Abstract
Current circulating tumor cells (CTC) detection methods have to compromise between sensitivity and throughput. High-throughput imaging cytometer based on serial time-encoded amplified microscopy (STEAM) facilitates CTC detection at single-cell sensitivity from abundant cells. However, this method lacks the information to spot heterogeneity of cells with high morphological similarity. Researches on cell biophysical properties suggest cell mechanotyping can be an indicator of phenotypic heterogeneity to improve classification ability of STEAM cytometer. Here, we present a high-throughput label-free acoustofluidic imaging cytometer for single-cell mechanotyping based on STEAM and acoustofluidic technology. The generated acoustic resonance field translocates cells to different transversal exit positions under continuous flow according to their intrinsic biophysical properties. Such displacements are recorded with images simultaneously using STEAM cytometry at approximately 2000 cells/s. We experimentally verified that our method accounting for both cell images and acoustic displacements can improve mechanotyping accuracy by 12% upon image-based phenotyping method. This new acoustofluidic imaging cytometer facilitates high-accuracy and high-throughput imaging cytometry for single-cell CTC mechanotyping.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
High sensitivity and fidelity detection of circulating tumor cells (CTC) is regarded a promising approach for the study of cancer disease, as non-invasive biopsy can be used as cancer diagnosis and prognosis biomarkers. Current gold standard of CTC liquid biopsy is immunoaffinity-based methods such as immunomagnetic affinity-based CTC capturing (Hoshino et al. 2011). But these methods suffer from low sensitivity due to insufficient binding and variance of cell surface biomarkers (Ghazani et al. 2013). Serial time-encoded amplified microscopy (STEAM) (Goda et al. 2009) is a continuous ultrafast imaging technology enabled by optical time-stretch (Goda and Jalali 2013) which achieves unprecedented imaging speed of millions of frames per second. It images, counts and phenotypes flowing cells with unprecedented speed at single-cell sensitivity and high-throughput, which appears to be a proper solution to highly sensitive detection of rare CTCs (Goda et al. 2012). Hence, researchers have explored extensively to further improve the performance of STEAM cytometer to meet clinical demands, such as improved image processing algorithm (Nitta et al. 2018; Chen et al. 2016; Zhao et al. 2018), higher resolution (Wu et al. 2017), lower system cost (Dong et al. 2018; Yan et al. 2018) and more diverse samples (Kobayashi et al. 2017; Jiang et al. 2017; Lei et al. 2016).
Nevertheless, STEAM cytometer which carries out cell phenotyping only based on two-dimensional cell images has limited utility in CTC applications. Characterization of tumor heterogeneity among CTCs at the single cell level is useful to study tumorigenesis, reduce drug resistance, and improve cancer therapies. So phenotyping ability is of great clinical and biological importance to single-cell CTC analysis. However, detection of CTC among blood cells using only two-dimensional images obtained by conventional STEAM cytometer suffers from high false positive rate when classifying various types of cancer cells with high morphological similarity, and unable to distinguish the origin and nature of CTC. Additional cell feature which can reflect phenotypic heterogeneity of cells is needed. While conventional cell phenotyping methods based on surface biomarkers are low-throughput and likely to affect cell viability (Hong and Zu 2013), cell biophysical property-based mechanotyping has proven to be an alternative approach for cancer cell classification, and has demonstrated its capacity in differentiating different cancer cell lines with various metastatic potential (Wirtz et al. 2011; Wang et al. 2019). Therefore, cell biophysical properties may be combined with STEAM imaging cytometer to improve cell mechanotyping ability, which provides multimodal identification of tumor cells to achieve high sensitivity and high-throughput for CTC detection.
