for VOoC, as well as the applicable optical imaging and TOC methods. Furthermore, we summarize the vascular analysis techniques employed in VOoC, including deep learning. Finally, we discuss the existing challenges in VOoC and vascular analysis methods and provide an outlook for future development.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Organ-on-a-chip (OoC) is a highly advanced approach combining microfluidics with cell culture, leading to significant advancements in simulating human microphysiological systems and functions [1,2,3]. The diffusion limit of nutrients, such as oxygen, ranges from 100 to 200 μm in the human body. Once the size of an organ or tissue surpasses this range, efficient substance exchange is achieved through the development of microvasculature [4]. Organoids cultured in vitro usually have diameters ranging from 50 to 1000 μm and rely solely on passive diffusion to receive nutrients and oxygen and to remove waste products [5]. When the size of an organoid exceeds the passive diffusion limit in hydrogel (~ 400 μm), its growth and function are significantly affected [6, 7]. Recently, numerous investigators have endeavored to construct microvascular networks in vitro using microfluidic chips, resulting in the vascularization of organoids and enhancing the authenticity of the OoC system. Vascularized organ-on-a-chip (VOoC) employs specialized microfluidic chips with specific structures to co-culture target organoids or spheroids with vascular endothelium and its supporting cell lines, leading to the formation of a perfusable vascular network (Fig. 1). Incorporating additional stimuli, such as flowing mechanical force and growth factor, can significantly improve the success rate of vascular perfusion.
The continuous advancement of VOoC has generated a growing need for advanced imaging techniques that can efficiently and accurately capture the relevant biological processes and the interaction between the organoid and its microenvironment. In this regard, confocal/multiphoton fluorescence microscopy (CFM/MPM) has gained prominence in VOoC imaging due to its exceptional imaging quality [8, 9]. Furthermore, with the progress of imaging technology, emerging microscopic techniques such as light-sheet fluorescence microscopy (LSFM) and fluorescence micro-optical sectioning tomography (fMOST) have generated attention and demonstrated results in achieving comprehensive 3D imaging of organoid structures [10, 11].
The enhanced substance exchange in the deep layer of organoids allows for further increases in their size. Concurrently, the co-culture of multiple cell types related to microvasculature leads to a significant increase in cellular complexity within organoid microenvironment. However, these factors, along with the heterogeneity of the chip material, can introduce challenges, such as light scattering and refractive index (RI) mismatches. Consequently, the CFM imaging depth is often limited to ~ 100 μm or even lower [8]. Given these limitations, obtaining comprehensive information about VOoC becomes challenging, hindering the analysis of the physiological activities within microvasculature and organoids. Recently, tissue optical clearing (TOC) methods, such as CLARITY and CUBIC, have been introduced to VOoC for obtaining the 3D structure of organoids [12,13,14]. Furthermore, the combination of TOC and LSFM enables rapid acquisition of the 3D images [15, 16], significantly improving imaging efficiency. While the efficient acquisition of comprehensive 3D data for the entire VOoC is essential, it is equally critical to process this data quickly, accurately, and automatically. Deep learning techniques have rapidly advanced in recent years [17] and have played a crucial role in cell identification, location tracking [18,19,20], and the analysis of microvascular networks [21, 22], greatly enhancing the automation level of VOoC systems (Fig. 1).
In this review, we will initially summarize the chip designs and vascularization culturing strategies employed in VOoC. Subsequently, we will discuss the imaging methods that apply to VOoC and briefly outline the advancements in TOC techniques within the context of the chip. Furthermore, we will present a comprehensive overview of the vascular analysis methods applicable to VOoC. Finally, we will delve into the future development prospects of VOoC and the ongoing advancements in vascular analysis methods.
Vascularized organoid-on-a-chip
Chip design
Microfluidic chips have emerged as promising platform for in vitro modeling of microvasculature due to several advantages. Firstly, microfluidic chips can construct perfusable microvessels with a diameter of 20–50 μm by controlling interstitial flow within the extracellular matrix (ECM) [23], closely resembling microvasculature’s function and size in the human body. Secondly, microfluidic technology enables the creation of hierarchical vessels with similar complexity to physiological vessels. This capability allows for a more realistic representation of the vascular network. Thirdly, microfluidic chips with relatively simple structural designs can be mass produced through techniques, such as laser cutting or injection molding [24, 25], which is conducive to high-throughput culturing and analysis of vascularized organoid models. Moreover, microfluidic chips offer precise control over various parameters, such as growth factor concentration, shear force, and other environmental conditions. This precise regulation facilitates optimal cell growth and angiogenesis by providing a controlled and tailored microenvironment [26]. One of key points in VOoC design involves incorporating hydrogel channels on the chip to facilitate substance exchange and provide structural support for vascular growth. Here, we categorized the design of VOoC into mainly two types, designs for self-assembly of endothelial cells (ECs) to form microvessels and designs for pre-patterned vessel lumens.
Designs for self-assembly of ECs
The designs for self-assembly of ECs on chip predominantly exploit the intrinsic hydrophobic properties of the chip material, along with the surface tension and capillary effects of the hydrogel liquid. These effects collectively contribute to effectively confining the hydrogel within the channels. Upon curing of the hydrogel under specific conditions, a durable solid–liquid interface is established. This interface facilitates the adhesion and proliferation of ECs, or alternatively, it offers structural support for the self-assembly of vascular ECs into microvascular network within a 3D hydrogel environment. Currently, there are several microstructures that can be employed, including micropillar, microridge, and capillary burst valve.
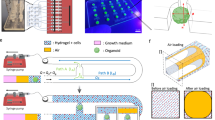
As depicted in Fig. 2a, in the micropillar design, multiple pillars are distributed at specific intervals along the sides of the hydrogel channels. The stability of the hydrogel channels can be improved by optimizing the hydrophobicity of the chip material and selecting suitable micropillar spacing (Rx) and channel height (Rz). These factors influence the maximum pressure perturbations a hydrogel channel can withstand.
where \(\Delta P\) is the pressure differential sustained by surface tension and \(\gamma\) is the surface tension coefficient. A trapezoidal micropillar structure can be utilized to enhance stability and promote a more suitable air–hydrogel interface during hydrogel injection. This structure effectively reduces the perturbation and ensures a more controlled hydrogel filling process [27]. Typically, the chip channel heights range from 100 to 200 μm [28], while lowering the channel height theoretically improves stability, channel heights below 100 μm may result in 3D vascular networks that lack sufficient depth. Micropillar-based OoC is commonly employed in various applications, including the construction of blood–brain barrier (BBB) chips [29], kidney chips [30], and VOoC [14]. However, it is essential to acknowledge that the hydrogel channel formed using micropillar structures can be susceptible to depression and local defects due to the inherent differences in properties between the chip material and the hydrogel, which may hinder the formation of a well-defined and functional vascular barrier.
Chip designs of VOoC. a Micropillars with specific spacing (Rx) and height (Rz) can bind the hydrogel in the central channel to provide the scaffold for microvasculature growth [27]. b Microridges can form a large area of flat hydrogel interface via the meniscus-pinning effect [75], making it suitable for vascular EC adhesion to form the vascular barrier. c Chambers are generally connected with the medium channel by the capillary burst valves to provide nutrients for vascular growth and regulate the interstitial flow [42]. d Microtubes construct the hydrogel lumen utilizing an artificially created vascular mold. Subsequently, a suspension containing vascular ECs is infused into this hydrogel lumen to create microvascular tubes [54]. e Mix ECs with hydrogel, inject into the channel, and place the device with gravity. Add low-viscosity culture medium at the inlet and collect at the outlet. This washes away high-viscosity hydrogel, forming symmetrical three-dimensional hydrogel lumens [51]. f The fugitive support material Pluronic F-127 is 3D printed on the chip’s hydrogel to create vascular molds. After removing Pluronic F-127, ECs are added to the resulting hollow vascular channels. Once the vessels form, organoids can be seeded into the hydrogel for further study [52]. Created with BioRender.com
The “microridge,” also known as “phaseguide™,” is a design that utilizes the meniscus-pinning effect, which is generated by a material line or a change in geometry, to form the liquid-hydrogel boundary (Fig. 2b) [31]. Generally, a higher and wider microridge structure contributes to increased hydrogel stability. However, it also leads to a reduced contact area between the hydrogel and the medium channels on both sides. To strike a balance, the height of the geometric change in microridge structures is typically set to be around 1/4 of the channel height, while the width is typically kept similar to the height [32,33,34,35]. The structural characteristics of microridge enable the formation of extensive and flat liquid–hydrogel interface, promoting tight adhesion of ECs for effective barrier functionality [36, 37]. Hence, this structure is highly suitable for constructing barrier-type organoids that demand high hydrogel interface integrity, as well as for observing the local formation of tip/stalk ECs [32, 34, 38]. Simultaneously, the higher surface area-to-volume ratio facilitates substance transport studies [39,40,41]. However, the hydrogel channel formed by microridge is prone to damage and leakage due to significant shear stress caused by fluid flow, particularly during the injection of the culture medium. Therefore, when using microridge chips, it may be advisable to reduce the height of the hydrogel channels, raise the height and width of the microridge, and increase the concentration of relevant proteins in the hydrogel to improve its stability during perfusion [31].
As shown in Fig. 2c, the capillary burst valve design is primarily employed in tissue chamber chips, serving to connect the central chamber with the culture medium channels on both sides [42]. Usually, the size of the capillary burst valve is 50 μm and the chamber is 1 × 2 × 0.1–0.12 mm (length × width × height). Due to surface tension and capillary effects, the hydrogel injected into the central chamber will not leak into the side channels. Upon the gelation of the hydrogel, the culture medium is introduced into the channels on both sides, with the medium channels typically connected to culture medium reservoirs. By adjusting the liquid level in the reservoirs, interstitial flow can be generated in the chip, thereby stimulating cell growth inside the tissue chambers [42,43,44]. The relatively simple structure of the tissue chamber chips makes them compatible with a 96-well plate [45], enabling high-throughput construction of VOoC. Furthermore, using multi-layer microfluidic chips connected by vertical perfusion channels holds significant potential for facilitating co-culture between different organoids [46]. However, it is important to note that the tissue chamber chip does impose certain limitations on the size (smaller than chamber channel’s height, typical 120 μm) and seeding sequence of co-cultured organoids. If researchers intend to seed larger organoids following the formation of microvasculature, alternative chip designs may need to be considered.
The microstructures mentioned above share a common characteristic in the formation of hydrogel channels, which are guided by capillary forces. They are primarily employed in VOoC to separate hydrogel and culture medium channels. One or more channels are used for injecting the hydrogel mixed with cell suspension, while the remaining channels serve for culturing, administering growth factors, or staining reagents. Interstitial flow can be achieved by creating a hydrostatic pressure difference, facilitating the delivery of nutrients and removal of cellular waste products [47].
Designs for pre-patterned lumens
The lumen design constructs microtube structures in hydrogels by artificially designing a series of tubular molds, facilitating the adhesion and growth of ECs to form blood vessels. Microneedle (typical diameter: 150–550 μm) is a commonly used vascular mold for simple structure [48, 49]. More intricate vascular structures can be created using techniques, such as viscous finger patterning (VFP) [50, 51] or 3D bioprinting [52, 53].
Figure 2d illustrates an example for constructing specified 3D vascular structures in vitro using a microneedle [54]. In specific, the process involves placing microneedle on the chip, then introducing the hydrogel into the chip to embed the mold. Once the hydrogel polymerizes, the mold can be removed, establishing a microtube with a controlled lumen size. The ECs suspension is then introduced into the hydrogel lumen and incubated for 15–30 min to promote adhesion. To prevent damage to the hydrogel structure during the molding process, the surface of the vascular mold is typically coated with bovine serum albumin [55]. What’s more, by utilizing various sizes of microneedles and multiple demolding steps, it is possible to achieve the construction of blood vessels with a multi-layered perivascular cell structure, which structurally appears more realistic compared to microvasculature composed of a single layer of ECs. However, this method is constrained by the diameter of the microneedles, making it challenging to construct capillaries that closely resemble real sizes [56]. In vascularization applications, vascularized tumor models can be achieved by introducing tumor organoids into parallel microtube or co-culturing tumor cells or organoids within hydrogel [54]. Moreover, the BBB function can be reconstructed by introducing cerebral vascular ECs and applying fluid flow stimulation [57,58,59]. It is evident that the generation of tens of micrometers level blood vessels using microneedle poses a considerable challenge, and the corresponding demolding process can be quite complex and damaging. Fortunately, VFP and 3D bioprinting show great potential in tackling this issue [52, 60,61,62].
VFP leverages the property of a less viscous fluid generating finger-like protrusions in more viscous fluids, providing an alternative to microneedle molding method (Fig. 2e). The specific methodology involves infusing a high-viscosity fluid (e.g., collagen I) into microchannels and prior to solidification, reinfusing the channels with a low-viscosity fluid (e.g., cell culture medium). At this juncture, the high-viscosity fluid within the channels gradually is replaced by the low-viscosity fluid, yet a layer of high-viscosity fluid persists on the inner walls of the channels, giving rise to finger-like microtubes [63, 64]. Some studies indicate that the perfusion pressure and flow angle of the low-viscosity fluid can influence the thickness and morphology of the resulting microtubes [50, 62]. Specifically, when the chip is vertically infused with low-viscosity fluid, a more uniformly thick lumen wall is established and an increase in pressure during the infusion of low-viscosity fluid results in a larger diameter of the formed lumens [51]. However, the resultant vessels commonly exhibit widths in the order of hundreds of microns, posing challenges in downscaling to the dimensions of typical capillaries. Therefore, Chen et al. utilized VFP to construct larger vessels on the side channels and employed EC self-assembly to create a microvascular network in the central hydrogel channel, achieving a chip with hierarchical microvascular structures [50].
3D bioprinting plays a crucial role in the field of tissue engineering, allowing for the precise placement of cells in 3D space to achieve the desired organ functions, particularly in the vascular construction (Fig. 2f). Currently, there are two main approaches: directly printing cells to form vessels with specific structures [53] and printing vascular mold to allow ECs to attach and grow into vascular tubes [52]. Both these two vascular fabrication approaches are essentially angiogenic remodeling strategies [65]. To achieve the construction of a capillary network, some researchers have adopted cartilage-derived ECM microfibers bioink [66] or techniques, such as laser bioprinting [67,68,69].
