Abstract
Erythrocytes undergo deformations when they transport O2 and CO2 across the membrane, yet the 3D nanomechanics of the skeletal network remains poorly understood. Expanding from our previous single isolated unit, we now simulate networks consisting of 1–10 concentric rings of repeating units in equibiaxial deformation. The networks are organized with (1) a 3D model for a single unit, (2) a wrap-around mode between Sp and actin protofilament in the intra-unit interaction, and (3) a random inter-unit connectivity. These assumptions permit efficient five-degrees-of-freedom (5DOF) simulations when up to 30 pN of radial forces are applied to the boundary spectrin (Sp) and the center and other units are analyzed. As 6 Sp balance their tensions, hexagonal units become irregular. While actin protofilaments remain tangent to the network, their yaw (Φ) angles change drastically with addition of neighboring units or an Sp unfolding. It is anticipated that during deformation, transmembrane complexes associated with the network move laterally through the lipid bilayer and increase the diffusion of molecules across the membrane. When protofilament/Sp sweeps under the lipid bilayer, they mix up the submembrane concentration gradient. Thus, the nanomechanics of actin protofilaments and Sp may enhance the transport of molecules during erythrocyte deformation.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the relationship between the structure of the cell membrane and its mechanical properties will be an important endeavor in cell biology and bioengineering in the next decade.8 An erythrocyte offers the simplest model to study cell membranes as it consists of only a lipid bilayer and an underlying protein skeleton. It is this structure that allows erythrocytes to repeatedly squeeze through small capillaries without compromising their structural integrity (Fig. 1a).
Deformation of erythrocytes in microcirculation and the network nanomechanics. (a) Erythrocytes deform (from their biconcave disk shape) extensively in microcirculation where gas exchange occurs in tissue capillaries.30 The flow is from left to right. (b) The dynamics of the membrane skeleton (e.g., the tension in Sp and the attitude of the protofilament) underneath the lipid bilayer may have a pivotal role in transport phenomena, facilitating the movement of some molecules through the lipid bilayer and protein carriers during membrane deformation. The presence of a JC (junctional complex), Sp (spectrin), and SC (suspension complex) in the middle of a Sp tetramer represents two SCs from adjacent units. The pointed end of the protofilament is marked with erythrocyte tropomodulin (E-Tmod, a sphere)
The erythrocyte membrane skeletal network (Fig. 1b) is composed of repeating units, consisting of two kinds of major protein complexes (Fig. 2b): junctional complexes (JC) and suspension complexes (SC). Within each unit, there are six peripheral SC, each is connected to a central JC by a flexible spectrin dimer (Sp). SC (mainly trans-membrane protein band 3, ankyrin, and protein 4.2) links the protein network to the lipid bilayer; JC (mainly an actin protofilament reinforced by tropomyosin, capped by erythrocyte tropomodulin, and stabilized by protein 4.1) functions as the hub for three pairs of Sp to converge. Between units, Sp dimers inter-connect to form tetramers (Sp-t), creating a network (Fig. 2c), consisting of mainly hexagons2,23,29 with a small number of pentagons and heptagons.23 In the study of Sung and Vera,36 we illustrated that it is the length of the actin filament that dictates the hexagonal topology of the network.
The network topology of the erythrocyte membrane skeleton with three levels of connectivity. (a) Actin/Sp connectivity: Sp wrapping around actin protofilament with α Sp from one side and β Sp from the other side. Actin binding site (a black dot) only exists at β Sp. Thus, tensional force exerted through flexible Sp in any direction will not produce torque (roll motion) of the actin protofilament. (b) Intra-unit connectivity: Sp/SC assignment is defined within each unit. Parenthesis near SC indicates the position of G actin on the protofilament and the connecting Sp. For detail see Sung and Vera, 2003.36 (c) Inter-unit connectivity: The head-to-head association of Sp from two neighboring units is random (six orientations/unit). The arrow points to the tetramerization site
Erythrocyte membranes have been the focus of many investigations. Experimentally, their local and global responses were characterized by micropipette aspiration,3,4,9,34,35,37 optical tweezers,16,32 and microrheology.19 The numerical models describing their mechanics, however, are not based on its 3D structure.1,6,7,14,15,17,21 As novel mechanical properties of several key components (e.g., Sp, ankyrin, and the lipid bilayer) have been characterized through atomic force microscopy18,25,28 and/or molecular dynamics simulations,12,13,24,33 a more comprehensive model that includes the detailed molecular architecture, single unit organization, and network topology would provide much deeper insights to the molecular basis of erythrocyte membrane mechanics.