The motivation behind this work is to provide a solution to apply the biophysical properties of cells to high-throughput STEAM imaging cytometer to improve its mechanotyping ability. Microfluidic technology has been extensively utilized to quantify cell deformability according to various output signals such as cell deformation, impedance changes, and cell transit time through constricting microchannels. Deng et al. (2017) presented a cell mechanotyping system characterizing large populations of single-cell deformability, where cell position was controlled in microchannels using inertial microfluidics and cell deformation was quantified. However, since these methods rely on the cell deformation caused by physical contact between cells and constricting geometries, they are prone to throughput limitation in general due to flow rate constrains. Moreover, cells may be damaged by direct contact with constricting microchannels. Another type of microfluidic methods to obtain biophysical properties relies on cell deformation generated by the flow. Otto et al. (2015) designed a narrow channel microstructure and Gossett et al. (2012) presented a cross-channel microdevice applying colliding fluids to cells, both to measure cell deformation at the cross-junction using a high-speed CCD camera and image processing module to image and analyze cell shape change. Recently, microfluidic acoustophoresis techniques have been utilized in non-contact cell and particle manipulation, separation, and concentration (Guo et al. 2016; Jakobsson et al. 2014a, b; Jakobsson et al. 2014a, b; Ku et al. 2018). Some studies have reported the application of bulk acoustic wave resonators to measure biophysical properties (Skowronek et al. 2015). The trajectories of cells moving in the applied acoustic resonant field were analyzed to obtain their compressibility parameter using reference cell densities in the literature (Hartono et al. 2011). Barnkob et al. (2010) used microbeads to calibrate the acoustic resonance field to extract density and compressibility parameters of particles and cells simultaneously. Moreover, Wang et al. (2019) incorporated flow focusing to control the entrance position of cells in the microchannel, and used cell sizes and their exit positions to improve cell mechanotyping accuracy, lower the setup cost and increase system throughput upon previous methods. This acoustofluidic technique is a non-contact, label-free, high-throughput method of cell mechanotyping which quantifies cell biophysical properties.
In this paper, a high-throughput label-free acoustofluidic imaging cytometer for single-cell mechanotyping based on STEAM is presented and demonstrated. We fabricated an acoustofluidic microchip and experimentally investigated its utility in acquiring biophysical features in ultrafast time stretch imaging. Using the developed system we have achieved ultrafast imaging at 50 million line frames per second, which is translated to 2000 cells per second under current experimental conditions. The acquired images have a field of view of 268 μm and resolution of 1.6 μm. Superior performance of the developed acoustofluidic imaging cytometer in cell mechanotyping as compared to conventional image-only STEAM cell mechanotyping method has been demonstrated. This acoustofluidic imaging cytometer technique has facilitated high-throughput time stretch imaging with additional biophysical feature to achieve more efficient and accurate cell detection, counting and mechanotyping.
2 Materials and methods
2.1 Principles and design of the acoustofluidic cytometer
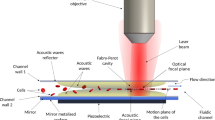
In this work, a continuous-flow acoustofluidic chip is constructed, as shown in Fig. 1, to acquire cell biophysical properties. On this chip, the cells could be introduced into the acoustic field at a constant position and then move in the transversal direction (Y-direction as in Fig. 1) in response to the acoustophoretic force, and have their exit positions recorded and analyzed. This method integrates a highly robust and accurate single-cell biophysical feature to STEAM mechanotyping at high throughput by recording cell exit positions. In this method cell exit positions and cell acoustic contrast factors are evaluated without the need for calculating the precise values of cell compressibilities and densities. The main force causing cell transversal displacement in an acoustic resonance field is primary acoustic radiation force. Its expression is as follows (Laurell et al. 2007):
where \(E_{{{\text{ac}}}}\) is the equivalent resonant wave acoustic field intensity, \(R\) is the radius of the cell, \(\lambda\) is the wavelength of the acoustic resonant wave, and \(F\) is the acoustic contrast factor given by the following equation:
Piezoelectric transducer attached to the bottom of the chip applies an acoustic resonance field in the main channel. Red spheres represent cells having a smaller acoustic contrast factor compared to the cells indicated as green spheres. When cells pass through the microchannel with acoustic resonance field, cells with different biophysical properties, such as size, density and compressibility, are subjected to different acoustophoretic forces and result in different exit positions. The scanning line spot records the exit positions of cells
where \(\rho_{{\text{p}}}\) and \(\rho_{{\text{o}}}\) are the densities of the cell and medium, respectively, and \(\gamma_{{\text{p}}}\) and \(\gamma_{{\text{o}}}\) are the compressibilities of the cell and medium, respectively. Besides, the cells are also subject to buoyant and gravitational forces as well as viscous drag force when moving in the microchannels.