Other designs
In recent years, some intriguing designs have significantly enhanced the diversity and functionality of VOoC. For example, Rajasekar et al. developed a microdevice called IFlowPlate based on a 384-well plate, applying it to vascularization in colon organoids and greatly improving the analytical efficiency of VOoC [70]. Additionally, certain studies have utilized leaf venation networks as molds to fabricate VOoC, allowing perfusion with enhanced physiological relevance [71, 72]. Furthermore, these designs often offer dedicated organoid compartments, streamlining the loading and co-culturing processes [24, 73, 74].
Vascularization culturing strategies on the chip
Vascularization has been successfully implemented in various organoids, including the brain, heart, kidney, liver, and intestine [5]. Additionally, a growing body of research focuses on the tumor microenvironment, which aims to establish microvasculature within tumor spheroid to simulate interactions between cells and the ECM [76, 77]. The initial formation of organoid/spheroid is crucial in most vascularization processes and dramatically influences their overall success and researchers often implement structural modifications to the chip to facilitate the cultivation and vascularization of organoids better.
Currently, the xenograft and VOoC models are two primary strategies for generating vascularized organoids. The xenograft model is presently considered the most physiologically relevant method for vascularization. In this approach, organoids are implanted into experimental animals, such as immunodeficiency mice, through the surgical process [78,79,80,81,82]. However, this organoid vascularization strategy necessitates better integration and cost reduction. Additionally, it is subjected to various factors, including animal-individual differences and ethical issues. Microfluidic systems, serving as carriers for VOoC, offer greater control than xenograft models, providing a valuable in vitro platform for studying the interaction between blood vessels and organoids [61, 83]. Based on the morphogenic processes of microvasculature, the VOoC model is specifically classified into vasculogenesis and angiogenesis.
Organoid/spheroid formation
Stem cells (SCs) can differentiate into both organoids and various supporting cells necessary for vascular generation by exogenous signals regulation (Fig. 3a) [84]. Consequently, regardless of whether they are employed in xenograft or VOoC models, SCs-differentiated organoids exhibit a relatively high successful rate in terms of vasculation. Among various SCs, human-induced pluripotent stem cells are predominantly utilized and have exhibited remarkable potential in establishing stable vascularized organoids, including those of the brain [78], kidney [79],intestine [81], skeletal muscle [85], and other tissues [70, 86].
Vascularization culturing strategies. a SCs can differentiate into organoid and vascular formation supporting cells (such as ECs and pericytes), significantly improving the success rate of vascularization. b Three typical spheroid formation strategies for vascularization. (c-f) VOoC model based on vasculogenesis and angiogenesis. Created with BioRender.com
Additionally, researchers commonly employ tumor cells or fibroblasts to establish mono-culture spheroids for studying on-chip vascularization (Fig. 3b) [87]. However, some studies have revealed the challenges associated with constructing a perfusable vascular network within a spheroid consisting solely of a single tumor cell line [88, 89], so this kind of spheroid is primarily employed for investigating tumor migration and invasion in the microvasculature [74, 90, 91]. Fibroblasts spheroids can secrete angiogenesis growth factors, such as basic fibroblast growth factor, thereby inducing the development of a perfusable vascular network and providing a convenient platform to observe substance exchange within spheroids [24, 74].
In contrast to the conventional mono-culture system of spheroids, numerous studies have implemented a proportional ratio of ECs, fibroblasts, and other vascular-supporting cells to enhance the success rate of vascularization and perfusibility. Fibroblasts play a crucial role in promoting microvascular sprouting by secreting related growth factors and providing basic physical cell–cell interactions [5, 92], while ECs contribute to the pre-formation of vascular structures within the spheroids (pre-vascularized) [87]. Typically, cell suspensions of experimental cells and vascular-supporting cells are combined in specific proportions and cultured in a low-adhesion U-shaped well plate. A low volume ratio of Matrigel (e.g., 1% v/v) or Methocel® A4M and various growth factors are added [14, 74, 88, 93] or the cells are cultured in gelled droplets to form spheroids [83]. The specific mixing ratio may vary depending on the cell lines utilized. Due to the higher aggregation ability of fibroblasts compared to other cells, they tend to gather in the center of the co-cultured spheroids [89], which hinders the diffusion of growth factors produced by fibroblasts. To address this, Wan et al. introduced a fibroblast suspension into tumor spheroids and observed that fibroblasts adhered to the surface of the spheroids, resulting in a more significant effect on enhanced vascularization compared to direct co-culture [94]. Additionally, a typical tri-culture approach involves culturing experiment cells, fibroblasts, and ECs in a specific proportion. Nashimoto et al. demonstrated that tri-culture spheroids with an MCF7:HLF:HUVEC ratio of 3:1:1 exhibited the highest success rate in vascularization and perfusibility, effectively reducing the necrosis within the inner core of spheroids [89].
Modification of chip for seeding organoid/spheroid
For the convenience of seeding organoids/spheroids into the chip, researchers typically make modifications to VOoC.
For VOoC based on self-assembly design, there are generally two modification approaches. The first involves creating an external seeding port at the central position of the chip’s hydrogel channel. The diameter of this seeding port typically ranges from 1 to 2 mm (Fig. 3c) [14, 38, 74, 83, 88, 89]. In theory, the larger the organoid/spheroid to be seeded, the larger the seeding port needed. To match the diameter of the seeding port, researchers typically trim the tips of 10-μL or 200-μL pipettes with a surgical knife. During the seeding process, hydrogel and organoid/spheroid should be slowly mixed to avoid the formation of additional bubbles. This method provides better control over the seeding location of organoid/spheroid but carries the risk of increased exposure to the external environment and may, to some extent, affect the fluid distribution within the chip. The second approach involves increasing the channel height of the chip’s hydrogel channel to allow organoid/spheroid to enter the chip through the inlet [91, 94,95,96,97,98]. This method ensures the integrity of the chip to the greatest extent, and the interstitial flow within the chip is not affected. However, the drawback is that the distribution of organoid/spheroid is relatively random, making it less conducive to subsequent imaging observations. Researchers can try to limit the movement of organoid/spheroid by designing micro pit structures inside the chip channels.
For VOoC based on lumen or other designs, it usually has a larger space (diameter: ~ 200 μm) and will be relatively convenient to seed organoid/spheroid into the lumen or directly into the hydrogel, thereby observing the subsequent vascularization effects (Fig. 2d–f) [52, 54, 61, 70, 86, 99].
Vasculogenesis
Vasculogenesis describes the de novo formation of new blood vessels and occurs mainly during the embryonic stage and during wound healing in adults [100, 101]. In microfluidic chips, vasculogenesis often refers to the process where ECs uniformly distributed in the ECM, under external stimuli, self-assemble into tube-like networks to form newly generated vessels [102]. Correspondingly, the vasculogenesis-based vascularization strategy involves co-culturing mature organoids/spheroids directly in hydrogels that contain ECs and supporting cell lines, such as fibroblasts and pericytes. Depending on the experimental designs, organoids/spheroids can be seeded into the chip together with ECs (Fig. 3d) [14, 90, 91, 94]. Alternatively, ECs can be initially seeded in the hydrogel, and the organoids/spheroids are placed on the chip once the ECs have self-assembled into a vascular bed. This approach allows for observing microvasculature invasion into the organoid/spheroid (Fig. 3e) [83]. While the latter method necessitates a longer culturing duration, it offers a temporal window for independent observation and evaluation of the microvascular network. This enables convenient pre- and post-comparison and validation of the effects of the organoids/spheroids on the microvascular network [24, 83, 103].
Angiogenesis
Different from vasculogenesis, angiogenesis characterizes the formation of new blood vessels from pre-existing ones, encompassing sprouting and intussusceptive angiogenesis. In sprouting angiogenesis, new vessels emerge as branches from existing ones, whereas intussusceptive angiogenesis involves the division of pre-existing blood vessels [104]. In the VOoC model based on angiogenesis, ECs are seeded into the medium channels on both sides of the hydrogel, allowing them to adhere to the hydrogel interface and form a “pre-existing blood vessel.” Under the stimulation of mechanical flow force and growth factors, the pre-existing blood vessels on either side grow and sprout, invading the hydrogels to establish microvasculature. Once the vascular bed is formed, the organoids/spheroids can be placed on the central hydrogel channel (Fig. 3f). The microvasculature on both sides gradually invades and vascularizes the organoids/spheroids under the influence of growth factors and flow stimulation [38]. Similar to vasculogenesis, ECs can be seeded into the medium channels simultaneously with the seeding of organoids/spheroids into the central hydrogel channel. Skipping the step of vascular bed formation can reduce the time required for the vascularization of organoids/spheroids significantly [88, 89]. Moreover, the specific seeding sequence can be adjusted based on different cell lines or experimental objectives. For instance, when using organoids and ECs from SCs, the organoids are first seeded into the chip under appropriate differentiation conditions. After organoid differentiation is complete, ECs are further seeded [86, 105].
Key factors for VOoC culturing
Regardless of whether the vascularization approach is based on vasculogenesis or angiogenesis, the processes of vascular growth and vascularization are influenced by several key factors, including interstitial flow, growth factors, the ECM environment, and supporting cell types.
It has been reported that interstitial flow can not only promote vasculogenesis and spheroid vascularization by regulating matrix metalloproteinase-2 [106] but also increase the sprout count and length of angiogenesis by providing proper shear stress to ECs, enhancing the success rate of anastomosis and perfusability in microvascular networks [47]. Generally, interstitial flow can be achieved on the chip by setting a hydrostatic pressure difference, and a range of 2–25 mmH2O pressure difference can typically generate interstitial flow rates of 0–10 μm/s within the ECM [102]. Researchers can calculate the distribution of interstitial flow rates in the chip by combining the channel size and ECM properties (e.g., permeability and porosity) of their VOoC, constructing hydrostatic pressure differences that conform to the physiological flow rate range (1.7–11 μm/s) [24]. Additionally, as shown in Table 1, 3D rocker and syringe pump can also generate controllable interstitial flow in the VOoC.
Vascular endothelial growth factor (VEGF) has played a crucial role in promoting vascular growth. It has been reported that a concentration of VEGF between 20 and 100 ng/mL can effectively promote vascular growth on the chip [102], with a concentration of 50 ng/mL being commonly used [24, 47, 104]. Furthermore, the combined use of various growth factors (such as bFGF, S1P, and PMA) can enhance the proliferative effect to some extent [34]. As shown in Table 1, the mostly used ECM in VOoC is primarily composed of fibrinogen and collagen I, with concentrations ranging from 1 to 10 mg/mL. Wan et al. found a negative correlation between the density and diameter of microvascular network on the chip and the concentration of the fibrinogen solution [107].
In addition, Ferrari et al. reported that the stiffness of ECM hydrogel controls the size of the patterned vasculature and the density of sprouting angiogenesis [104]. Recently, some studies have demonstrated the spontaneous formation of microvascular networks using synthetically ECM, such as soft gelatin methacrylate and a mixture of calcium alginate and decellularized ECM [98, 108,109,110].
Multicellular interactions should be involved in the complex vascular growth process. As mentioned earlier, fibroblasts are the commonly used supporting cells in VOoC. Fibroblasts can help to prevent early network regression, prolong the lifespan of the vascular network, and enhance open lumens formation [92]. Wan et al. reported that as the proportion of co-cultured fibroblasts with ECs increased in the chip, the diameter and perfusability of the formed microvasculature decreased, but the complexity of overall network increased [107]. Additionally, depending on the specific VOoC being constructed, the corresponding supporting cells may be adjusted. For example, when constructing neurovascular unit (NVU) chip, astrocytes, oligodendrocytes, and neuronal cells may be selected as co-cultured supporting cells [29, 36].
Optical imaging methods applicable to VOoC
To characterize the structural and functional information of the vascularization, advanced optical imaging methods are needed that span different spatial and temporal scales [111]. In the routine culture of VOoC, researchers commonly rely on simple inverted fluorescence microscopy (IFM) to assess the growth status of the microvasculature. However, for more accurate imaging and analysis of 3D settings, other imaging techniques, such as CFM/MPM, fMOST, and LSFM, have become mainstream choices. The quality and accuracy of the acquired image data significantly influence the subsequent data analysis.
CFM/MPM
CFM and MPM techniques are favored in practical applications for their excellent imaging quality and compatibility with VOoC systems (Fig. 4a). CFM not only allows the acquisition of 3D vascular data in VOoC but also enables the tracking of fluorescent particles/cells (Fig. 4b-II) and the assessment of microvascular structure and function through perfusion of FITC-Dextran solutions (Fig. 4b-V) [14, 83]. However, traditional CFM has a limited imaging depth of approximately 100 µm [112]. In contrast, MPM overcomes this limitation by enabling deeper and higher-quality imaging using longer-wavelength excitation lasers, leveraging non-linear optical effects, reducing scattering, and minimizing fluorescence excitation outside the region of interest. Two-photon fluorescence microscopy has been widely employed for imaging deep layers of organoids, including the brain [113], intestine [114], and pancreas [115]. And three-photon microscopy has achieved intact imaging of label-free and non-transparent tissue in cerebral organoids at a depth of approximately 2 mm, providing a valuable option for tracking organoid growth [116].