The first 3D model for JC reported by Sung and Vera in 2003,36 predicted the connectivity among 6 Sp bound to the protofilament. This connectivity was then used to develop a mathematical model to determine the equilibrium condition for a variety of initial configurations. For the first time 3D nanomechanics of a single isolated unit during equibiaxial and anisotropic deformations was predicted in 2005 by Vera et al.38 Later, the modeling approach for the lipid bilayer developed by Granek11 and Lin and Brown22 was included to develop a hybrid model of a single unit coupled with lipid bilayer.40 Both models provided a unique glimpse as to how a single unit (~3650 nm2) of the membrane skeleton may behave in 3D. This is significant since its response to mechanical stress is difficult to obtain experimentally.
The model developed here further characterizes the mechanical behavior of a multiunit network during equibiaxial deformation. It provides insights as to how a unit connected in the network may be influenced by neighboring units. Interesting new phenomena were revealed when random connectivity among units was simulated. In the future, this model will also be used to simulate membrane skeletal networks with molecular defects found in human patients or animal models, since mutations, damages, or cross-links often result in alterations of the structure of the units, their connectivity, and mechanical properties of the elements of the units. These issues may be incorporated in the new simulations.
Methods
The purpose of this paper is to study the mechanical equilibrium of networks formed by concentric rings of repeating units based on the 3D model of a single JC in the erythrocyte membrane skeleton36 (Fig. 2). This approach focuses on the average behavior of the ensemble of units in the network as we vary the number of units, the connectivity, the forces applied, and other factors that we shall soon describe. We single out the behavior of a characteristic unit embedded in the network, in contrast with the study of a single unit in isolation, which was done previously.38,40
Model Assumptions
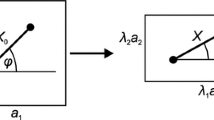
The calculations of the network nanomechanics depend on several assumptions chosen as close as possible to the physiological conditions, similar to those in Vera et al.38 In order to simulate large networks in reasonable computational time several simplifications were made (Fig. 3a). These include assuming a rigid body model with 5DOF for the protofilament, which preserves its three translational and two rotational motions (pitch and yaw) in 3D space (Fig. 3b). The protofilament mass and damping coefficients were selected to minimize the time in which the network reached equilibrium (by dynamic relaxation). Here Sp-t was modeled as a nonlinear massless elastic string with a force–extension curve based on the worn-like-chain (WLC) model of Rief et al.28 (Fig. 3c). The WLC model allows Sp to unfold. Sp-t is modeled as two Sp in series (we do not simulate the separation of Sp-t to two Sp) and the contour length for each Sp used is 163.4 nm.28 We also used 30 pN as the maximal tension for any Sp before unfolding, and every time it unfolds the length increases by 31.7 nm. A major difference is that previously, when a single unit was modeled,38 the coordinates of 6 SC were fixed corresponding to specific stretch ratios. Here we allowed them to move in response to the peripheral force applied.
A 5DOF multiunit model of the erythrocyte membrane skeleton. (a) A 5-ring network of the erythrocyte membrane skeleton. Actin protofilaments are represented as green cylinders; Pointed-end with a sphere; Sp-t a black string connecting protofilaments. The cartoon represented the steady state reached after a radial force of 2.5 pN applied at every free Sp on all PU. (b) Motion of the protofilament in 5DOF model. It is allowed to freely translate in the x, y, and z directions with two rotations: pitch angle (θ) out of the plane of the membrane and yaw angle (Φ) in the plane of the membrane, producing a 5 DOF model. (c) Modular elongation behavior of Sp modeled by the WLC paradigm according to AFM.26 (d) How a pair of Sp may wrap around the actin protofilament.36 Block: G actin; Circle: Sp; Arrow: orientation of α or β Sp; (e) How Sp domains may wrap around the actin filament40
Network Connectivity and Applied Forces
Hexagonal networks (each representing a repeating unit) have been constructed with varying number of them arranged in concentric rings (Fig. 4). In this study, we define three connectivity levels for the network from small to large in the scale: (a) Sp/Actin connectivity: Each Spi wraps around the actin protofilament (Fig. 2a); (b) Intra-unit connectivity: Each Spi connects to the nearby corresponding SCi (e.g., Sp1/SC1) (Fig. 2b); and (c) Inter-unit connectivity: Random head-to-head association between any 2 Spi (or any SCi) from neighboring units in forming Sp-t (e.g., Sp1 from unit 1 to Sp2 in unit 2) (Fig. 2c). Membrane deformation was simulated in two ways: (a) Radial force: variable radial forces applied to all peripheral Sp as in Fig. 4b; (b) Fixed boundaries: the final coordinates of the peripheral SC have been defined corresponding to specific stretch ratios.