Figure 1 illustrates the microfluidic chip design and working principle. This microfluidic chip shown in Fig. 1 is fabricated in glass/silicon. A photoresist layer (AZ4620, thickness: 12 μm) is patterned on a silicon substrate to form the etch mask. The microchannel is then etched in silicon by deep reactive ion etching to a depth of 40 μm. The fabricated silicon microchip is then anodically bonded to a glass substrate with inlet/ outlet holes pre-drilled. The fluidic connection is provided by flat-bottom ferrules and Tygon tubing. The main microchannel width of the fabricated microfluidic chip is 375 μm and the side channel width is 75 μm. The flow rates in each channel are set to: center main inlet 800 μl/h and side inlet 200 μl/h. This chip has a piezoelectric transducer attached to its bottom to form an acoustic resonance field inside the main microchannel to generate acoustophoretic forces on cells passing through. This piezoelectric ceramic plate is stimulated with an amplified sinusoidal wave at 1.980 MHz frequency generated by an arbitrary waveform generator and a 50 dB power amplifier. The arbitrary waveform generator generates sinusoidal waves to form the acoustic resonance field in the main channel. This structure of the microfluidic chip is generated from the microfluidic chip published by Wang et al. (2019). Several optimizations are made to adapt to the high flow rate and the setup of STEAM imaging system.
When cells enter the main channel, they are subjected to primary acoustic radiation force which moves them towards the channel center which is the transversal first harmonics pressure node when applied with the first order resonant frequency corresponding to the main channel width. Also, cells are subjected to size-dependent viscous drag force in the opposite direction of motion at the same time. The Y-direction positions of the cells as they exit the acoustic resonance field can be adjusted within the range between the main channel sidewall and the channel center by changing acoustic resonant wave field intensity, so that the cells with higher acoustic contrast factors move closer to channel center pressure node, while the cells with lower contrast factors remain near the channel sidewall. Since cells with different acoustic contrast factors are subjected to different acoustophoretic forces, the difference in Y-direction exit positions can be correlated to their difference in biophysical properties caused by the variation in size, density or compressibility, which adds to the cellular features obtained by STEAM. Finally, their displacements in the transversal Y-direction towards the channel center are scanned and recorded by STEAM scanning line spot as well as cell images for mechanotyping.
2.2 Imaging system design and setup
In our ultra-fast imaging system based on STEAM (Fig. 2), a broadband pulsed laser is employed as the light source with a pulse repetition rate of 50 MHz. After propagating through a section of dispersion compensating fiber, the optical pulses are dispersed in the time domain, which leads to the mapping between time domain and wavelength domain. Then, the optical pulse is cast into free space from a collimator. A combination of 1/2 and 1/4 wave plates is used to adjust the polarization state. The laser beam is then spatially dispersed by a diffraction grating. The two cylindrical lenses reduce the beam’s diameter to fit the size of objective lens (NA = 0.65). When the dispersed laser beam is focused onto the above-mentioned acoustofluidic chip, a laterally distributed focused scanning line spot is formed on the focal plane. Along this line spot, different wavelengths are located at different positions, indicating that the wavelength-to-space mapping is established. Target cells flow through and are illuminated by this one-dimensional scanning beam so that the intensity information of pulses forms a two-dimensional image containing cell images and transversal exit positions. Finally, the light beam travels back through the diffraction grating to a photodiode so that spatial information is encoded into time-domain waveform and received in sequence.
The parameters of optical components are optimized to meet the imaging requirements of the acoustofluidic chip. Since the width of the main microchannel is 375 μm, the imaging field of the scanning line spot should be more than 200 μm. The incident angle, diffraction angle of the diffraction grating and cylindrical lenses’ scaling factor are adjusted according to the following equations:
where \({\text{FOV}}\) is the imaging field of the line spot, \(f\) (4 mm) is the focal length of objective lens, \(n\) is the cylindrical lenses’ scaling factor, \(\lambda_{{{\text{band}}}}\) (10 nm) is the bandwidth of laser source, \(\lambda_{{{\text{center}}}}\) (1558 nm) is the central wavelength of laser source, \(d\) (1/1200 mm) is the grating constant, \(\phi_{z}\) (7 mm) is the diameter of collimator, \(\phi_{{\text{r}}}\) (6 mm) is the diameter of objective lens, and \(\alpha\) and \(\beta\) are the incident angle and diffraction angle of diffraction grating. Finally the component parameters are determined as follows: the incident angle 77.9°, the diffraction angle 63.4°, the scaling factor 2.5, and the imaging field 268 μm.