Optical imaging methods applicable to VOoC. a Schematic representation of CFM for imaging VOoC. b-I Co-culture of HepG2 organoid and vascular network on-a-chip. Reproduced with permission [14]. Copyright 2022, Elsevier. b-II Tumor cells are arrested in two microvasculatures in VOoC. Reproduced under CC-BY-NC-ND license [107]. Copyright 2022, The Authors, published by Wiley–VCH GmbH. b-III Vascularized kidney organoids on-a-chip. Reproduced under exclusive license to Springer Nature America, Inc [136]. Copyright 2019, The Authors, published by Springer Nature. b-IV Immunofluorescence staining for CD31 of VOoC. Reproduced under exclusive license to Springer Nature America, Inc. [33]. Copyright 2022, The Authors, published by Springer Nature. b-V Perfusion testing with 70-kDa FITC-Dextran solution in VOoC. Reproduced with permission [24]. Copyright 2022, Royal Society of Chemistry. c Schematic representation of fMOST applied to cerebral organoids. d Z-projection of cerebral organoids showing GFAP + cells (d-I) and PI-labeled cytoarchitecture (d-II). Reproduced under terms of the CC-BY license [11]. Copyright 2022, The Authors, published by MDPI, Basel, Switzerland. e Schematic representation of open-top LSFM applied to VOoC. f-I Microvasculature images captured by LSFM and its 3D reconstruction. Reproduced with permission [131]. Copyright 2020, Elsevier. f-II 3D digital reconstruction of immunofluorescence on CLARITY-processed bioprinted tissue stained for CD31. Reproduced under terms of the CC-BY-NC-ND license [128]. Copyright 2018, The Authors, published by Elsevier. f-III Tracing the process of microvascular sprouting. Reproduced under the terms of the Optica Open Access Publishing Agreement [122]. Copyright 2022, The Authors, published by Optica Publishing Group. f-IV Quantifying the absolute displacement near the surface of the vascular sprout. Reproduced under terms of the CC-BY license [133]. The Authors, published by Public Library of Science. f-V 3D reconstruction of tumor spheroid. Reproduced under the terms of the Optica Open Access Publishing Agreement [129]. Copyright 2023, The Authors, published by Optica Publishing Group. Created with BioRender.com
Classic confocal systems, utilizing point scanning and photomultiplier tube detectors, are relatively time-consuming and can result in photobleaching [117]. Although techniques like MPM or spinning disk confocal microscopy can enhance imaging depth and speed, there is still a demand for further advancements in efficient and accurate 3D imaging acquisition of entire organoids. To address this challenge, a combination of optical sectioning and mechanical slicing can be employed for large samples, such as sequential two-photon tomography and fMOST.
fMOST
fMOST is a technique that combines fluorescence microscopy imaging with a microtome to achieve high-resolution full-size 3D imaging. It is widely used for reconstructing large-scale samples. The process involves embedding the entire sample in resin, followed by fluorescence imaging using a laser scanning microscope to provide high sensitivity. To ensure imaging stability and speed, a non-mechanical acousto-optical deflector is employed as the scanner, with the scan mode set to rapid frequency swap. Additionally, the sample can be immersed in a fluorescent dye solution, facilitating real-time staining (Fig. 4c) [118,119,120]. Ma et al. carried out high-resolution 3D imaging of cerebral organoids using fMOST, achieving 3D visualization of GFAP + signal and continuous spatial distribution of neurons. The authors revealed regional nuclear size and density difference within cerebral organoids (Fig. 4d) [11]. While the majority of VOoC substrates are presently composed of glass, limiting direct application to fMOST, the vascularized organoids can be retrieved from the VOoC for subsequent fixing and embedding and subjected to fMOST for imaging and analysis. Furthermore, the chip’s substrate can be substituted with a flexible material, such as PDMS, to facilitate sectioning [121]. In summary, fMOST provides a powerful imaging platform that offers a new option for visualizing VOoC in future.
LSFM
The LSFM generates a thin light sheet to selectively excite samples at a specific plane, while the emitted fluorescence is detected orthogonally. This enables 3D imaging with higher acquisition efficiency and reduced photobleaching and phototoxicity compared to other imaging techniques [122]. LSFM has been extensively used for 3D imaging of various real tissues and organs [123,124,125], cerebral organoids [126, 127], colon organoids [10], tumor spheroids [128, 129], pancreas organoids [130], etc.
In the context of organoids, LSFM and similar imaging techniques play a crucial role in characterizing the angiogenesis and vasculogenesis processes [122]. For example, Atlas et al. employed digital scanned light-sheet microscopy to observe the formation process of a dense microvascular network in a collagen hydrogel and quantified relevant generation parameters (Fig. 4f-I) [131]. Sebastian et al. used LSFM to visualize a pre-vascularized biological scaffold derived from rat intestines with intact arterio-venous capillary loops, allowing analysis of the distribution of typical endothelial markers CD31 and vWF factor (Fig. 4f-II) [132]. Bissardon et al. utilized LSFM to observe and track the angiogenesis process on a microfluidic chip, providing critical analytical parameters for understanding microvasculature formation (Fig. 4f-III) [122]. Steuwe et al. applied LSFM to quantitatively measure the subcellular level displacement of angiogenic sprouts near the surface (Fig. 4f-IV) [133].
LSFM systems with conventional orthogonal setups require special sample mounting methods to accommodate their orthogonal dual-objective configurations. Therefore, they are not well suited for imaging samples contained within a flat microchip. Additionally, in traditional microfluidic chips using materials like PDMS, the RI of PDMS itself differs significantly from that of the samples in the hydrogel channels, which is unfavorable for light-sheet imaging systems. To address this issue, Zhu et al. developed an open-top optical system and combined it with a novel chip material, Bio 133, allowing better imaging results in tumor spheroids and enabling high-throughput screening of anti-tumor drugs (Fig. 4f-V) [129].
LSFM is a powerful technique for observing the dynamic process of microvasculature growth in VOoC with speed and accuracy. This capability is crucial for studying and understanding the interaction between tissues and blood vessels in vitro [134]. Additionally, when combined with optical clearing (see section below), LSFM provides an essential means to obtain the 3D structure of tissue samples [135].
Tissue optical clearing on the chip
With the advancement of microfluidic and OoC technologies, tissue on chips increasingly resembles the structures and functions of real organs. However, these 3D cultures face several imaging challenges, such as adjusting the sample focal distance, handling tissue refraction variability, and addressing reduced light penetration caused by tissue opacity [121].
In general, light attenuation in biological tissue primarily arises from the absorption and scattering of light by its internal components. For instance, heme, abundant in residual blood within tissues, exhibits high light absorption within the visible light range (400–600 nm) and is a primary contributor to light intensity attenuation in tissues. High scattering is the leading cause of tissue turbidity, mainly caused by the difference in the RI between tissue components. On a macro scale, most biological tissues consist of water with a low RI, along with lipids, proteins, and other substances with a high RI. The primary objective of emerging TOC technology is to reduce tissue absorption and scattering (Fig. 5a) [15]. This can be achieved through dehydration, lipid removal, decalcification, decolorization, RI matching, and other approaches while also aiming to preserve the fluorescence signal and target protein molecules within the tissue to the greatest extent possible [137, 138].
TOC on the chip. a TOC on the chip greatly improves the imaging depth and quality of vascularized organoids by reducing scattering through refractive index matching. b Schematic diagram of the XY- and Z-views of vascularized HepG2 spheroids before and after clearing. Reproduced with permission [14]. Copyright 2022, Elsevier. c Schematic of on-chip clearing, as well as the tumor spheroids before and after clearing. Reproduced with permission [8]. Copyright 2016, National Academy of Science. d The TOC process utilizes mLSI chips and depicts the cleared hACS spheroids in bright-field imaging. Reproduced with permission [13]. Copyright 2017, Royal Society of Chemistry
Currently, existing TOC methods, can be categorized into ex vivo clearing methods [139] and in vivo clearing methods [140,141,142]. However, most of these methods involve relatively complex reagent exchange steps, which are labor intensive and can easily result in damage or loss of microtissues. Microfluidic systems, on the other hand, possess excellent fluid control capabilities, enabling efficient and rapid liquid exchange during the clearing process. Therefore, microfluidic systems are advantageous for achieving fully automatic clearing. However, it is worth noting that microfluidic systems also have certain limitations. For instance, the size of the chip limits the number of access ports, making it challenging to achieve complex clearing processes. Additionally, some reagents used in solvent-based clearing methods may have corrosive properties, potentially causing damage to the tubes connecting the microfluidic system and the chip itself.
With the advancement of organoid and OoC technology, obtaining deep tissue information from organoids has become a prominent area of research. Ahn et al. improved the CUBIC method, allowing for observing vascular marker signal CD31 at the depth of the HepG2 tumor spheroid (Fig. 5b) [14]. Chen et al. integrated spheroid formation, CLARITY clearing, and imaging into a spiral-structured chip for quick labeling, clearing, and analysis and showed significantly enhanced imaging depth for spheroids (Fig. 5c) and successful visualization of Langerhans’ vascular structure from islets [8]. Santisteban et al. developed a microfluidic large-scale integration (mLSI) chip platform for high-throughput, integrated CLARITY clearing (Fig. 5d). The extraction time for lipids from adipose-derived stem cell (hASC) spheroids was reduced by utilizing pH change osmotic pumping [13]. Briana et al. optimized the Scale clearing method in combination with vibratome sectioning to visualize thick neural layers in the spinal cord chips [121]. Grist et al. utilized SeeDB, ClearT2, and ScaleSQ with microfluidic chips for high-throughput and fast optical clearing of breast cancer spheroids [9]. Zhang et al. employed a simplified CLARITY for passive tissue clearing and lens-free microscopy for low-cost and efficient imaging [12]. Oh et al. developed a transfer-free 3D micro-tumor culture and visualization system, effectively improving imaging depth and facilitating high-content tumor phenotypic analysis [143]. Further, Yu et al. proposed an on-chip clearing method for imaging live 3D cell cultures, offering a new strategy for dynamically monitoring deep tissue in VOoC [144].
Vascular analysis methods of VOoC
Classic analysis tools
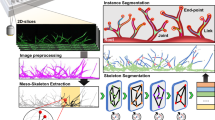
Vascular analysis of VOoC involves quantifying key parameters, such as vascular area, number of branches, and diameter; moreover, it involves tracking the movement trajectory of fluorescent particles to assess vascular perfusibility and connectivity. The standard pipeline for investigating microvascular networks comprises image preprocessing, image binarization, skeletonization of the vascular network, and analysis of characteristic parameters (Fig. 6a). The vascular data in VOoC often deviate significantly from real vessels in terms of morphology and structure. Researchers often employ analysis tools with intuitive user interfaces and real-time feedback on results, such as Fiji, Imaris, and AngioTool, which are widely used for biological image analysis. These tools allow researchers to adjust parameters based on the results to ensure analysis reliability. Moreover, some researchers develop analysis algorithms specifically designed for the characteristics of developing VOoC models, thereby improving the efficiency and accuracy of the analysis for microvasculature. In the following, we present several examples showcasing the application of these classical tools for the analysis of VOoC.
Classic vascular analysis methods of VOoC. a Classic vascular network analysis pipeline. b Fiji (ImageJ) software was employed to investigate vasculature growth and demonstrate the improved connectivity of microvascular networks cultured under flow stimulation. Reproduced with permission [47]. Copyright 2021, Royal Society of Chemistry. c Imaris was utilized for 3D reconstruction of blood vessels ((c-I) Reproduced with permission [151]. Copyright 2021, American Chemical Society and (c-II) Reproduced under the CC-BY license [149]. Copyright 2022, The Authors, published by Elsevier), as well as particle tracking of microbeads or fluorescent particles ((c-III) Reproduced with permission [150]. Copyright 2014, Society for Laboratory Automation and Screening, and (c-IV) Reproduced with permission [14]. Copyright 2022, Elsevier). d Schematic representation of AngioTool analysis, wherein the yellow line denotes the outline of the blood vessel, the red line indicates the centerline of the blood vessel, and the blue point represents the branch point of the blood vessel. Reproduced under the Creative Commons CC0 public domain dedication [152]. Copyright 2011, The Authors, published by Public Library of Science. e Schematic representation of angiogenic sprout tracking on the chip, using disk fitting (j disks) to model the angiogenic process with the disk radius representing the blood vessel radius. Reproduced with permission [154]. Copyright 2013, Elsevier
ImageJ/Fiji
Fiji, a distribution of the open-source software ImageJ specifically designed for biological image analysis, offers rapid algorithm design and application through its powerful software library and a broad range of scripting languages [145]. The software’s robust image processing capabilities and plug-in libraries make it convenient for analyzing microvascular networks formed on the chip. For instance, Winkelman et al. utilized ImageJ to project the maximum value of confocal imaging data, binarize the result to analyze the vascular area, and skeletonize the binary image to assess parameters, such as the number of branches, branch length, and diameter. This analysis demonstrated the promotion of angiogenesis under flow stimulation (Fig. 6b) [47]. However, the traditional binarization method used after maximum projection can introduce errors along the Z-axis. To address this, Lee et al. manually divided the image data into upper and lower parts before projection and transformed it into 2D binary mask images to measure vascular morphology (diameter, area, branch) and evaluate differences in vascular growth in diverse culture environments [146]. Additionally, Fiji plug-ins such as Analyze Particles, Analyze Skeleton (2D/3D), and Automated Sprout Analysis significantly enhance the accuracy and efficiency of vascular analysis [147, 148].
Imaris
Imaris is a powerful software tool designed for the 3D reconstruction and analysis of biomedical images. It provides a convenient image preprocessing tool for vascular analysis in scientific research, including deconvolution, signal enhancement, denoising, and more. It also enables the reconstruction of the 3D structure of blood vessels (Fig. 6c-I, II) to analyze spatial relationships between vessels and can be used for the analysis of volume changes in vessels or organoids. With the powerful 3D reconstruction capability, Imaris facilitates intuitive visualization of fluorescent particles within microvascular lumen based on the z-stack images (Fig. 6c-III) [149,150,151]. Ahn et al. used Imaris software to 3D render and visualize the vascularized tumor spheroid structures. The ‘surface’ tool in Imaris was used to reconstruct the blood vessels and the spheroids, while the ‘spot’ tool was employed to reconstruct the flow path of fluorescent beads (Fig. 6c-IV) [14].
AngioTool
AngioTool is a lightweight and user-friendly software developed by Zutaire et al. It is specifically designed to quantitatively analyze vascular networks with repeatability and ease. The software employs a series of algorithms to process the input image. First, the image is convolved with the fast recursive Gaussian kernel and tube-like structures to segment the blood vessel. Then, based on a combination of the eigenvalues of the 2D Hessian matrix, the software computes the vascular structures. AngioTool automatically calculates and outputs various parameters, such as vascular area, length, and density, according to user-defined vascular diameter. It also provides desktop applications, is particularly suitable for entry-level researchers as it displays real-time segmentation and recognition results on the image, allowing users to adjust parameters easily (Fig. 6d) [152]. This software has demonstrated impressive performance in the quantitative analysis of VOoC, including colon organoids [70], co-cultured embryoid bodies—tumor spheroids organoids [153], and NVU organoids [86].