A multiunit network of erythrocytes subjected to equibiaxial deformation. (a) A network of three rings before deformation: one ring network is a CU, a two-ring network is a CU plus six PU, and a three-ring network is a two-ring network plus 12 PU. (b) A network after equibiaxial deformation. Small arrows represent the radial forces applied to free Sp on PU. Large empty arrows represent the directions of network extension during deformation. The nanomechanics of CU is compared with that of a single isolated unit in previous models (view supplemental materials for animations of the simulations performed)
Due to computational constraints, we simulated an isolated network up to 10 concentric rings with a maximum of 271 units, as opposed to the entire erythrocyte network of ~106 concentric rings with ~33,000 units. Simulating the largest network of 10 rings corresponds to the solution of an ordinary differential equation of order 13,008. We ignored the coupling of the protein network to the lipid bilayer and maintained the same intra-unit connectivity (between JC and SC) as proposed by Sung and Vera36 (Fig. 2b). We allowed for each unit in the network to be connected in one of the six possible different orientations, which were chosen at random as expected in the physiological condition (Fig. 2c). Starting from a single unit in the center (CU), we simulated progressively complex networks by adding one ring of peripheral units (PU) at a time connected to the distal (head) ends of Sp in the existing network.
Attachment Modes and DOF
We previously proposed a wrap-around model as an attachment mode between Sp and actin protofilament36 in addition to the point-attachment mode. In our previous single-unit simulation, we treated the point-attachment as a simple mechanical ball-joint located at the surface of the protofilament producing a 6DOF model.38 More recently, we also simulated the nanomechanics of a single unit coupled with lipid bilayer using both point-attachment (6DOF) and the wrap-around modes (5DOF).40 The extension of α and/or β Sp wrapping around the protofilament may provide a mechanism to minimize the torque applied to the protofilament during deformation, and eliminate the roll motion, effectively producing a 5DOF system, which can be simulated efficiently in this paper.
We devoted a whole paper in 2003 to address the question as to where 6 Sp may be positioned along an actin protofilament.36 After extensive analysis, we discovered that the only way 6 Sp may span 360° (top view) around the protofilament is for them to form three pairs at the top, middle, and bottom of the filament (Fig. 2b), and for each pair to wrap the filament in a back-to-back fashion (Fig. 3d). The biochemistry data further allowed us to propose in greater detail how domains in α and β Sp may wrap around the actin protofilament (Fig. 3e).40 Even though it remains to be proven, we believe that the proposed wrap-around model is likely to be the functional and physiological mode of attachment. The wrap-around and the flexibility of α and β Sp make the 5DOF assumption reasonable, allowing the simulation of large networks to be possible in reasonable computational time.
Network Simulations
We have generated 1950 random instances of the multiunit networks consisting of one to ten rings of units. We performed two types of simulation. In one type, we subjected the networks to an equibiaxial deformation by applying an increasing radial force to all peripheral Sp (Fig. 4b). Forces were applied in such a way as to not produce any rigid motion on the network, i.e., global translations or rotations. In another type, we defined a set of fixed final coordinates for peripheral SC (corresponding to specific stretch ratios) and allowed the dynamic system to reach equilibrium. We found equivalent mechanical behaviors between these two types of simulation. In any given series of simulations, we first applied a force of 2.5 pN to a network of a specified number of subunits with an initial θ = 90° and let the dynamic network reach its equilibrium. We then used this equilibrium condition as the initial condition for the next simulation, in which the force was increased by another 2.5 pN. We followed this scheme until the force reached 30 pN.
Analysis
For each simulation, we obtained the attitude (pitch (θ) and yaw (Φ) angles) for all the actin protofilaments and the length and tension for each Sp in the network, with which we computed some average properties of the ensembles. We analyzed and reported in greater detail the behavior of the central unit (CU), which is the unit that is most isolated from potential effects of the boundary conditions.
Mathematical Model of a Single Unit
The mathematical model used here for a single unit is discussed in detail by Skelton and de Oliveira.31 Under the assumptions discussed above, the protofilament is a rigid rod with 5 DOF of mass m and length \( \ell \). Following Skelton and de Oliveira,31 the equations of motion for such rigid body can be described by:
where r is a vector pointing to the center of mass, m, of the protofilament, b is a unit vector in the direction of the protofilament, and J is the moment of inertia of the rigid protofilament. As we are interested mostly in the static equilibrium, we have chosen \( J = m\ell^{2} /12 \) which corresponds to the assumption that mass is being uniformly distributed along the protofilament length. The generalized forces f r and f b are calculated as:
where the vector f i represents a force applied at a node located at distance \( \ell_{i} \) measured from the center of mass of the protofilament. In a typical unit, these forces arise from elongation of Sp, which were modeled as nonlinear elastic elements following the force vs. elongation curve of the WLC model of Rief et al.28 as discussed before. The contour of Sp and its potential collisions with other molecules were not modeled.