2.3 Sample culture and treatments
The presented acoustofluidic imaging cytometer was tested with four samples: BT474 cells, CACO2 cells, polystyrene beads of 10 μm diameter and polystyrene beads of 3 μm diameter. Both of the two cell lines were cultured in Dulbecco’s modified eagle medium (DMEM), with 10% fetal bovine serum and 1% penicillin and streptomycin. The cell lines were incubated in a 5% CO2 incubator at 37 °C. When the cell culture reached about 80% confluency, cells were washed with phosphate-buffered saline and treated with trypsin–EDTA for 3 min. Fresh medium was added to the cell culture mixture and the cells were gently pipetted from the bottom of the petri dish and centrifuged. The final single-cell suspension was prepared at the concentration of 107 cells/ml for use.
To calibrate the acoustic resonance field, 10 μm polystyrene beads with known density and compressibility were first introduced into the microchannel and used to calculate the equivalent acoustic resonance field intensity. Following this calibration, all these samples were tested for mechanotyping.
2.4 Automated cellular parameter extraction and classification algorithm
To obtain cellular parameters from raw data obtained by this acoustofluidic imaging system, we first extract cell images and their transversal exit positions. A threshold segmentation method binarizes the original image to separate cells from the background initially, because the obtained image has a uniform gray value within background area and a large contrast between background and cells. Then, we use closing operation and opening operation on the binary images obtained. Opening and closing operations are morphological image operations, where opening operation consists of an image erosion followed by an image dilation, and closing operation consists of a dilation followed by an erosion. They are applied to smooth edges, reduce noise, and eliminate the isolated background areas surrounded by cell areas. Next, we mark all the connected cell regions individually. For tiny inclusion removal, a threshold of the number of pixels under each area in the image is set according to the approximate sizes of cells flowing in our system. Then, we record the positions of all the cell regions left and extract grayscale cell images and their Y-direction positions (transversal displacement feature) accordingly.
As transversal displacement features of cells are extracted, it is time to extract the features representing cell morphological characteristics. According to the cell positions obtained above, we can cut out the cells in each image. Next, to avoid the impact of error image samples such as bubble images on our classification result, density based spatial clustering of applications with noise (DBSCAN) algorithm is used to remove outlier samples from acquired cell images (Ester et al. 1996). Then, principal component analysis (PCA) algorithm is employed to extract image features (image PCA features) to fit classification models instead of the cell images (Jolliffe 2002). The main idea of PCA is to reduce dimensionality of images having numerous interrelated variables while keeping the maximum possible variations within sample set. From the high-dimensional vector representation of the images, PCA finds a low-dimensional subspace whose basis vectors correspond to the maximum variance direction in the original image space. All images are projected onto the new subspace to find a set of weights that describes the contribution of each vector. The weights form the feature vector for the cell image.
Finally, extreme gradient boosting (XGBoost) algorithm (Chen and Guestrin 2016), which is a widely-used high-speed accurate classification algorithm, is used for cell mechanotyping based on transversal displacement features and image PCA features obtained above.
3 Results
3.1 Transversal displacement detection and image acquisition
Using the STEAM based acoustofluidic imaging cytometer, we imaged, analyzed and mechanotyped BT474 cells, CACO2 cells, polystyrene beads of 10 μm diameter and polystyrene beads of 3 μm diameter while using the 10 μm polystyrene beads for calibration. The laser signal containing samples' exit positions and images were recorded with an oscilloscope and then analyzed using a MATLAB program. The flow rate tested was 1000 μl/h, making the throughput in the range of 2000 cells/s.
The viability of cells in the experiment would not be affected. The power of the objective lens focal spot we set is 11 dBm. As the flow speed of cells is about 1.85 × 10–2 m/s, a cell with a radius of 20 μm flowing through the acoustic chip would absorb up to 7.96 × 10–7 J of energy. Referring to the 50% lethal dose of MCF-7 cells provided by Garsha (2003), the lethal energy of exposure is approximately 108 times the exposure energy in our experiment. Therefore, the cell viability is not affected by the STEAM imaging system.
Exit position detection and image acquisition of cells are shown in Figs. 3 and 4. Each sample image consists of approximately 6,000,000 pixels (approximately 80 pixels in the lateral direction and approximately 75,000 pixels in the flow direction). This excessively large number of pixels in the flow direction indicates our microscope’s ability to acquire images of cells that flow at a much higher speed. The maximum possible throughput is found to be about 1.5 × 106 cells/s theoretically (about 750 times higher throughput than 2000 cells/s we achieved in this experiment), assuming that the microfluidic device can endure such a high normal pressure in its microchannel and the sinusoidal wave amplifier has large enough output power.