Other methods
Unlike the analysis of microvascular networks, the angiogenesis process is highly dynamic and relatively complex, requiring high-performance algorithms. Common processing steps include image stitching, registration, vascular segmentation, vascular skeletonization, and branch tracking. The Bayesian filtering framework is particularly useful in determining the correlation between ECs in the sprouting process and treating the tip/stalk cell, lumen, and filopodia as separate parts to predict sprout motion (Fig. 6e) [154]. Some algorithms combine convolutional neural network (CNN) and support vector machine (SVM) to improve vessel segmentation and skeletonization [155, 156]. To enhance the algorithm’s robustness, some research groups have explored using the total cell cytosolic integral and the total nuclear integral invading the gel region above the monolayer to quantify the sprouting activity and directed migration. This approach has shown low sensitivity to signal noise, outliers, and diverse shapes and sizes of sprouts [27]. Furthermore, utilizing differential equations or mathematical models to simulate the process of angiogenesis regulated by tumor or growth factors can help strengthen experimental parameter control and provide new insights into the angiogenesis process [157, 158].
In summary, classical vascular analysis methods predominantly utilize software or traditional image processing and analysis algorithms, sometimes in combination. Manual debugging and proofreading can yield relatively accurate results but are limited in achieving fully automatic analysis. Although there have been efforts to incorporate deep learning into vascular analysis, the current focus has primarily been on tasks such as image segmentation and parameter classification. Therefore, a more effective combination of deep learning and VOoC analysis methods will likely be the future development trend of this field.
Deep learning-assisted vascular analysis
With the increasing popularity and development of deep learning, powerful toolkits such as TensorFlow and PyTorch have emerged. Furthermore, in recent years, MATLAB has provided a comprehensive deep learning network toolbox, which has alleviated the environmental set-up difficulties and simplified the usage of deep learning. In general, the application of a deep learning network involves the following key steps: (1) data acquisition, (2) data preprocessing, (3) design requirements, (4) network selection and training model, (5) model evaluation, (6) hyperparameter optimization, and (7) model deploying (Fig. 7). In different scenarios, each step may require adjustment. Notably, the data acquisition phase often requires considerable time and effort. In deep learning tasks, careful attention must be given to data processing, network model selection, and training. During hyperparameter optimization, the choice of an appropriate loss function and adjusting other influential hyperparameters, such as learning rate and batch size, can notably enhance the performance of the model [17, 21].
Currently, many studies have successfully integrated deep learning with vascular analysis [159], particularly focusing on the vasculature of real organs [160,161,162]. Diverse approaches were developed, including VesSAP, DeepVess, and TubeMAP, for segmenting and quantifying brain blood vessels in different disease models [125, 163, 164]; Singapore I vessel assessment system for measuring the diameter of human retinal blood vessels [161]; and topology and width aware generative adversarial network for retinal artery/vein classification [165]. Moreover, deep learning networks were used to differentiate vessel invasion and non-invasion in various cancers and identify tumor angiogenesis [166,167,168].
Deep learning relies on a substantial amount of data to achieve optimal performance, and OoC technology represents a cost-effective and high-throughput approach to generating the necessary training samples for deep learning networks. This combination between deep learning and OoC enables automated and intelligent data analysis [21, 169, 170]. The requisite data volume for deep learning on OoC varies based on the specific task and the type of network employed. Common image processing tasks and CNNs typically utilize datasets ranging from hundreds to thousands of images [171,172,173,174]. In the case of OoC generated from precious samples, where obtaining large datasets may be challenging, subsequent enhancements in network performance can be achieved through data augmentation methods (e.g., flipping, rotation, scaling, and noise injection). In recent years, there have been notable advancements in the field of integrating deep learning with OoC [175], with a primary focus on applications, such as recognition, classification [176, 177], target tracking [173, 178], aid imaging [179, 180], drug analysis, and disease diagnosis [172, 181].
Recently, deep learning techniques have been used in the vascular analysis of VOoC. For instance, Urban et al. developed a deep learning model capable of effectively classifying image data before and after drug treatment on the chip, enabling the screening of compounds that can disrupt the vascular network [171]. Irisa et al. developed a deep learning-based visual methodology in conjunction with a vasculature-on-a-chip model to recognize and classify developmental phases of vascular networks [182]. Tronolone et al. employed a random forest model to predict the linear relationship between oxygen transport and microvascular network characteristic parameters in VOoC [183]. For tracking angiogenesis, Wang et al. developed an automated tracking system for blood vessels that can extract quantitative information about the blood vessels and track their formation. They utilized machine learning techniques to enhance the effectiveness of blood vessel skeletonization, making the system achieved a high precision of 97.3% and a recall of 93.9% compared to the ground truth [156]. Subsequently, Wang et al. further optimized the efficiency of EC tracking using CNN and combined it with a Kalman filter to rapidly track and accurately analyze the expansion and migration of ECs during angiogenesis [155].
In summary, deep learning has emerged as a valuable tool in vascular analysis across various fields. However, its application specifically in the context of VOoC is still limited. Influenced by various factors, including cell types, seeding density, and hydrogel strength, microvascular data on VOoC tends to exhibit relative instability. Moreover, the throughput of current VOoC microphysiological models is relatively low, making it challenging to acquire a large volume of data suitable for training purposes. Establishing a versatile deep learning dataset and analysis network applicable to diverse situations poses a challenge. To tackle this issue, a potential solution involves designing a deep learning tool for VOoC analysis that accommodates both training and analysis, which should allow users to integrate new data during usage, facilitating the update of analysis network weights. This approach may enhance the tool’s adaptability to diverse VOoC data, thereby improving the robustness of the network. As VOoC continues to advance, there is significant potential for leveraging deep learning techniques to detect the growth status of microvasculature, diagnose diseases and test drugs (Fig. 7). By combining deep learning with VOoC, researchers can unlock new opportunities for automated analysis, improved disease understanding, and more effective drug development. Further exploration and development of deep learning methods in the context of VOoC hold promise for advancing our understanding of vascular biology and enhancing healthcare outcomes.
Discussion and future perspectives
In this review, we provide an overview of various chip designs, vascularization culturing strategies, and imaging methods utilized in VOoC, as well as classic and deep learning-based vascular analysis methods. The essential information is summarized in Table 1.
The complex vascular environment in vivo is characterized by intricate cell–cell and cell–matrix signaling networks, coupled with diverse flow status (e.g., flow rate, flow direction, and shear stress), and it varies across different organs and vascular structures [184]. For instance, the vascular walls in the arterial system are thicker, enabling them to withstand higher blood pressure and maintain a relatively unidirectional flow, while the distal capillaries typically exhibit slow and multidirectional blood flow, facilitating efficient substance exchange; whereas lymphatic vessels incorporate blind-ended structures that permit only one-way flow, optimizing waste removal and supporting immune functions [185]. However, the reported VOoC encounters some limitations in mimicking human physiological conditions and the environment, for example, the hydrogel channel height of reported VOoC is typically 100–200 μm, the flow pattern is generally simple, and the types of cells used for vascularization are also limited. Therefore, constructing a more realistic vascular network remains challenging, requiring targeted adjustments to the biochemical and mechanical environment based on the types of vascularized organoids. Common approaches involve regulating concentrations and types of growth factors, establishing appropriate flow status, or selecting co-cultured cell types [186]. Additionally, some researchers are addressing this challenge from the perspective of microvascular structure, aiming to build microvascular networks with complex hierarchical structures and lymphatic vessels featuring blind-ended structures [97, 187].
Furthermore, the increased thickness of tissues poses challenges in terms of imaging time and information acquisition in deep tissue. LSFM and TOC techniques have provided valuable assistants in imaging thick tissues. However, LSFM’s performance can be influenced by the chip’s structure and the on-chip TOC is primarily applicable to fixed samples, limiting their applications in real-time dynamic observation of microvascular networks. Therefore, optimizing the design of VOoC to enhance compatibility with imaging systems and realizing TOC for imaging living organoids are directions that warrant further exploration.
In terms of data processing, classic methods that rely on much manual work are predominantly employed for VOoC analysis currently. Consequently, the integration of deep learning is expected to improve vascular analysis efficiency and accuracy in VOoC significantly. Continued advances in VOoC technology and deep learning analysis methods will contribute to a better understanding of microvasculature and its role in human health and disease.
The human body is a complex and dynamic system that relies on the coordinated interactions of multiple cells and tissues. Therefore, it is of great importance to explore suitable cell interactions and develop a multi-cell co-culture OoC system for drug development and disease diagnosis. The incorporation of a vascular network into a microphysiological system on chips to build VOoC can further enhance the authenticity of OoC platforms. However, the structure of the VOoC system is relatively simple due to limitations in micro/nanofabrication technology and cell culture, typically with a standard thickness of 100–200 μm. While a simple monolayer vascular network can be constructed within this thickness, constructing a thicker and more complex vascular layer that closely resembles human physiology is a crucial object in this direction [83]. However, realizing an ideal thick vascular network presents challenges in imaging and data analysis. Researchers should consider how to strike a balance between these aspects, which is a topic worthy of careful consideration.
In conclusion, it is crucial to establish a comprehensive and efficient pipeline encompassing VOoC fabrication, culturing, clearing, imaging, and analysis methods to study microvasculature growth and enable quantitative analysis of related diseases.
References
Ma C, Peng Y, Li H, Chen W (2021) Organ-on-a-chip: a new paradigm for drug development. Trends Pharmacol Sci 42(2):119–133. https://doi.org/10.1016/j.tips.2020.11.009
Ingber DE (2022) Human organs-on-chips for disease modelling, drug development and personalized medicine. Nat Rev Genet 23(8):467–491. https://doi.org/10.1038/s41576-022-00466-9