For a PU, there might also be forces associated with membrane forces or boundary constraints, as already discussed. SC is modeled as simple point mass. For convergence to an equilibrium configuration, diagonal damping is added with mass and damping coefficient tuned for faster convergence.
Results and Discussion
Actin Protofilaments Are Tangent to the Network
Our 1950 simulations revealed that θ angles of actin protofilaments in the CU were largely ~0° when radial forces of 2.5–30 pN were applied to all PU in the network. Therefore, the protofilaments are predicted to be mostly tangent to the network during equibiaxial deformations.
Actin protofilament tangent to the membrane is consistent with experimental results using erythrocyte ghosts labeled with rhodamine–phalloidin visualized by fluorescence polarization microscopy (FPM),27 where more than 60% of the protofilaments had a pitch angle within 0° to 22° (mean error of 17%). The agreement suggests that parameters used here are likely to be near the physiological values. Simulation results also suggest that this 3D network has a molecular organization designed to minimize the intrusion/disturbance of the protofilaments into the lipid bilayer, thus maximizing the stability of the membrane during whole cell deformation. The results of our simulations in which the network reaches equilibrium states where θ is zero or near zero further suggests that the protofilaments have enough freedom to rotate and translate to achieve these lower energy equilibrium states.
Tensions in Sp Are Balanced
In all of our simulations, the tensions among 6 Sp in a CU are very similar, regardless of the magnitude of force and the number of rings. It is also insensitive to the position of Sp along the actin protofilament (Fig. 2b), despite the fact that the protofilament is 37 nm long (not a node). Similar results are also found in PU. This result indicates that the skeletal network of erythrocytes balances the forces around each JC and prevents a build up of greater tension in any particular Sp. This simulation result implies that an erythrocyte has the ability to redistribute the load, minimizing the damage to any Sp in any given areas that are subjected to equibiaxial deformation during circulation.
Repeating Units Become Irregular Hexagons
In our simulations, each unit is initiated at a perfectly symmetric hexagonal state. After application of the forces, each unit has become an irregular hexagon when the final equilibrium state is reached. This occurs because when 6 Sp (attached to various points along the protofilament) balance their tensions, each unit deforms its boundary into an irregular hexagon (Fig. 5). Such behavior would not happen if a JC were treated as a dimensionless node.
Basic repeating units become irregular hexagons after equibiaxial deformation of the network. (a) A 5-ring network deformation after the application of 2.5 pN equibiaxially. Note the irregularity of the hexagonal units (shaded gray) and their (nonmaterial, imaginary) boundaries in the network. Each shaded small hexagon occupies the center portion of a large hexagon which is defined by 6 JC in the corner and bordered by 6 Sp-t. (b) The same network represented by all shaded units (without structural elements)
We derived a hexagon irregularity (Ir) index, which is calculated as:
where Li is the length of the individual boundary and Avg(Li) is the average length of Li within the same hexagon. The corner of the hexagon is assumed by the midpoint between 2 SC. For example, in Fig. 5, after a force of 2.5 pN was applied to each PU in a network the Ir index became 2.21 ± 0.3 SD as the network underwent equibiaxial extension. The Ir index is a useful measure of local deformation at the resolution of the single-unit level. However, it can be summarized (mean ± SD) to provide insights into how much interconnected units may undergo uneven deformation to achieve a global equibiaxial deformation of the entire network. We anticipate that this index in the future would become more indicative for the analysis of diseased networks with molecular defects (e.g., with reduced Sp number) or networks undergoing anisotropic deformation.
Sp Unfolding May Be Rare in the Network
To determine Sp unfolding frequency, we performed 35 simulations and used the increase of Sp length (by ~31.7 nm) as the indication for unfolding (Fig. 3c). We noted that when a radial force 27.5 pN was applied to each PU, a small number of Sp in CU began to unfold (even though the value of 30 pN was set to be the threshold for Sp unfolding). At 25 pN or below, none did. Among the 6 Sp in each unit, Sp1 and Sp6 appear to have slightly lower instances of first unfolding. A second unfolding (two domains of Sp unfolded) occurred even less often. As only about 0.15 ± 0.16% of Sp in all units had unfolded once among 35 simulations (with 2–10 rings), Sp unfolding may not be a frequent event (without considering other factors, e.g., thermal fluctuation). The simulation results suggest that overall the network has an excellent design to distribute and balance the tension (by reshaping the basic repeating units); but minor degrees of heterogeneity in tension (e.g., due to the different connectivity) may lead to small numbers of Sp unfolding, even though the applied force itself to the network has not reached the threshold. Thus, a small number of Sp unfolding or local hemolysis may happen if erythrocytes are subjected to near threshold mechanical stress.