3.2 Image-based mechanotyping
The image-based information provided by our time-stretch microscope of BT474 cells, CACO2 cells, 10 μm polystyrene beads and 3 μm polystyrene beads are shown in Fig. 4, respectively. In Fig. 4, it can be seen that the sizes and morphology of 3 μm polystyrene beads and 10 μm polystyrene beads have great difference from other sample groups, while the images of the two groups of cell samples are very similar. To further classify these four groups of samples, 50% cell images are picked randomly from all sample groups as training set to fit classification model and the 50% left as test set. Image PCA features of training sets are extracted as described above and XGBoost models are trained accordingly. Finally, the PCA features of test sets are calculated and grouped by the XGBoost models, respectively.
An overall mechanotyping accuracy of 91.6%, as shown in Table 1, is achieved. However, the mechanotyping ability between the two groups of cell samples deteriorates a lot with a correctly classified rate of 84.3% as their images have high similarity. To improve the detection specificity, additional cell biophysical parameters are needed for multi-dimensional characterization of cell samples.
3.3 Combined mechanotyping of acoustofluidic events and images
When no acoustophoretic force was applied, all samples showed similar exit positions as expected. Transversal displacement feature of each sample shown in Fig. 3 is obtained by their exit positions with acoustic resonance field on minis average exit positions of each sample group with acoustic resonance field off. This transversal displacement feature can be fused with the image PCA feature to reduce the classification error rate. Therefore, another XGBoost model is fit and predicted by the same samples we have just grouped. The only difference is that the image PCA features are combined with transversal displacement features this time.
The results in Fig. 5 are based on the sample size vs. transversal displacement, which can help to easily visualize differences in their biophysical properties, and thus can be used for cell mechanotyping. The sample sizes are calculated using a MATLAB program to analyze their images. As can be seen in Fig. 5, BT474 cells and CACO2 cells have similar size distributions, while CACO2 shows longer travelling distance in the transversal direction, which can be interpreted as CACO2 experiencing a stronger acoustic radiation force, an indicator of a higher acoustic contrast factor. As for the two groups of bead samples, they have the same acoustic contrast factors. However the smaller 3 μm beads showed less transversal displacement than larger 10 μm beads, due to their size difference and resulted acoustic radiation force and viscous drag force. Their accelerations generated by acoustic radiation force are equal, while viscous drag force accelerations working against acoustic radiation force are more significant for smaller beads. Therefore, 3 μm beads result in less transversal displacements than 10 μm beads.
To calculate the acoustic contrast factor of a cell according to its transversal displacement, cell size and equivalent acoustic resonance field intensity is needed. Cell size can be extracted from its image. The acoustic resonance field intensity is calibrated by the transversal displacements of 10 μm polystyrene beads with known density and compressibility (the acoustic contrast factor of polystyrene beads is 0.472). Each bead help calculate a field intensity. The final field intensity used to determine the acoustic contrast factors of other samples is the average of all imaged 10 μm beads. Therefore, based on the displacement results in Fig. 5, the acoustic contrast factors of these samples are calculated and shown in Fig. 6, after taking into account all forces the cells are subjected to (i.e., acoustophoretic force, viscous drag force, gravitational force, and buoyancy force). As shown in Eq. (2), the acoustic contrast factor reflecting the cell biophysical properties is determined by cell density and compressibility. From Fig. 6, it can be clearly seen that CACO2 group has higher acoustic contrast factor than BT474 group, and 3 μm polystyrene beads have similar acoustic contrast factors to 10 μm beads due to their same material. The order of the acoustic contrast factors of these samples is: 3 μm polystyrene beads (0.499) ≈ 10 μm polystyrene beads (0.472) > CACO2 (0.185) > BT474 (0.122). It is also shown in the figure that acoustic contrast factors are independent of sample sizes. This biophysical property makes the discrimination between the two cell sample sets more significant. In other words, the displacements contain a biophysical feature not included in pure cell imaging, which could help discriminate cells from an additional perspective.