Bhatia SN, Ingber DE (2014) Microfluidic organs-on-chips. Nat Biotechnol 32(8):760–772. https://doi.org/10.1038/nbt.2989
Jain RK, Au P, Tam J, Duda DG, Fukumura D (2005) Engineering vascularized tissue. Nat Biotechnol 23(7):821–823. https://doi.org/10.1038/nbt0705-821
Shirure VS, Hughes CCW, George SC (2021) Engineering vascularized organoid-on-a-chip models. Annu Rev Biomed Eng 23:141–167. https://doi.org/10.1146/annurev-bioeng-090120-094330
Park SE, Georgescu A, Huh D (2019) Organoids-on-a-chip. Science 364(6444):960–965. https://doi.org/10.1126/science.aaw7894
Sonntag F, Schilling N, Mader K, Gruchow M, Klotzbach U, Lindner G, Horland R, Wagner I, Lauster R, Howitz S, Hoffmann S, Marx U (2010) Design and prototyping of a chip-based multi-micro-organoid culture system for substance testing, predictive to human (substance) exposure. J Biotechnol 148(1):70–75. https://doi.org/10.1016/j.jbiotec.2010.02.001
Chen YY, Silva PN, Syed AM, Sindhwani S, Rocheleau JV, Chan WC (2016) Clarifying intact 3D tissues on a microfluidic chip for high-throughput structural analysis. Proc Natl Acad Sci U S A 113(52):14915–14920. https://doi.org/10.1073/pnas.1609569114
Grist SM, Nasseri SS, Poon T, Roskelley C, Cheung KC (2016) On-chip clearing of arrays of 3-D cell cultures and micro-tissues. Biomicrofluidics 10(4):044107. https://doi.org/10.1063/1.4959031
Dekkers JF, Alieva M, Wellens LM, Ariese HCR, Jamieson PR, Vonk AM, Amatngalim GD, Hu H, Oost KC, Snippert HJG, Beekman JM, Wehrens EJ, Visvader JE, Clevers H, Rios AC (2019) High-resolution 3D imaging of fixed and cleared organoids. Nat Protoc 14(6):1756–1771. https://doi.org/10.1038/s41596-019-0160-8
Ma H, Chen J, Deng Z, Sun T, Luo Q, Gong H, Li X, Long B (2022) Multiscale analysis of cellular composition and morphology in intact cerebral organoids. Biology (Basel) 11(9):1270. https://doi.org/10.3390/biology11091270
Zhang Y, Shin Y, Sung K, Yang S, Chen H, Wang H, Teng D, Rivenson Y, Kulkarni RP, Ozcan A (2017) 3D imaging of optically cleared tissue using a simplified CLARITY method and on-chip microscopy. Sci Adv 3(8):e1700553. https://doi.org/10.1126/sciadv.1700553
Silva Santisteban T, Rabajania O, Kalinina I, Robinson S, Meier M (2017) Rapid spheroid clearing on a microfluidic chip. Lab Chip 18(1):153–161. https://doi.org/10.1039/c7lc01114h
Ahn J, Kim DH, Koo DJ, Lim J, Park TE, Lee J, Ko J, Kim S, Kim M, Kang KS, Min DH, Kim SY, Kim Y, Jeon NL (2023) 3D microengineered vascularized tumor spheroids for drug delivery and efficacy testing. Acta Biomater 165:153–167. https://doi.org/10.1016/j.actbio.2022.10.009
Susaki EA, Ueda HR (2016) Whole-body and whole-organ clearing and imaging techniques with single-cell resolution: toward organism-level systems biology in mammals. Cell Chem Biol 23(1):137–157. https://doi.org/10.1016/j.chembiol.2015.11.009
Zhao S, Todorov MI, Cai R, Maskari RA, Steinke H, Kemter E, Mai H, Rong Z, Warmer M, Stanic K, Schoppe O, Paetzold JC, Gesierich B, Wong MN, Huber TB, Duering M, Bruns OT, Menze B, Lipfert J, Puelles VG, Wolf E, Bechmann I, Erturk A (2020) Cellular and molecular probing of intact human organs. Cell 180(4):796–812e719. https://doi.org/10.1016/j.cell.2020.01.030
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Mencattini A, Di Giuseppe D, Comes MC, Casti P, Corsi F, Bertani FR, Ghibelli L, Businaro L, Di Natale C, Parrini MC, Martinelli E (2020) Discovering the hidden messages within cell trajectories using a deep learning approach for in vitro evaluation of cancer drug treatments. Sci Rep 10(1):7653. https://doi.org/10.1038/s41598-020-64246-3
Bannon D, Moen E, Schwartz M, Borba E, Kudo T, Greenwald N, Vijayakumar V, Chang B, Pao E, Osterman E, Graf W, Van Valen D (2021) DeepCell Kiosk: scaling deep learning-enabled cellular image analysis with Kubernetes. Nat Methods 18(1):43–45. https://doi.org/10.1038/s41592-020-01023-0
Haberl MG, Churas C, Tindall L, Boassa D, Phan S, Bushong EA, Madany M, Akay R, Deerinck TJ, Peltier ST, Ellisman MH (2018) CDeep3M-Plug-and-Play cloud-based deep learning for image segmentation. Nat Methods 15(9):677–680. https://doi.org/10.1038/s41592-018-0106-z
Rota A, Possenti L, Offeddu GS, Senesi M, Stucchi A, Venturelli I, Rancati T, Zunino P, Kamm RD, Costantino ML (2023) A three-dimensional method for morphological analysis and flow velocity estimation in microvasculature on-a-chip. Bioeng Transl Med 8(5):e10557. https://doi.org/10.1002/btm2.10557
Tetteh G, Efremov V, Forkert ND, Schneider M, Kirschke J, Weber B, Zimmer C, Piraud M, Menze BH (2020) DeepVesselNet: vessel segmentation, centerline prediction, and bifurcation detection in 3-D angiographic volumes. Front Neurosci 14:592352. https://doi.org/10.3389/fnins.2020.592352
Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF, Lam WA (2018) Microvasculature-on-a-chip for the long-term study of endothelial barrier dysfunction and microvascular obstruction in disease. Nat Biomed Eng 2:453–463. https://doi.org/10.1038/s41551-018-0224-z
Li Q, Niu K, Wang D, Xuan L, Wang X (2022) Low-cost rapid prototyping and assembly of an open microfluidic device for a 3D vascularized organ-on-a-chip. Lab Chip 22(14):2682–2694. https://doi.org/10.1039/d1lc00767j
Kim Y, Ko J, Shin N, Park S, Lee SR, Kim S, Song J, Lee S, Kang KS, Lee J, Jeon NL (2022) All-in-one microfluidic design to integrate vascularized tumor spheroid into high-throughput platform. Biotechnol Bioeng 119(12):3678–3693. https://doi.org/10.1002/bit.28221
Myers DR, Lam WA (2021) Vascularized microfluidics and their untapped potential for discovery in diseases of the microvasculature. Annu Rev Biomed Eng 23:407–432. https://doi.org/10.1146/annurev-bioeng-091520-025358
Farahat WA, Wood LB, Zervantonakis IK, Schor A, Ong S, Neal D, Kamm RD, Asada HH (2012) Ensemble analysis of angiogenic growth in three-dimensional microfluidic cell cultures. PLoS ONE 7(5):e37333. https://doi.org/10.1371/journal.pone.0037333
Zeinali S, Bichsel CA, Hobi N, Funke M, Marti TM, Schmid RA, Guenat OT, Geiser T (2018) Human microvasculature-on-a chip: anti-neovasculogenic effect of nintedanib in vitro. Angiogenesis 21(4):861–871. https://doi.org/10.1007/s10456-018-9631-8
Kim J, Lee KT, Lee JS, Shin J, Cui B, Yang K, Choi YS, Choi N, Lee SH, Lee JH, Bahn YS, Cho SW (2021) Fungal brain infection modelled in a human-neurovascular-unit-on-a-chip with a functional blood–brain barrier. Nat Biomed Eng 5(8):830–846. https://doi.org/10.1038/s41551-021-00743-8
Wang L, Tao T, Su W, Yu H, Yu Y, Qin J (2017) A disease model of diabetic nephropathy in a glomerulus-on-a-chip microdevice. Lab Chip 17(10):1749–1760. https://doi.org/10.1039/c7lc00134g
Vulto P, Podszun S, Meyer P, Hermann C, Manz A, Urban GA (2011) Phaseguides: a paradigm shift in microfluidic priming and emptying. Lab Chip 11(9):1596–1602. https://doi.org/10.1039/c0lc00643b
Kramer B, Corallo C, van den Heuvel A, Crawford J, Olivier T, Elstak E, Giordano N, Vulto P, Lanz HL, Janssen RAJ, Tessari MA (2022) High-throughput 3D microvessel-on-a-chip model to study defective angiogenesis in systemic sclerosis. Sci Rep 12(1):16930. https://doi.org/10.1038/s41598-022-21468-x
Hajal C, Offeddu GS, Shin Y, Zhang S, Morozova O, Hickman D, Knutson CG, Kamm RD (2022) Engineered human blood–brain barrier microfluidic model for vascular permeability analyses. Nat Protoc 17(1):95–128. https://doi.org/10.1038/s41596-021-00635-w
van Duinen V, Zhu D, Ramakers C, van Zonneveld AJ, Vulto P, Hankemeier T (2019) Perfused 3D angiogenic sprouting in a high-throughput in vitro platform. Angiogenesis 22(1):157–165. https://doi.org/10.1007/s10456-018-9647-0
Soragni C, Queiroz K, Ng CP, Stok A, Olivier T, Tzagkaraki D, Heijmans J, Suijker J, de Ruiter SPM, Olczyk A, Bokkers M, Schavemaker F, Trietsch SJ, Lanz HL, Vulto P, Joore J (2023) Phenotypic screening in Organ-on-a-Chip systems: a 1537 kinase inhibitor library screen on a 3D angiogenesis assay. Angiogenesis. https://doi.org/10.1007/s10456-023-09888-3
Wevers NR, Nair AL, Fowke TM, Pontier M, Kasi DG, Spijkers XM, Hallard C, Rabussier G, van Vught R, Vulto P, de Vries HE, Lanz HL (2021) Modeling ischemic stroke in a triculture neurovascular unit on-a-chip. Fluids Barriers CNS 18(1):59. https://doi.org/10.1186/s12987-021-00294-9
Lyu Z, Park J, Kim KM, Jin HJ, Wu H, Rajadas J, Kim DH, Steinberg GK, Lee W (2021) A neurovascular-unit-on-a-chip for the evaluation of the restorative potential of stem cell therapies for ischaemic stroke. Nat Biomed Eng 5(8):847–863. https://doi.org/10.1038/s41551-021-00744-7
Bonanini F, Kurek D, Previdi S, Nicolas A, Hendriks D, de Ruiter S, Meyer M, ClapesCabrer M, Dinkelberg R, Garcia SB, Kramer B, Olivier T, Hu H, Lopez-Iglesias C, Schavemaker F, Walinga E, Dutta D, Queiroz K, Domansky K, Ronden B, Joore J, Lanz HL, Peters PJ, Trietsch SJ, Clevers H, Vulto P (2022) In vitro grafting of hepatic spheroids and organoids on a microfluidic vascular bed. Angiogenesis 25(4):455–470. https://doi.org/10.1007/s10456-022-09842-9
Trietsch SJ, Naumovska E, Kurek D, Setyawati MC, Vormann MK, Wilschut KJ, Lanz HL, Nicolas A, Ng CP, Joore J, Kustermann S, Roth A, Hankemeier T, Moisan A, Vulto P (2017) Membrane-free culture and real-time barrier integrity assessment of perfused intestinal epithelium tubes. Nat Commun 8(1):262. https://doi.org/10.1038/s41467-017-00259-3
Wong JF, Mohan MD, Young EWK, Simmons CA (2020) Integrated electrochemical measurement of endothelial permeability in a 3D hydrogel-based microfluidic vascular model. Biosens Bioelectron 147:111757. https://doi.org/10.1016/j.bios.2019.111757
Wevers NR, Kasi DG, Gray T, Wilschut KJ, Smith B, van Vught R, Shimizu F, Sano Y, Kanda T, Marsh G (2018) A perfused human blood–brain barrier on-a-chip for high-throughput assessment of barrier function and antibody transport. Fluids Barriers CNS 15(1):1–12. https://doi.org/10.1186/s12987-018-0108-3
Sobrino A, Phan DT, Datta R, Wang X, Hachey SJ, Romero-Lopez M, Gratton E, Lee AP, George SC, Hughes CC (2016) 3D microtumors in vitro supported by perfused vascular networks. Sci Rep 6:31589. https://doi.org/10.1038/srep31589
Phan DTT, Wang X, Craver BM, Sobrino A, Zhao D, Chen JC, Lee LYN, George SC, Lee AP, Hughes CCW (2017) A vascularized and perfused organ-on-a-chip platform for large-scale drug screening applications. Lab Chip 17(3):511–520. https://doi.org/10.1039/c6lc01422d
Wang X, Phan DT, Sobrino A, George SC, Hughes CC, Lee AP (2016) Engineering anastomosis between living capillary networks and endothelial cell-lined microfluidic channels. Lab Chip 16(2):282–290. https://doi.org/10.1039/c5lc01050k
Hachey SJ, Movsesyan S, Nguyen QH, Burton-Sojo G, Tankazyan A, Wu J, Hoang T, Zhao D, Wang S, Hatch MM, Celaya E, Gomez S, Chen GT, Davis RT, Nee K, Pervolarakis N, Lawson DA, Kessenbrock K, Lee AP, Lowengrub J, Waterman ML, Hughes CCW (2021) An in vitro vascularized micro-tumor model of human colorectal cancer recapitulates in vivo responses to standard-of-care therapy. Lab Chip 21(7):1333–1351. https://doi.org/10.1039/d0lc01216e
Yue T, Zhao D, Phan DTT, Wang X, Park JJ, Biviji Z, Hughes CCW, Lee AP (2021) A modular microfluidic system based on a multilayered configuration to generate large-scale perfusable microvascular networks. Microsyst Nanoeng 7:4. https://doi.org/10.1038/s41378-020-00229-8
Winkelman MA, Kim DY, Kakarla S, Grath A, Silvia N, Dai G (2021) Interstitial flow enhances the formation, connectivity, and function of 3D brain microvascular networks generated within a microfluidic device. Lab Chip 22(1):170–192. https://doi.org/10.1039/d1lc00605c
Pauty J, Nakano S, Usuba R, Nakajima T, Johmura Y, Omori S, Sakamoto N, Kikuchi A, Nakanishi M, Matsunaga YT (2021) A 3D tissue model-on-a-chip for studying the effects of human senescent fibroblasts on blood vessels. Biomater Sci 9(1):199–211. https://doi.org/10.1039/d0bm01297a
Pauty J, Usuba R, Cheng IG, Hespel L, Takahashi H, Kato K, Kobayashi M, Nakajima H, Lee E, Yger F, Soncin F, Matsunaga YT (2018) A vascular endothelial growth factor-dependent sprouting angiogenesis assay based on an in vitro human blood vessel model for the study of anti-angiogenic drugs. EBioMedicine 27:225–236. https://doi.org/10.1016/j.ebiom.2017.12.014
Chen SW, Blazeski A, Zhang S, Shelton SE, Offeddu GS, Kamm RD (2023) Development of a perfusable, hierarchical microvasculature-on-a-chip model. Lab Chip 23(20):4552–4564. https://doi.org/10.1039/d3lc00512g