Φ Angles Are Greatly Affected by Inter-Unit Connectivity
Increasing force applied to the networks from 5.0 to 25 pN did not alter Φ of the CU significantly (e.g., see R5 in Fig. 6a). However, addition of rings (Fig. 4), drastically changed Φ to a new value (e.g., from R6 to R7 in Fig. 6a). Such change may be attributed to the random inter-unit connectivity with several new PU, indicating the drastic influence exerted by the neighboring units. These predictions imply that the inter-unit connectivity may dominate the Φ values of the actin protofilaments in each unit, especially during the construction phase of the skeletal network. Once connections are made and the network is completed; however, the Φ values may be largely maintained. Since the yaw angle distributions in networks of 5, 7, and 10 rings are similar, consistent behaviors may be expected in larger networks consisting of more than 100 rings as found in whole erythrocytes.
The behavior of the yaw (Φ) angle in CU in response to forces applied at PU and/or the addition of neighboring units. (a) Φ in CU when the network grows from 1 ring (R1) to 10 rings (R10). Each network is subjected to increasing forces sequentially from 2.5 to 25 pN; an extra ring of PU is added to the existing network and the same range of forces is applied again. Φ remains relatively constant within the range of radial forces (e.g., within R5), but jumps each time an additional ring is added (e.g., from R1 to R2). (b) Diagram following one example of protofilament, where its Φ jumps from one of the preferred six angles to another preferred angles when each ring is added. A single unit is shown with Φ = 30°
Six Preferred Φ Angles
The value of Φ in a single unit (one ring network structure) was ~30°. The values of Φ in the networks of multiple rings spanned 360°. The heterogeneity of Φ angles (and thus JC) in the network may be advantageous in the case of unsustainable deformation, as a given failure will not be extended linearly throughout the entire network.
However, there seemed to be six preferred Φ values with an interval of ~60°. This was not only found in CU, but also in all units. For example, Fig. 7a shows a histogram of Φ in 1500 simulations and six preferred Φ values of about −150°, −90°, −30°, 30°, 90°, and 150° were predicted. This is surprising considering the number of units involved in the networks (e.g., 271 units in 10 rings, Fig. 7b). Remarkably, as a ring of units was added, Φ of CU drastically changed from one to another preferred angle (Fig. 6b).
Six preferential Φ angles of JC in the network. (a) Histogram of Φ angles in all units or CU from 1500 simulations. Six preferred Φ angles (−150°, −90°, −30°, 30°, 90°, and 150°) in the networks of various size are present when a step 2.5 pN was sequentially applied. (b) A 5-ring network simulation after 5 pN was applied. Note the orientation of protofilament in all units. (c) Histogram of Φ angles from all units, showing the variation from the mean value of preferred angles (the vertical axis is in thousands)
It is not yet clear exactly why six preferred angles exist. Perhaps it is related to the organization of a JC, where three pairs of Sp wrap around a 37-nm long filament at its top, middle, and bottom, so that a protofilament may tend to align with one of three pairs of Sp. We assume all units have the same intra-unit connectivity (Fig. 2b), meaning there is a given orientation for each Sp, due to its binding site coordinates, head-to-tail directionality, and relative rigidity. We also assume all repeating units have 6 Sp without any heterogeneity (e.g., pentagons or heptagons which exist in erythrocytes in small percentages).23 Therefore, the Φ values in erythrocytes may be slightly more random than predicted here.
There has been no experimental data reporting, or other 3D models predicting, Φ values in erythrocytes or networks under equibiaxial extension. Using a 2D network, Picart et al. simulated anisotropic deformation and predicted a random distribution of yaw angles.26 There was a transmission electron micrograph of the erythrocyte membrane skeleton5 from which we also obtained similar six preferred Φ values. However, the sample was prepared under a hypertonic condition (not in equibiaxial extension), so more studies are needed to confirm our predictions experimentally at the whole cell level.
Single Sp Unfolding Sets off Global Changes of Φ in the Network
The Φ values of protofilaments in the network do not significantly change with increasing forces (in the range of 5–25 pN). However, when any Sp unfolds (e.g., at 27.5 pN or higher), then the Φ of every JC over the entire network changes significantly (Fig. 8). This result suggests that when one Sp unfolds anywhere in the network, the entire network readjusts and reaches a new steady state. A force of 27.5 pN corresponds to a λ of 3.4, which is far beyond the sustainable level (λ = 2.67).20 This result implies that under physiological conditions, the orientations of the actin protofilaments in the network remain relatively unchanged during equibiaxial deformation (except at the very low tension, see Fig. 8). That is, until Sp unfolds.