By combining the image PCA features and transversal displacement features, we can classify the groups of BT474 cells and CACO2 cells with higher accuracy. As indicated in Table 2, the error rate of the image-only-based mechanotyping method is about 15.7%, while the combined mechanotyping of acoustic transversal displacements and images method as low as 3.2% (calculated from 32 error events over the total population of 1000). In other words, STEAM-based cell classification is less erroneous by a factor of 5 using the two feature dimensions simultaneously, which validates the effectiveness of our combined mechanotyping of acoustofluidic events and images. Some other methods were also translocating cells to separate them (Yamada et al. 2004; Geislinger and Franke 2013). However, their displacement differences were caused by cell sizes purely. Without cell biophysical properties not included in cell images, the displacement feature would not improve the classification capability of STEAM.
4 Conclusion
In conclusion, we present a high-throughput label-free acoustofluidic imaging cytometer for single-cell CTC mechanotyping based on STEAM and acoustofluidic technology. The acoustophoretic force is utilized to translocate cells to different transversal exit positions under continuous flow according to their intrinsic biophysical properties. Such movement can be analyzed to reflect differences of various cells to improve the cell mechanotyping capability of STEAM. We experimentally verify that the presented system which accounts for both the cell images and cell acoustic transversal displacements can improve cell mechanotyping accuracy significantly upon previous image-based STEAM method. The throughput of the presented method can be further increased by applying higher flow rate and higher actuation power. The label-free, non-contact and high processing speed characteristics of our cytometer ensures its potential to provide real-time mechanotyping and further CTC sorting applications. We expect that the developed system can be used in a variety of applications, such as phenotyping of cancer cells with different metastatic potential.
Availability of data and material
Not applicable.
References
Barnkob R, Augustsson P, Laurell T, Bruus H (2010) Measuring the local pressure amplitude in microchannel acoustophoresis. Lab Chip 10(5):563–570
Chen T, Guestrin C (2016) Xgboost: a scalable tree boosting system. In: Proceedings of the 22nd acm sigkdd international conference on knowledge discovery and data mining ACM, pp 785–794
Chen CL, Mahjoubfar A, Tai LC, Blaby IK, Huang A, Niazi KR, Jalali B (2016) Deep learning in label-free cell classification. Sci Rep 6:21471
Deng Y, Davis SP, Yang F, Paulsen KS, Kumar M, Sinnott DeVaux R, Chung AJ (2017) Inertial microfluidic cell stretcher (iMCS): fully automated, high-throughput, and near real-time cell mechanotyping. Small 13(28):1700705
Dong X, Zhou X, Kang J, Chen L, Lei Z, Zhang C, Zhang X (2018) Ultrafast time-stretch microscopy based on dual-comb asynchronous optical sampling. Opt Lett 43(9):2118–2121
Ester M, Kriegel HP, Sander J, Xu X (1996) A density-based algorithm for discovering clusters in large spatial databases with noise. KDD 96(34):226–231
Garsha K (2003) Effects of femtosecond pulse dispersion precompensation on average power damage thresholds for live cell imaging: implications for relative roles of linear and nonlinear absorption in live cell imaging. Proc SPIE 4963:134–142
Geislinger TM, Franke T (2013) Sorting of circulating tumor cells (MV3-melanoma) and red blood cells using non-inertial lift. Biomicrofluidics 7(4):044120
Ghazani AA, McDermott S, Pectasides M, Sebas M, Mino-Kenudson M, Lee H, Castro CM (2013) Comparison of select cancer biomarkers in human circulating and bulk tumor cells using magnetic nanoparticles and a miniaturized micro-NMR system. Nanomed Nanotechnol Biol Med 9(7):1009–1017
Goda K, Ayazi A, Gossett DR, Sadasivam J, Lonappan CK, Sollier E, Wang C (2012) High-throughput single-microparticle imaging flow analyzer. Proc Natl Acad Sci 109(29):11630–11635
Goda K, Jalali B (2013) Dispersive Fourier transformation for fast continuous single-shot measurements. Nat Photonics 7(2):102