Selahi A, Fernando T, Chakraborty S, Muthuchamy M, Zawieja DC, Jain A (2022) Lymphangion-chip: a microphysiological system which supports co-culture and bidirectional signaling of lymphatic endothelial and muscle cells. Lab Chip 22(1):121–135. https://doi.org/10.1039/d1lc00720c
Nothdurfter D, Ploner C, Coraca-Huber DC, Wilflingseder D, Muller T, Hermann M, Hagenbuchner J, Ausserlechner MJ (2022) 3D bioprinted, vascularized neuroblastoma tumor environment in fluidic chip devices for precision medicine drug testing. Biofabrication 14(3):035002. https://doi.org/10.1088/1758-5090/ac5fb7
Andrique L, Recher G, Alessandri K, Pujol N, Feyeux M, Bon P, Cognet L, Nassoy P, Bikfalvi A (2019) A model of guided cell self-organization for rapid and spontaneous formation of functional vessels. Sci Adv 5(6):eaau6562. https://doi.org/10.1126/sciadv.aau6562
Kim D, Hwang KS, Seo EU, Seo S, Lee BC, Choi N, Choi J, Kim HN (2022) Vascularized lung cancer model for evaluating the promoted transport of anticancer drugs and immune cells in an engineered tumor microenvironment. Adv Healthc Mater 11(12):e2102581. https://doi.org/10.1002/adhm.202102581
Paek J, Park SE, Lu Q, Park KT, Cho M, Oh JM, Kwon KW, Yi YS, Song JW, Edelstein HI, Ishibashi J, Yang W, Myerson JW, Kiseleva RY, Aprelev P, Hood ED, Stambolian D, Seale P, Muzykantov VR, Huh D (2019) Microphysiological engineering of self-assembled and perfusable microvascular beds for the production of vascularized three-dimensional human microtissues. ACS Nano 13(7):7627–7643. https://doi.org/10.1021/acsnano.9b00686
Liu J, Zheng H, Dai X, Poh PSP, Machens HG, Schilling AF (2020) Transparent PDMS bioreactors for the fabrication and analysis of multi-layer pre-vascularized hydrogels under continuous perfusion. Front Bioeng Biotechnol 8:568934. https://doi.org/10.3389/fbioe.2020.568934
Zhao N, Guo Z, Kulkarni S, Norman D, Zhang S, Chung TD, Nerenberg RF, Linville R, Searson P (2022) Engineering the human blood–brain barrier at the capillary scale using a double-templating technique. Adv Funct Mater 32(30):2110289. https://doi.org/10.1002/adfm.202110289
Peng B, Tong Z, Tong WY, Pasic PJ, Oddo A, Dai Y, Luo M, Frescene J, Welch NG, Easton CD, Thissen H, Voelcker NH (2020) In Situ surface modification of microfluidic blood–brain–barriers for improved screening of small molecules and nanoparticles. ACS Appl Mater Interfaces 12(51):56753–56766. https://doi.org/10.1021/acsami.0c17102
Seo S, Jang M, Kim H, Sung JH, Choi N, Lee K, Kim HN (2022) Neuro-glia-vascular-on-a-chip system to assess aggravated neurodegeneration via brain endothelial cells upon exposure to diesel exhaust particles. Adv Funct Mater 33(12):2210123. https://doi.org/10.1002/adfm.202210123
Orellano I, Thomas A, Herrera A, Brauer E, Wulsten D, Petersen A, Kloke L, Duda GN (2022) Engineering vascular self-assembly by controlled 3D-printed cell placement. Adv Funct Mater 32(52):2208325. https://doi.org/10.1002/adfm.202208325
Meng F, Meyer CM, Joung D, Vallera DA, McAlpine MC, Panoskaltsis-Mortari A (2019) 3D bioprinted in vitro metastatic models via reconstruction of tumor microenvironments. Adv Mater 31(10):e1806899. https://doi.org/10.1002/adma.201806899
Bischel LL, Young EW, Mader BR, Beebe DJ (2013) Tubeless microfluidic angiogenesis assay with three-dimensional endothelial-lined microvessels. Biomaterials 34(5):1471–1477. https://doi.org/10.1016/j.biomaterials.2012.11.005
Bischel LL, Lee S-H, Beebe DJ (2012) A practical method for patterning lumens through ECM hydrogels via viscous finger patterning. J Lab Autom 17(2):96–103. https://doi.org/10.1177/2211068211426694
de Graaf MNS, Vivas A, Kasi DG, van den Hil FE, van den Berg A, van der Meer AD, Mummery CL, Orlova VV (2022) Multiplexed fluidic circuit board for controlled perfusion of 3D blood vessels-on-a-chip. Lab Chip 23(1):168–181. https://doi.org/10.1039/d2lc00686c
Jiang H, Li X, Chen T, Liu Y, Wang Q, Wang Z, Jia J (2023) Bioprinted vascular tissue: assessing functions from cellular, tissue to organ levels. Mater Today Bio 23:100846. https://doi.org/10.1016/j.mtbio.2023.100846
Terpstra ML, Li J, Mensinga A, de Ruijter M, van Rijen MH, Androulidakis C, Galiotis C, Papantoniou I, Matsusaki M, Malda J (2022) Bioink with cartilage-derived extracellular matrix microfibers enables spatial control of vascular capillary formation in bioprinted constructs. Biofabrication 14(3):034104. https://doi.org/10.1088/1758-5090/ac6282
Koch L, Deiwick A, Chichkov B (2021) Capillary-like formations of endothelial cells in defined patterns generated by laser bioprinting. Micromachines (Basel). https://doi.org/10.3390/mi12121538
Nikolaev M, Mitrofanova O, Broguiere N, Geraldo S, Dutta D, Tabata Y, Elci B, Brandenberg N, Kolotuev I, Gjorevski N (2020) Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 585(7826):574–578. https://doi.org/10.1038/s41586-020-2724-8
Enrico A, Voulgaris D, Ostmans R, Sundaravadivel N, Moutaux L, Cordier A, Niklaus F, Herland A, Stemme G (2022) 3D microvascularized tissue models by laser-based cavitation molding of collagen. Adv Mater 34(11):e2109823. https://doi.org/10.1002/adma.202109823
Rajasekar S, Lin DSY, Abdul L, Liu A, Sotra A, Zhang F, Zhang B (2020) IFlowPlate—a customized 384-well plate for the culture of perfusable vascularized colon organoids. Adv Mater 32(46):e2002974. https://doi.org/10.1002/adma.202002974
Mao M, He J, Lu Y, Li X, Li T, Zhou W, Li D (2018) Leaf-templated, microwell-integrated microfluidic chips for high-throughput cell experiments. Biofabrication 10(2):025008. https://doi.org/10.1088/1758-5090/aaa900
Mao M, Chen P, He J, Zhu G, Li X, Li D (2022) Deciphering fluid transport within leaf-inspired capillary networks based on a 3D computational model. Small 18(16):e2108102. https://doi.org/10.1002/smll.202108102
Yu J, Lee S, Song J, Lee SR, Kim S, Choi H, Kang H, Hwang Y, Hong YK, Jeon NL (2022) Perfusable micro-vascularized 3D tissue array for high-throughput vascular phenotypic screening. Nano Converg 9(1):16. https://doi.org/10.1186/s40580-022-00306-w
Ko J, Ahn J, Kim S, Lee Y, Lee J, Park D, Jeon NL (2019) Tumor spheroid-on-a-chip: a standardized microfluidic culture platform for investigating tumor angiogenesis. Lab Chip 19(17):2822–2833. https://doi.org/10.1039/c9lc00140a
Riddle RB, Jennbacken K, Hansson KM, Harper MT (2022) Endothelial inflammation and neutrophil transmigration are modulated by extracellular matrix composition in an inflammation-on-a-chip model. Sci Rep 12(1):6855. https://doi.org/10.1038/s41598-022-10849-x
Pampaloni F, Reynaud EG, Stelzer EH (2007) The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 8(10):839–845. https://doi.org/10.1038/nrm2236
Rodrigues J, Heinrich MA, Teixeira LM, Prakash J (2021) 3D in vitro model (r)evolution: unveiling tumor–stroma interactions. Trends Cancer 7(3):249–264. https://doi.org/10.1016/j.trecan.2020.10.009
Mansour AA, Goncalves JT, Bloyd CW, Li H, Fernandes S, Quang D, Johnston S, Parylak SL, Jin X, Gage FH (2018) An in vivo model of functional and vascularized human brain organoids. Nat Biotechnol 36(5):432–441. https://doi.org/10.1038/nbt.4127
Garreta E, Prado P, Tarantino C, Oria R, Fanlo L, Marti E, Zalvidea D, Trepat X, Roca-Cusachs P, Gavalda-Navarro A, Cozzuto L, Campistol JM, Izpisua Belmonte JC, Hurtado Del Pozo C, Montserrat N (2019) Fine tuning the extracellular environment accelerates the derivation of kidney organoids from human pluripotent stem cells. Nat Mater 18(4):397–405. https://doi.org/10.1038/s41563-019-0287-6
Tamai M, Adachi E, Kawase M, Tagawa YI (2022) Syngeneic implantation of mouse hepatic progenitor cell-derived three-dimensional liver tissue with dense collagen fibrils. World J Gastroenterol 28(14):1444–1454. https://doi.org/10.3748/wjg.v28.i14.1444
Cortez AR, Poling HM, Brown NE, Singh A, Mahe MM, Helmrath MA (2018) Transplantation of human intestinal organoids into the mouse mesentery: A more physiologic and anatomic engraftment site. Surgery 164(4):643–650. https://doi.org/10.1016/j.surg.2018.04.048
Shankar AS, Du Z, Tejeda Mora H, Boers R, Cao W, van den Bosch TPP, Korevaar SS, Boers J, van IJcken WFJ, Bindels EMJ, Eussen B, de Klein A, Pan Q, Oudijk L, Clahsen-van Groningen MC, Hoorn EJ, Baan CC, Gribnau J, Hoogduijn MJ (2022) Kidney organoids are capable of forming tumors, but not teratomas. Stem Cells 40(6):577–591. https://doi.org/10.1093/stmcls/sxac009
Hu Z, Cao Y, Galan EA, Hao L, Zhao H, Tang J, Sang G, Wang H, Xu B, Ma S (2022) Vascularized tumor spheroid-on-a-chip model verifies synergistic vasoprotective and chemotherapeutic effects. ACS Biomater Sci Eng 8(3):1215–1225. https://doi.org/10.1021/acsbiomaterials.1c01099
Fang G, Chen YC, Lu H, Jin D (2023) Advances in spheroids and organoids on a chip. Adv Funct Mater 33(19):2215043. https://doi.org/10.1002/adfm.202215043
Maffioletti SM, Sarcar S, Henderson ABH, Mannhardt I, Pinton L, Moyle LA, Steele-Stallard H, Cappellari O, Wells KE, Ferrari G, Mitchell JS, Tyzack GE, Kotiadis VN, Khedr M, Ragazzi M, Wang W, Duchen MR, Patani R, Zammit PS, Wells DJ, Eschenhagen T, Tedesco FS (2018) Three-dimensional human iPSC-derived artificial skeletal muscles model muscular dystrophies and enable multilineage tissue engineering. Cell Rep 23(3):899–908. https://doi.org/10.1016/j.celrep.2018.03.091
Salmon I, Grebenyuk S, Abdel Fattah AR, Rustandi G, Pilkington T, Verfaillie C, Ranga A (2022) Engineering neurovascular organoids with 3D printed microfluidic chips. Lab Chip 22(8):1615–1629. https://doi.org/10.1039/d1lc00535a
Zhang S, Wan Z, Kamm RD (2021) Vascularized organoids on a chip: strategies for engineering organoids with functional vasculature. Lab Chip 21(3):473–488. https://doi.org/10.1039/d0lc01186j
Nashimoto Y, Hayashi T, Kunita I, Nakamasu A, Torisawa YS, Nakayama M, Takigawa-Imamura H, Kotera H, Nishiyama K, Miura T, Yokokawa R (2017) Integrating perfusable vascular networks with a three-dimensional tissue in a microfluidic device. Integr Biol (Camb) 9(6):506–518. https://doi.org/10.1039/c7ib00024c
Nashimoto Y, Okada R, Hanada S, Arima Y, Nishiyama K, Miura T, Yokokawa R (2020) Vascularized cancer on a chip: the effect of perfusion on growth and drug delivery of tumor spheroid. Biomaterials 229:119547. https://doi.org/10.1016/j.biomaterials.2019.119547
Park J, Kim S, Hong J, Jeon JS (2022) Enabling perfusion through multicellular tumor spheroids promoting lumenization in a vascularized cancer model. Lab Chip 22(22):4335–4348. https://doi.org/10.1039/d2lc00597b
Haase K, Offeddu GS, Gillrie MR, Kamm RD (2020) Endothelial regulation of drug transport in a 3D vascularized tumor model. Adv Funct Mater 30(48):2002444. https://doi.org/10.1002/adfm.202002444
Tan SY, Jing Q, Leung Z, Xu Y, Cheng LKW, Tam SST, Wu AR (2022) Transcriptomic analysis of 3D vasculature-on-a-chip reveals paracrine factors affecting vasculature growth and maturation. Lab Chip 22(20):3885–3897. https://doi.org/10.1039/d2lc00570k
Nashimoto Y, Teraoka Y, Banan Sadeghian R, Nakamasu A, Arima Y, Hanada S, Kotera H, Nishiyama K, Miura T, Yokokawa R (2018) Perfusable vascular network with a tissue model in a microfluidic device. J Vis Exp 134:57242. https://doi.org/10.3791/57242
Wan Z, Floryan MA, Coughlin MF, Zhang S, Zhong AX, Shelton SE, Wang X, Xu C, Barbie DA, Kamm RD (2023) New strategy for promoting vascularization in tumor spheroids in a microfluidic assay. Adv Healthc Mater 12(14):e2201784. https://doi.org/10.1002/adhm.202201784
Li C, Li S, Du K, Li P, Qiu B, Ding W (2021) On-chip replication of extremely early-stage tumor behavior. ACS Appl Mater Interfaces 13(17):19768–19777. https://doi.org/10.1021/acsami.1c03740
Wang HF, Liu Y, Wang T, Yang G, Zeng B, Zhao CX (2020) Tumor-microenvironment-on-a-chip for evaluating nanoparticle-loaded macrophages for drug delivery. ACS Biomater Sci Eng 6(9):5040–5050. https://doi.org/10.1021/acsbiomaterials.0c00650
Zhou Y, Wu Y, Paul R, Qin X, Liu Y (2023) Hierarchical vessel network-supported tumor model-on-a-chip constructed by induced spontaneous anastomosis. ACS Appl Mater Interfaces 15(5):6431–6441. https://doi.org/10.1021/acsami.2c19453
Wu Y, Zhao Y, Zhou Y, Islam K, Liu Y (2023) Microfluidic droplet-assisted fabrication of vessel-supported tumors for preclinical drug discovery. ACS Appl Mater Interfaces 15(12):15152–15161. https://doi.org/10.1021/acsami.2c23305