The effect of a single Sp unfolding in all Φ angles of the network. The change of the absolute value of Φ angle is plotted against the level of forces. The average is from all Φ in a 10-ring network in a single simulation. After an initial variation induced by increasing force from 2.5 to 5 pN (left bar), additional force resulted in less and less variations in the Φ angle, until the first Sp begins to unfold (25–27.5 pN) (right bar). At that time, a simultaneous change in Φ angle of all units is observed. The drawing illustrates the Φ changes (arrows) in the network before and after a single Sp unfolding. The bars: SD
3D Modeling: From Single-Unit to Multiunit
Table 1 summarizes the nanomechanics of Sp and actin protofilament during equibiaxial deformation using our three mathematical simulations, our previous dynamic relaxation model38 and our hybrid model40 for a single isolated unit, and the current network model for a multiunit network.
Regarding the θ angle: In our previous 6DOF single-unit simulations,38 a vertical stiffness modulus Km (representing the interaction between a JC and the lipid bilayer) was introduced. With this, a θ of ~18° or less was predicted within a “fixed” JC. Single-unit models coupled with the lipid bilayer produced a θ of ~28° (6DOF) and ~0° (5DOF).40 In our current 5DOF multiunit simulations, when the boundary of each unit is allowed to move in the network in response to the tension, a “flat” network was generated without the participation of the lipid bilayer. This implies that a network organized with (1) the proposed 3D model of each unit, (2) a wrap-around mode between Sp and protofilament in the intra-unit interaction, and (3) a random inter-unit connectivity may be close to the physiological condition.
Regarding the Φ angle: Our previous simulations for a single isolated unit reported Φ = 28°–33°, and in this network simulation we report a 360° distribution with six preferential values. These simulation results demonstrate the importance of inter-unit connectivity of the network in the orientation of the protofilaments in each unit. When a single Sp unfolds, all Φ angles in the network change significantly.
Regarding Sp tension: Tension in 6 Sp of a single isolated unit is similar to that in the network, but the latter has a smaller SD. While the former were forced to maintain a perfect hexagon, the latter was allowed to move, resulting in irregular topology of individual units.
Regarding Sp unfolding: In our previous 6DOF single-unit simulations,38 the equibiaxial extension ratio (λ) required to produce the first Sp unfolding was ~3.6; in the current 5DOF multiunit simulations, it was 27.5 pN, corresponding to a λ of ~3.4. Therefore, all models predicted Sp unfolding only at very high equibiaxial deformation ratios (λ = 3.4–3.6). Since intravascular lysis of erythrocytes normally does not happen, erythrocytes must undergo sustainable equibiaxial deformation (λ < 2.67)9 in circulation. Therefore, Sp unfolding may be a rare event. This implies that the remarkable elasticity of erythrocytes may be a function of the nonlinear behavior of Sp (Fig. 3c), and not the modular elongation produced by unfolding. The latter may only happen at a very large equibiaxial deformation, e.g., at spleen sinusoids, where old and diseased erythrocytes are caught and eliminated from circulation.
In the future, we will use the current 5DOF multiunit model to simulate networks (e.g., of mouse knockout/mutant models and human patients) with specific defects in their membrane skeletal network. Many known defects are at the molecular level and are expected to affect the unit structure, the connection between units, their mechanical properties, or combination thereof. Therefore, it is possible to vary these in the current model to explore their consequences at the network level.
Network Nanomechanics in Membrane Transport
Simulations of the nanomechanics of membrane skeletal network may provide some insights into why deformations of erythrocytes in small capillaries may be important or essential for their O2 and CO2 transport function across the cell membrane (Fig. 1): (1) as SC and JC (both contain transmembrane proteins) move laterally during equibiaxial deformation through the lipid bilayer back and forth vertically. Such disturbances may function to increase the diffusion of molecules (e.g., O2) across the membrane; and (2) as JC move or change yaw angles during equibiaxial deformation (e.g., at very low level, see Fig. 8), the protofilament and six associated Sp sweep horizontally back and forth under the lipid bilayer. Such disturbance may mix the submembrane concentration gradient of molecules. The combination of the two may facilitate the transport of molecules across the erythrocyte membrane.