Goda K, Tsia KK, Jalali B (2009) Serial time-encoded amplified imaging for real-time observation of fast dynamic phenomena. Nature 458(7242):1145
Gossett DR, Tse HTK, Lee SA, Ying Y, Lindgren AG, Yang OO, Di Carlo D (2012) Hydrodynamic stretching of single cells for large population mechanical phenotyping. Proc Natl Acad Sci 109(20):7630–7635
Guo F, Mao Z, Chen Y, Xie Z, Lata JP, Li P, Ren L, Liu J, Yang J, Dao M, Suresh S, Huang TJ (2016) Three-dimensional manipulation of single cells using surface acoustic waves. Proc Natl Acad Sci 113(6):201524813
Hartono D, Liu Y, Tan PL, Then XYS, Yung LYL, Lim KM (2011) On-chip measurements of cell compressibility via acoustic radiation. Lab Chip 11(23):4072–4080
Hong B, Zu Y (2013) Detecting circulating tumor cells: current challenges and new trends. Theranostics 3(6):377
Hoshino K, Huang YY, Lane N, Huebschman M, Uhr JW, Frenkel EP, Zhang X (2011) Microchip-based immunomagnetic detection of circulating tumor cells. Lab Chip 11(20):3449–3457
Jakobsson O, Antfolk M, Laurell T (2014) Continuous flow two-dimensional acoustic orientation of nonspherical cells. Anal Chem 86(12):6111–6114
Jakobsson O, Grenvall C, Nordin M, Evander M, Laurell T (2014) Acoustic actuated fluorescence activated sorting of microparticles. Lab Chip 14(11):1943–1950
Jiang Y, Lei C, Yasumoto A, Kobayashi H, Aisaka Y, Ito T, Nakagawa A (2017) Label-free detection of aggregated platelets in blood by machine-learning-aided optofluidic time-stretch microscopy. Lab Chip 17(14):2426–2434
Jolliffe IT (2002) Principal component analysis. J Mark Res 87(4):513
Kobayashi H, Lei C, Wu Y, Mao A, Jiang Y, Guo B, Goda K (2017) Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning. Sci Rep 7(1):12454
Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S, Ceder Y (2018) Acoustic enrichment of extracellular vesicles from biological fluids. Anal Chem 90(13):8011–8019
Laurell T, Petersson F, Nilsson A (2007) Chip integrated strategies for acoustic separation and manipulation of cells and particles. Chem Soc Rev 36(3):492–506
Lei C, Ito T, Ugawa M, Nozawa T, Iwata O, Maki M, Tsumura N (2016) High-throughput label-free image cytometry and image-based classification of live Euglena gracilis. Biomed Opt Express 7(7):2703–2708
Nitta N, Sugimura T, Isozaki A, Mikami H, Hiraki K, Sakuma S, Fukuzawa H (2018) Intelligent image-activated cell sorting. Cell 175(1):266–276
Otto O, Rosendahl P, Mietke A, Golfier S, Herold C, Klaue D, Wobus M (2015) Real-time deformability cytometry: on-the-fly cell mechanical phenotyping. Nat Methods 12(3):199
Skowronek V, Rambach RW, Franke T (2015) Surface acoustic wave controlled integrated band-pass filter. Microfluid Nanofluidics 19(2):1–7
Wang H, Liu Z, Shin DM, Chen ZG, Cho Y, Kim YJ, Han A (2019) A continuous-flow acoustofluidic cytometer for single-cell mechanotyping. Lab Chip 19(3):387–393
Wirtz D, Konstantopoulos K, Searson PC (2011) The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 11(7):512
Wu JL, Xu YQ, Xu JJ, Wei XM, Chan AC, Tang AH, Wong KK (2017) Ultrafast laser-scanning time-stretch imaging at visible wavelengths. Light Sci Appl 6(1):e16196
Yamada M, Nakashima M, Seki M (2004) Pinched flow fractionation: continuous size separation of particles utilizing a laminar flow profile in a pinched microchannel. Anal Chem 76(18):5465–5471
Yan W, Wu J, Wong KK, Tsia KK (2018) A high-throughput all-optical laser-scanning imaging flow cytometer with biomolecular specificity and subcellular resolution. J Biophotonics 11(2):e201700178
Zhao W, Wang C, Chen H, Chen M, Yang S (2018) High-speed cell recognition algorithm for ultrafast flow cytometer imaging system. J Biomed Opt 23(4):046001
Funding
This study was funded by National Natural Science Foundation of China (NSFC) (61771284, 81801791); Beijing Municipal Natural Science Foundation (BMNSF) (L182043); and National Key Research and Development Program of China (2019YFB1803501).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Code availability
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhao, W., Wang, H., Guo, Y. et al. A high-throughput label-free time-stretch acoustofluidic imaging cytometer for single-cell mechanotyping. Microfluid Nanofluid 24, 88 (2020). https://doi.org/10.1007/s10404-020-02395-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10404-020-02395-7