Sotra A, Jozani KA, Zhang B (2023) A vascularized crypt-patterned colon model for high-throughput drug screening and disease modelling. Lab Chip 23(15):3370–3387. https://doi.org/10.1039/d3lc00211j
Pollet A, den Toonder JMJ (2020) Recapitulating the vasculature using organ-on-chip technology. Bioengineering (Basel). https://doi.org/10.3390/bioengineering7010017
Henderson AR, Choi H, Lee E (2020) Blood and lymphatic vasculatures on-chip platforms and their applications for organ-specific in vitro modeling. Micromachines (Basel). https://doi.org/10.3390/mi11020147
Tronolone JJ, Jain A (2021) Engineering new microvascular networks on-chip: ingredients, assembly, and best practices. Adv Funct Mater. https://doi.org/10.1002/adfm.202007199
Shaji M, Tamada A, Fujimoto K, Muguruma K, Karsten SL, Yokokawa R (2024) Deciphering potential vascularization factors of on-chip co-cultured hiPSC-derived cerebral organoids. Lab Chip 24(4):680–696. https://doi.org/10.1039/d3lc00930k
Ferrari D, Sengupta A, Heo L, Petho L, Michler J, Geiser T, de JesusPerez VA, Kuebler WM, Zeinali S, Guenat OT (2023) Effects of biomechanical and biochemical stimuli on angio- and vasculogenesis in a complex microvasculature-on-chip. iScience 26(3):106198. https://doi.org/10.1016/j.isci.2023.106198
Carvalho MR, Barata D, Teixeira LM, Giselbrecht S, Reis RL, Oliveira JM, Truckenmuller R, Habibovic P (2019) Colorectal tumor-on-a-chip system: a 3D tool for precision onco-nanomedicine. Sci Adv 5(5):eaaw1317. https://doi.org/10.1126/sciadv.aaw1317
Zhang S, Wan Z, Pavlou G, Zhong AX, Xu L, Kamm RD (2022) Interstitial flow promotes the formation of functional microvascular networks in vitro through upregulation of matrix metalloproteinase-2. Adv Funct Mater 32(43):2206767. https://doi.org/10.1002/adfm.202206767
Wan Z, Zhong AX, Zhang S, Pavlou G, Coughlin MF, Shelton SE, Nguyen HT, Lorch JH, Barbie DA, Kamm RD (2022) A robust method for perfusable microvascular network formation in vitro. Small Methods 6(6):e2200143. https://doi.org/10.1002/smtd.202200143
Wang X, Liu X, Liu W, Liu Y, Li A, Qiu D, Zheng X, Gu Q (2023) 3D bioprinting microgels to construct implantable vascular tissue. Cell Prolif 56(5):e13456. https://doi.org/10.1111/cpr.13456
Liu T, Zhou C, Ji J, Xu X, Xing Z, Shinohara M, Sakai Y, Sun T, Feng X, Yu Z (2023) Spheroid on-demand printing and drug screening of endothelialized hepatocellular carcinoma model at different stages. Biofabrication 15(4):044102. https://doi.org/10.1088/1758-5090/ace3f9
Le HT, Phan HL, Lenshof A, Duong VT, Choi C, Cha C, Laurell T, Koo KI (2023) Ultrasound standing wave spatial patterning of human umbilical vein endothelial cells for 3D micro-vascular networks formation. Biofabrication. https://doi.org/10.1088/1758-5090/ad03be
Kumar M, Rauthan R, Chakraborty D, Arumugam S (2023) Visualization of 3D organoids through the latest advancements in microscopy. In: Gopalakrishnan J (ed) Brain organoid research. Neuromethods. Springer, New York, pp 43–66. https://doi.org/10.1007/978-1-0716-2720-4_3
Ji N (2017) Adaptive optical fluorescence microscopy. Nat Methods 14(4):374–380. https://doi.org/10.1038/nmeth.4218
Rakotoson I, Delhomme B, Djian P, Deeg A, Brunstein M, Seebacher C, Uhl R, Ricard C, Oheim M (2019) Fast 3-D imaging of brain organoids with a new single-objective planar-illumination two-photon microscope. Front Neuroanat 13:77. https://doi.org/10.3389/fnana.2019.00077
Hushka EA, Yavitt FM, Brown TE, Dempsey PJ, Anseth KS (2020) Relaxation of extracellular matrix forces directs crypt formation and architecture in intestinal organoids. Adv Healthc Mater 9(8):e1901214. https://doi.org/10.1002/adhm.201901214
Lai Benjamin FL, Lu Rick X, Hu Y, Davenport HL, Dou W, Wang EY, Radulovich N, Tsao MS, Sun Y, Radisic M (2020) Recapitulating pancreatic tumor microenvironment through synergistic use of patient organoids and organ-on-a-chip vasculature. Adv Funct Mater 30(48):2000545. https://doi.org/10.1002/adfm.202000545
Yildirim M, Delepine C, Feldman D, Pham VA, Chou S, Ip J, Nott A, Tsai LH, Ming GL, So PTC, Sur M (2022) Label-free three-photon imaging of intact human cerebral organoids for tracking early events in brain development and deficits in Rett syndrome. eLife 11:e78079. https://doi.org/10.7554/eLife.78079
Stelzer EH (2015) Light-sheet fluorescence microscopy for quantitative biology. Nat Methods 12(1):23–26. https://doi.org/10.1038/nmeth.3219
Gong H, Xu D, Yuan J, Li X, Guo C, Peng J, Li Y, Schwarz LA, Li A, Hu B, Xiong B, Sun Q, Zhang Y, Liu J, Zhong Q, Xu T, Zeng S, Luo Q (2016) High-throughput dual-colour precision imaging for brain-wide connectome with cytoarchitectonic landmarks at the cellular level. Nat Commun 7:12142. https://doi.org/10.1038/ncomms12142
Li A, Gong H, Zhang B, Wang Q, Yan C, Wu J, Liu Q, Zeng S, Luo Q (2010) Micro-optical sectioning tomography to obtain a high-resolution atlas of the mouse brain. Science 330(6009):1404–1408. https://doi.org/10.1126/science.1191776
Gong H, Zeng S, Yan C, Lv X, Yang Z, Xu T, Feng Z, Ding W, Qi X, Li A, Wu J, Luo Q (2013) Continuously tracing brain-wide long-distance axonal projections in mice at a one-micron voxel resolution. Neuroimage 74:87–98. https://doi.org/10.1016/j.neuroimage.2013.02.005
Ondatje BN, Sances S, Workman MJ, Svendsen CN (2022) Tissue clearing of human iPSC-derived organ-chips enables high resolution imaging and analysis. Lab Chip 22(21):4246–4255. https://doi.org/10.1039/d2lc00116k
Bissardon C, Mermet X, Quintard C, Sanjuan F, Fouillet Y, Bottausci F, Carriere M, Rivera F, Blandin P (2022) Selective plane illumination microscope dedicated to volumetric imaging in microfluidic chambers. Biomed Opt Express 13(10):5261–5274. https://doi.org/10.1364/BOE.455377
Rusch H, Brammerloh M, Stieler J, Sonntag M, Mohammadi S, Weiskopf N, Arendt T, Kirilina E, Morawski M (2022) Finding the best clearing approach—towards 3D wide-scale multimodal imaging of aged human brain tissue. Neuroimage 247:118832. https://doi.org/10.1016/j.neuroimage.2021.118832
Schneidereit D, Brollochs A, Ritter P, Kreiss L, Mokhtari Z, Beilhack A, Kronke G, Ackermann JA, Faas M, Gruneboom A, Schurmann S, Friedrich O (2021) An advanced optical clearing protocol allows label-free detection of tissue necrosis via multiphoton microscopy in injured whole muscle. Theranostics 11(6):2876–2891. https://doi.org/10.7150/thno.51558
Kirst C, Skriabine S, Vieites-Prado A, Topilko T, Bertin P, Gerschenfeld G, Verny F, Topilko P, Michalski N, Tessier-Lavigne M, Renier N (2020) Mapping the fine-scale organization and plasticity of the brain vasculature. Cell 180(4):780–795e725. https://doi.org/10.1016/j.cell.2020.01.028
He Z, Maynard A, Jain A, Gerber T, Petri R, Lin HC, Santel M, Ly K, Dupre JS, Sidow L, Sanchis Calleja F, Jansen SMJ, Riesenberg S, Camp JG, Treutlein B (2022) Lineage recording in human cerebral organoids. Nat Methods 19(1):90–99. https://doi.org/10.1038/s41592-021-01344-8
Cho AN, Jin Y, An Y, Kim J, Choi YS, Lee JS, Kim J, Choi WY, Koo DJ, Yu W, Chang GE, Kim DY, Jo SH, Kim J, Kim SY, Kim YG, Kim JY, Choi N, Cheong E, Kim YJ, Je HS, Kang HC, Cho SW (2021) Microfluidic device with brain extracellular matrix promotes structural and functional maturation of human brain organoids. Nat Commun 12(1):4730. https://doi.org/10.1038/s41467-021-24775-5
Langer EM, Allen-Petersen BL, King SM, Kendsersky ND, Turnidge MA, Kuziel GM, Riggers R, Samatham R, Amery TS, Jacques SL, Sheppard BC, Korkola JE, Muschler JL, Thibault G, Chang YH, Gray JW, Presnell SC, Nguyen DG, Sears RC (2019) Modeling tumor phenotypes in vitro with three-dimensional bioprinting. Cell Rep 26(3):608–623e606. https://doi.org/10.1016/j.celrep.2018.12.090
Zhu T, Nie J, Yu T, Zhu D, Huang Y, Chen Z, Gu Z, Tang J, Li D, Fei P (2023) Large-scale high-throughput 3D culture, imaging, and analysis of cell spheroids using microchip-enhanced light-sheet microscopy. Biomed Opt Express 14(4):1659–1669. https://doi.org/10.1364/BOE.485217
Rafiei N, Moghadam MG, Au A, Regeenes R, Chidambaram S, Liang T, Wang Y, Yip CM, Gaisano H, Rocheleau JV (2022) Design of a versatile microfluidic device for imaging precision-cut-tissue slices. Biofabrication 14(4):041001. https://doi.org/10.1088/1758-5090/ac7eea
Atlas Y, Gorin C, Novais A, Marchand MF, Chatzopoulou E, Lesieur J, Bascetin R, Binet-Moussy C, Sadoine J, Lesage M, Opsal-Vital S, Peault B, Monnot C, Poliard A, Girard P, Germain S, Chaussain C, Muller L (2021) Microvascular maturation by mesenchymal stem cells in vitro improves blood perfusion in implanted tissue constructs. Biomaterials 268:120594. https://doi.org/10.1016/j.biomaterials.2020.120594
Kress S, Baur J, Otto C, Burkard N, Braspenning J, Walles H, Nickel J, Metzger M (2018) Evaluation of a Miniaturized Biologically Vascularized Scaffold in vitro and in vivo. Sci Rep 8(1):4719. https://doi.org/10.1038/s41598-018-22688-w
Steuwe C, Vaeyens MM, Jorge-Penas A, Cokelaere C, Hofkens J, Roeffaers MBJ, Van Oosterwyck H (2020) Fast quantitative time lapse displacement imaging of endothelial cell invasion. PLoS ONE 15(1):e0227286. https://doi.org/10.1371/journal.pone.0227286
Kaushik G, Gil DA, Torr E, Berge ES, Soref C, Uhl P, Fontana G, Antosiewicz-Bourget J, Edington C, Schwartz MP, Griffith LG, Thomson JA, Skala MC, Daly WT, Murphy WL (2019) Quantitative Label-Free Imaging of 3D Vascular Networks Self-Assembled in Synthetic Hydrogels. Adv Healthc Mater 8(2):e1801186. https://doi.org/10.1002/adhm.201801186
Ueda HR, Dodt HU, Osten P, Economo MN, Chandrashekar J, Keller PJ (2020) Whole-brain profiling of cells and circuits in mammals by tissue clearing and light-sheet microscopy. Neuron 106(3):369–387. https://doi.org/10.1016/j.neuron.2020.03.004
Homan KA, Gupta N, Kroll KT, Kolesky DB, Skylar-Scott M, Miyoshi T, Mau D, Valerius MT, Ferrante T, Bonventre JV, Lewis JA, Morizane R (2019) Flow-enhanced vascularization and maturation of kidney organoids in vitro. Nat Methods 16(3):255–262. https://doi.org/10.1038/s41592-019-0325-y
Ueda HR, Erturk A, Chung K, Gradinaru V, Chedotal A, Tomancak P, Keller PJ (2020) Tissue clearing and its applications in neuroscience. Nat Rev Neurosci 21(2):61–79. https://doi.org/10.1038/s41583-019-0250-1
Zhu J, Liu X, Deng Y, Li D, Yu T, Zhu D (2021) Tissue optical clearing for 3D visualization of vascular networks: a review. Vascul Pharmacol 141:106905. https://doi.org/10.1016/j.vph.2021.106905
Yu T, Zhu J, Li D, Zhu D (2021) Physical and chemical mechanisms of tissue optical clearing. Science 24(3):102178. https://doi.org/10.1016/j.isci.2021.102178
Xia Q, Li D, Yu T, Zhu J, Zhu D (2023) In vivo skin optical clearing for improving imaging and light-induced therapy: a review. J Biomed Opt 28(6):060901. https://doi.org/10.1117/1.JBO.28.6.060901
Xu C (2022) Optical clearing of the mouse skull. Light Sci Appl 11(1):284. https://doi.org/10.1038/s41377-022-00989-0
Li D, Hu Z, Zhang H, Yang Q, Zhu L, Liu Y, Yu T, Zhu J, Wu J, He J, Fei P, Xi W, Qian J, Zhu D (2022) A Through-Intact-Skull (TIS) chronic window technique for cortical structure and function observation in mice. eLight 2(1):15. https://doi.org/10.1186/s43593-022-00022-2
Oh MS, Khawar IA, Lee DW, Park JK, Kuh HJ (2020) Three-dimensional imaging for multiplex phenotypic analysis of pancreatic microtumors grown on a minipillar array chip. Cancers (Basel) 12(12):3662. https://doi.org/10.3390/cancers12123662
Yu T, Zhong X, Yang Q, Gao C, Chen W, Liu X, Liu Z, Zhu T, Li D, Fei P, Chen Z, Gu Z, Zhu D (2023) On-chip clearing for live imaging of 3D cell cultures. Biomed Opt Express 14(6):3003–3017. https://doi.org/10.1364/BOE.489219
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682. https://doi.org/10.1038/nmeth.2019
Lee S, Chung M, Lee SR, Jeon NL (2020) 3D brain angiogenesis model to reconstitute functional human blood–brain barrier in vitro. Biotechnol Bioeng 117(3):748–762. https://doi.org/10.1002/bit.27224
Haase K, Gillrie MR, Hajal C, Kamm RD (2019) Pericytes contribute to dysfunction in a human 3D model of placental microvasculature through VEGF-Ang-Tie2 signaling. Adv Sci (Weinh) 6(23):1900878. https://doi.org/10.1002/advs.201900878
Kempers L, van der Bijl I, van Stalborch AD, Ponsioen B, Margadant C (2021) Fast in vitro protocol for the visualization and quantitative high-throughput analysis of sprouting angiogenesis by confocal microscopy. STAR Protoc 2(3):100690. https://doi.org/10.1016/j.xpro.2021.100690