Normally physical and chemical factors (e.g., lipid solubility, size, and charge) govern the diffusion coefficient (D) of a molecule across a steady lipid bilayer. Further, the overall diffusion rate is a function of D and the concentration gradient across the membrane. The nanomechanics of the network may increase the efficiency of the diffusion. It is also known that erythrocytes undergo anisotropic deformation in addition to equibiaxial deformation. Therefore, a transition from one mode of deformation to the other10 (e.g., inside the capillaries and/or in and out of capillaries) may further enhance the transport function of erythrocytes. Finally, there are proteins associated with the network function as transporter (e.g., anion exchanger in SC). Therefore, mechano-regulation of ion transports (e.g., HCO3 − and Cl−) during the network deformation is a possibility and would also be an interesting mechanism to study.
References
Boey, S. K., D. H. Boal, and D. E. Discher. Simulations of the erythrocyte cytoskeleton at large deformation. I. Microscopic models. Biophys. J. 75:1573–1583, 1998.
Byers, T. J., and D. Branton. Visualization of the protein associations in the erythrocyte membrane skeleton. Proc. Natl Acad. Sci. USA 82:6153–6157, 1985.
Chabanel, A., M. Flamm, K. L. Sung, M. M. Lee, D. Schachter, and S. Chien. Influence of cholesterol content on red cell membrane viscoelasticity and fluidity. Biophys. J. 44:171–176, 1983.
Chien, S., K.-L. P. Sung, R. Skalak, S. Usami, and A. Tozeren. Theoretical and experimental studies on viscoelastic properties of erythrocyte membrane. Biophys. J. 24:463–487, 1978.
Coleman, T. R., D. J. Fishkind, M. S. Mooseker, and J. S. Morrow. Functional diversity among spectrin isoforms. Cell Motil. Cytoskeleton 12:225–247, 1989.
Discher, D., D. Boal, and S. Boey. Phase transitions and anisotropic responses of planar triangular nets under large deformation. Phys. Rev. E 55:4762–4772, 1997.
Discher, D. E., D. H. Boal, and S. K. Boey. Simulations of the erythrocyte cytoskeleton at large deformation. II. Micropipette aspiration. Biophys. J. 75:1584–1597, 1998.
Discher, D., C. Dong, J. J. Fredberg, F. Guilak, D. Ingber, P. Janmey, R. D. Kamm, G. W. Schmid-Schonbein, and S. Weinbaum. Biomechanics: cell research and applications for the next decade. Ann. Biomed. Eng. 37:847–859, 2009.
Discher, D. E., N. Mohandas, and E. A. Evans. Molecular maps of red cell deformation: hidden elasticity and in situ connectivity. Science 266:1032–1035, 1994.
Fischer, T. M., M. Stohr-Lissen, and H. Schmid-Schonbein. The red cell as a fluid droplet: tank tread-like motion of the human erythrocyte membrane in shear flow. Science 202:894–896, 1978.
Granek, R. From semi-flexible polymers to membranes: anomalous diffusion and reptation. J. Phys. III 7:1761–1788, 1997.
Gullingsrud, J., and K. Schulten. Lipid bilayer pressure profiles and mechanosensitive channel gating. Biophys. J. 86:3496–3509, 2004.
Gumbart, J., Y. Wang, A. Aksimentiev, E. Tajkhorshid, and K. Schulten. Molecular dynamics simulations of proteins in lipid bilayers. Curr. Opin. Struct. Biol. 15:423–431, 2005.
Hansen, J. C., R. Skalak, S. Chien, and A. Hoger. An elastic network model based on the structure of the red blood cell membrane skeleton. Biophys. J. 70:146–166, 1996.
Hansen, J., R. Skalak, S. Chien, and A. Hoger. Spectrin properties and the elasticity of the red blood cell membrane skeleton. Biorheology 34:327–348, 1997.
Henon, S., G. Lenormand, A. Richert, and F. Gallet. A new determination of the shear modulus of the human erythrocyte membrane using optical tweezers. Biophys. J. 76:1145–1151, 1999.
Jay, A. W., and P. B. Canham. Viscoelastic properties of the human red blood cell membrane. II. Area and volume of individual red cells entering a micropipette. Biophys. J. 17:169–178, 1977.
Law, R., G. Liao, S. Harper, G. Yang, D. W. Speicher, and D. E. Discher. Pathway shifts and thermal softening in temperature-coupled forced unfolding of spectrin domains. Biophys. J. 85:3286–3293, 2003.
Lee, J. C., and D. E. Discher. Deformation-enhanced fluctuations in the red cell skeleton with theoretical relations to elasticity, connectivity, and spectrin unfolding. Biophys. J. 81:3178–3192, 2001.