Orlova VV, Nahon DM, Cochrane A, Cao X, Freund C, van den Hil F, Westermann CJJ, Snijder RJ, Ploos van Amstel JK, Ten Dijke P, Lebrin F, Mager HJ, Mummery CL (2022) Vascular defects associated with hereditary hemorrhagic telangiectasia revealed in patient-derived isogenic iPSCs in 3D vessels on chip. Stem Cell Rep 17(7):1536–1545. https://doi.org/10.1016/j.stemcr.2022.05.022
Ryu H, Oh S, Lee HJ, Lee JY, Lee HK, Jeon NL (2015) Engineering a blood vessel network module for body-on-a-chip applications. J Lab Autom 20(3):296–301. https://doi.org/10.1177/2211068214562831
Lee S, Kim S, Koo DJ, Yu J, Cho H, Lee H, Song JM, Kim SY, Min DH, Jeon NL (2021) 3D microfluidic platform and tumor vascular mapping for evaluating anti-angiogenic RNAi-based nanomedicine. ACS Nano 15(1):338–350. https://doi.org/10.1021/acsnano.0c05110
Zudaire E, Gambardella L, Kurcz C, Vermeren S (2011) A computational tool for quantitative analysis of vascular networks. PLoS ONE 6(11):e27385. https://doi.org/10.1371/journal.pone.0027385
Rodoplu D, Matahum JS, Hsu CH (2022) A microfluidic hanging drop-based spheroid co-culture platform for probing tumor angiogenesis. Lab Chip 22(7):1275–1285. https://doi.org/10.1039/d1lc01177d
Ong LL, Dauwels J, Ang MH Jr, Asada HH (2014) A Bayesian filtering approach to incorporate 2D/3D time-lapse confocal images for tracking angiogenic sprouting cells interacting with the gel matrix. Med Image Anal 18(1):211–227. https://doi.org/10.1016/j.media.2013.10.008
Wang M, Ong LS, Dauwels J, Asada HH (2018) Multicell migration tracking within angiogenic networks by deep learning-based segmentation and augmented Bayesian filtering. J Med Imaging (Bellingham) 5(2):024005. https://doi.org/10.1117/1.JMI.5.2.024005
Wang M, Ong LS, Dauwels J, Asada HH (2017) Automated tracking and quantification of angiogenic vessel formation in 3D microfluidic devices. PLoS ONE 12(11):e0186465. https://doi.org/10.1371/journal.pone.0186465
Kuzmic N, Moore T, Devadas D, Young EWK (2019) Modelling of endothelial cell migration and angiogenesis in microfluidic cell culture systems. Biomech Model Mechanobiol 18(3):717–731. https://doi.org/10.1007/s10237-018-01111-3
Akbarpour Ghazani M, Nouri Z, Saghafian M, Soltani M (2020) Mathematical modeling reveals how the density of initial tumor and its distance to parent vessels alter the growth trend of vascular tumors. Microcirculation 27(1):e12584. https://doi.org/10.1111/micc.12584
Bates R, Irving B, Markelc B, Kaeppler J, Brown G, Muschel RJ, Brady SM, Grau V, Schnabel JA (2019) Segmentation of vasculature from fluorescently labeled endothelial cells in multi-photon microscopy images. IEEE Trans Med Imaging 38(1):1–10. https://doi.org/10.1109/TMI.2017.2725639
Solovyev R, Kalinin AA, Gabruseva T (2022) 3D convolutional neural networks for stalled brain capillary detection. Comput Biol Med 141:105089. https://doi.org/10.1016/j.compbiomed.2021.105089
Cheung CY, Xu D, Cheng CY, Sabanayagam C, Tham YC, Yu M, Rim TH, Chai CY, Gopinath B, Mitchell P, Poulton R, Moffitt TE, Caspi A, Yam JC, Tham CC, Jonas JB, Wang YX, Song SJ, Burrell LM, Farouque O, Li LJ, Tan G, Ting DSW, Hsu W, Lee ML, Wong TY (2021) A deep-learning system for the assessment of cardiovascular disease risk via the measurement of retinal-vessel calibre. Nat Biomed Eng 5(6):498–508. https://doi.org/10.1038/s41551-020-00626-4
Li X, Tang Q, Yu J, Wang Y, Shi Z (2019) Microvascularity detection and quantification in glioma: a novel deep-learning-based framework. Lab Invest 99(10):1515–1526. https://doi.org/10.1038/s41374-019-0272-3
Todorov MI, Paetzold JC, Schoppe O, Tetteh G, Shit S, Efremov V, Todorov-Volgyi K, During M, Dichgans M, Piraud M, Menze B, Erturk A (2020) Machine learning analysis of whole mouse brain vasculature. Nat Methods 17(4):442–449. https://doi.org/10.1038/s41592-020-0792-1
Haft-Javaherian M, Fang L, Muse V, Schaffer CB, Nishimura N, Sabuncu MR (2019) Deep convolutional neural networks for segmenting 3D in vivo multiphoton images of vasculature in Alzheimer disease mouse models. PLoS ONE 14(3):e0213539. https://doi.org/10.1371/journal.pone.0213539
Chen W, Yu S, Ma K, Ji W, Bian C, Chu C, Shen L, Zheng Y (2022) TW-GAN: topology and width aware GAN for retinal artery/vein classification. Med Image Anal 77:102340. https://doi.org/10.1016/j.media.2021.102340
Zhou Y, Sun SW, Liu QP, Xu X, Zhang Y, Zhang YD (2022) TED: Two-stage expert-guided interpretable diagnosis framework for microvascular invasion in hepatocellular carcinoma. Med Image Anal 82:102575. https://doi.org/10.1016/j.media.2022.102575
Jiang X, Li J, Kan Y, Yu T, Chang S, Sha X, Zheng H, Luo Y, Wang S (2021) MRI based radiomics approach with deep learning for prediction of vessel invasion in early-stage cervical cancer. IEEE/ACM Trans Comput Biol Bioinform 18(3):995–1002. https://doi.org/10.1109/TCBB.2019.2963867
Kuri PM, Pion E, Mahl L, Kainz P, Schwarz S, Brochhausen C, Aung T, Haerteis S (2022) Deep learning-based image analysis for the quantification of tumor-induced angiogenesis in the 3D In vivo tumor model-establishment and addition to laser speckle contrast imaging (LSCI). Cells 11(15):2321. https://doi.org/10.3390/cells11152321
Yan J, Li Z, Guo J, Liu S, Guo J (2022) Organ-on-a-chip: a new tool for in vitro research. Biosens Bioelectron 216:114626. https://doi.org/10.1016/j.bios.2022.114626
Riordon J, Sovilj D, Sanner S, Sinton D, Young EWK (2019) Deep learning with microfluidics for biotechnology. Trends Biotechnol 37(3):310–324. https://doi.org/10.1016/j.tibtech.2018.08.005
Urban G, Bache KM, Phan D, Sobrino A, Shmakov AK, Hachey SJ, Hughes C, Baldi P (2019) Deep learning for drug discovery and cancer research: automated analysis of vascularization images. IEEE/ACM Trans Comput Biol Bioinform 16(3):1029–1035. https://doi.org/10.1109/TCBB.2018.2841396
Chen Z, Huang J, Zhang J, Xu Z, Li Q, Ouyang J, Yan Y, Sun S, Ye H, Wang F, Zhu J, Wang Z, Chao J, Pu Y, Gu Z (2022) A storm in a teacup—a biomimetic lung microphysiological system in conjunction with a deep-learning algorithm to monitor lung pathological and inflammatory reactions. Biosens Bioelectron 219:114772. https://doi.org/10.1016/j.bios.2022.114772
Bian X, Li G, Wang C, Liu W, Lin X, Chen Z, Cheung M, Luo X (2021) A deep learning model for detection and tracking in high-throughput images of organoid. Comput Biol Med 134:104490. https://doi.org/10.1016/j.compbiomed.2021.104490
Matthews JM, Schuster B, Kashaf SS, Liu P, Ben-Yishay R, Ishay-Ronen D, Izumchenko E, Shen L, Weber CR, Bielski M, Kupfer SS, Bilgic M, Rzhetsky A, Tay S (2022) OrganoID: a versatile deep learning platform for tracking and analysis of single-organoid dynamics. PLoS Comput Biol 18(11):e1010584. https://doi.org/10.1371/journal.pcbi.1010584
Li J, Chen J, Bai H, Wang H, Hao S, Ding Y, Peng B, Zhang J, Li L, Huang W (2022) An overview of organs-on-chips based on deep learning. Research. https://doi.org/10.34133/2022/9869518
Abdul L, Xu J, Sotra A, Chaudary A, Gao J, Rajasekar S, Anvari N, Mahyar H, Zhang B (2022) D-CryptO: deep learning-based analysis of colon organoid morphology from brightfield images. Lab Chip 22(21):4118–4128. https://doi.org/10.1039/d2lc00596d
Trossbach M, Akerlund E, Langer K, Seashore-Ludlow B, Joensson HN (2023) High-throughput cell spheroid production and assembly analysis by microfluidics and deep learning. SLAS Technol. https://doi.org/10.1016/j.slast.2023.03.003
Kassis T, Hernandez-Gordillo V, Langer R, Griffith LG (2019) OrgaQuant: human intestinal organoid localization and quantification using deep convolutional neural networks. Sci Rep 9(1):12479. https://doi.org/10.1038/s41598-019-48874-y
Beghin A, Grenci G, Sahni G, Guo S, Rajendiran H, Delaire T, Raffi SBM, Blanc D, de Mets R, Ong HT, Galindo X, Monet A, Acharya V, Racine V, Levet F, Galland R, Sibarita JB, Viasnoff V (2022) Automated high-speed 3D imaging of organoid cultures with multi-scale phenotypic quantification. Nat Methods 19(7):881–892. https://doi.org/10.1038/s41592-022-01508-0
de Medeiros G, Ortiz R, Strnad P, Boni A, Moos F, Repina N, Challet Meylan L, Maurer F, Liberali P (2022) Multiscale light-sheet organoid imaging framework. Nat Commun 13(1):4864. https://doi.org/10.1038/s41467-022-32465-z
Benning L, Peintner A, Finkenzeller G, Peintner L (2020) Automated spheroid generation, drug application and efficacy screening using a deep learning classification: a feasibility study. Sci Rep 10(1):11071. https://doi.org/10.1038/s41598-020-67960-0
Irisa T, Zhou H, Fujimoto K, Yokokawa R (2023) Developmental phases of on-chip vasculogenesis classified using a deep learning visual model. Paper presented at the 2023 IEEE 36th international conference on micro electro mechanical systems (MEMS), 15–19 January 2023. https://doi.org/10.1109/MEMS49605.2023.10052548
Tronolone JJ, Mathur T, Chaftari CP, Jain A (2023) Evaluation of the morphological and biological functions of vascularized microphysiological systems with supervised machine learning. Ann Biomed Eng 51(8):1723–1737. https://doi.org/10.1007/s10439-023-03177-2
Strobel HA, Moss SM, Hoying JB (2022) Methods for vascularization and perfusion of tissue organoids. Mamm Genome 33(3):437–450. https://doi.org/10.1007/s00335-022-09951-2
O’Connor C, Brady E, Zheng Y, Moore E, Stevens KR (2022) Engineering the multiscale complexity of vascular networks. Nat Rev Mater 7(9):702–716. https://doi.org/10.1038/s41578-022-00447-8
Morss Clyne A, Swaminathan S, Diaz Lantada A (2019) Biofabrication strategies for creating microvascular complexity. Biofabrication 11(3):032001. https://doi.org/10.1088/1758-5090/ab0621
Cao X, Ashfaq R, Cheng F, Maharjan S, Li J, Ying G, Hassan S, Xiao H, Yue K, Zhang YS (2019) A tumor-on-a-chip system with bioprinted blood and lymphatic vessel pair. Adv Funct Mater. https://doi.org/10.1002/adfm.201807173
Fujimoto K, Erickson S, Nakayama M, Ihara H, Sugihara K, Nashimoto Y, Nishiyama K, Miura T, Yokokawa R (2023) Pericytes and shear stress each alter the shape of a self-assembled vascular network. Lab Chip 23(2):306–317. https://doi.org/10.1039/d2lc00605g
Adjei-Sowah EA, O’Connor SA, Veldhuizen J, Lo Cascio C, Plaisier C, Mehta S, Nikkhah M (2022) Investigating the interactions of glioma stem cells in the perivascular niche at single-cell resolution using a microfluidic tumor microenvironment model. Adv Sci (Weinh) 9(21):e2201436. https://doi.org/10.1002/advs.202201436
Liu Y, Li J, Zhou J, Liu X, Li H, Lu Y, Lin B, Li X, Liu T (2022) Angiogenesis and functional vessel formation induced by interstitial flow and vascular endothelial growth factor using a microfluidic chip. Micromachines (Basel) 13(2):225. https://doi.org/10.3390/mi13020225
Pradhan S, Smith AM, Garson CJ, Hassani I, Seeto WJ, Pant K, Arnold RD, Prabhakarpandian B, Lipke EA (2018) A microvascularized tumor-mimetic platform for assessing anti-cancer drug efficacy. Sci Rep 8(1):3171. https://doi.org/10.1038/s41598-018-21075-9
Cui X, Morales RT, Qian W, Wang H, Gagner JP, Dolgalev I, Placantonakis D, Zagzag D, Cimmino L, Snuderl M, Lam RHW, Chen W (2018) Hacking macrophage-associated immunosuppression for regulating glioblastoma angiogenesis. Biomaterials 161:164–178. https://doi.org/10.1016/j.biomaterials.2018.01.053
Acknowledgements
This work was supported by the National Key Research and Development Program of China (Grant No. 2017YFA0700501); the National Natural Science Foundation of China (Grant Nos. 61860206009, 62375096, 82361138569, 82372012); the Key Research and Development Project of Hubei Province (Grant No. 2022BCA023); and the Open Research Fund of WNLO (Grant No. 2022WNLOKF009); the Open Competition Project of Wuhan East Lake High-tech Development Zone (Grant Nos. 2023KJB224, 2023KJB213).
Author information
Authors and Affiliations
Contributions
TY: Conceptualization; funding acquisition; visualization; writing—original draft; writing—review and editing of the manuscript. QY: Conceptualization; visualization; writing—original draft; writing—review and editing of the manuscript. Bo Peng: Funding acquisition; writing—review and editing of the manuscript. ZG: Supervision; writing—review and editing of the manuscript. DZ: conceptualization; funding acquisition; supervision; writing—review and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yu, T., Yang, Q., Peng, B. et al. Vascularized organoid-on-a-chip: design, imaging, and analysis. Angiogenesis 27, 147–172 (2024). https://doi.org/10.1007/s10456-024-09905-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10456-024-09905-z