Lee, J. C., D. T. Wong, and D. E. Discher. Direct measures of large, anisotropic strains in deformation of the erythrocyte cytoskeleton. Biophys. J. 77:853–864, 1999.
Li, J., M. Dao, C. T. Lim, and S. Suresh. Spectrin-level modeling of the cytoskeleton and optical tweezers stretching of the erythrocyte. Biophys. J. 88:3707–3719, 2005.
Lin, L. C., and F. L. Brown. Dynamic simulations of membranes with cytoskeletal interactions. Phys. Rev. E Stat. Nonlin. Soft Matter. Phys. 72:011910, 2005.
Liu, S. C., L. H. Derick, and J. Palek. Visualization of the hexagonal lattice in the erythrocyte membrane skeleton. J. Cell Biol. 104:527–536, 1987.
Meyer, G. R., J. Gullingsrud, K. Schulten, and B. Martinac. Molecular dynamics study of MscL interactions with a curved lipid bilayer. Biophys. J. 91:1630–1637, 2006.
Paramore, S., G. S. Ayton, D. T. Mirijanian, and G. A. Voth. Extending a spectrin repeat unit. I: linear force-extension response. Biophys. J. 90:92–100, 2006.
Picart, C., P. Dalhaimer, and D. E. Discher. Actin protofilament orientation in deformation of the erythrocyte membrane skeleton. Biophys. J. 79:2987–3000, 2000.
Picart, C., and D. E. Discher. Actin protofilament orientation at the erythrocyte membrane. Biophys. J. 77:865–878, 1999.
Rief, M., J. Pascual, M. Saraste, and H. E. Gaub. Single molecule force spectroscopy of spectrin repeats: low unfolding forces in helix bundles. J. Mol. Biol. 286:553–561, 1999.
Shen, B. W., R. Josephs, and T. L. Steck. Ultrastructure of the intact skeleton of the human erythrocyte membrane. J. Cell Biol. 102:997–1006, 1986.
Skalak, R., and P. I. Branemark. Deformation of red blood cells in capillaries. Science 164:717–719, 1969.
Skelton, R., and M. de Oliveira. Tensegrity Systems. Berlin: Springer Verlag, 2009.
Sleep, J., D. Wilson, R. Simmons, and W. Gratzer. Elasticity of the red cell membrane and its relation to hemolytic disorders: an optical tweezers study. Biophys. J. 77:3085–3095, 1999.
Sotomayor, M., and K. Schulten. Single-molecule experiments in vitro and in silico. Science 316:1144–1148, 2007.
Sung, K. L., and S. Chien. Influence of temperature on rheology of human erythrocytes. Chin. J. Physiol. 35:81–94, 1992.
Sung, K. L., J. Freedman, A. Chabanel, and S. Chien. Effect of complement on the viscoelastic properties of human erythrocyte membrane. Br. J. Haematol. 61:455–466, 1985.
Sung, L. A., and C. Vera. Protofilament and hexagon: a three-dimensional mechanical model for the junctional complex in the erythrocyte membrane skeleton. Ann. Biomed. Eng. 31:1314–1326, 2003.
Tozeren, A., R. Skalak, K. L. Sung, and S. Chien. Viscoelastic behavior of erythrocyte membrane. Biophys. J. 39:23–32, 1982.
Vera, C., R. Skelton, F. Bossens, and L. A. Sung. 3-D nanomechanics of an erythrocyte junctional complex in equibiaxial and anisotropic deformations. Ann. Biomed. Eng. 33:1387–1404, 2005.
Zhu, Q., and R. J. Asaro. Spectrin folding versus unfolding reactions and RBC membrane stiffness. Biophys. J. 94:2529–2545, 2008.
Zhu, Q., C. Vera, R. J. Asaro, P. Sche, and L. A. Sung. A hybrid model for erythrocyte membrane: a single unit of protein network coupled with lipid bilayer. Biophys. J. 93:386–400, 2007.
Acknowledgments
Pierre Valdez conducted his research at UCSD when he was supported by the “Bridges to the Future Program” from the San Diego Mesa College funded by the National Institute of General Medical Sciences (NIGMS), the National Center for Research on Minority Health (NCRMH), and National Institutes of Health (NIH). Carlos Vera and Mauricio de Oliveira were supported in part by NIH Research Grant No. PO1HL-43026 (Project 2, LA Sung).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Associate Editor Cheng Dong oversaw the review of this article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
de Oliveira, M., Vera, C., Valdez, P. et al. Nanomechanics of Multiple Units in the Erythrocyte Membrane Skeletal Network. Ann Biomed Eng 38, 2956–2967 (2010). https://doi.org/10.1007/s10439-010-0040-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10439-010-0040-4