Abstract
A xylanase gene (thxyn11A) from the Thermobifida halotolerans strain YIM 90462T was cloned and expressed in Escherichia coli. The open reading frame (ORF) of thxyn11A has 1,008 bp encoding a mature xylanase with a high degree of similarity (80 %) to the xylanase from Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111. This enzyme (Thxyn11A) also possesses a glycosyl hydrolases family 11 (GH11) domain and a high isoelectric point (pI = 9.1). However, Thxyn11A varies from most GH11 xylanases, due to its large molecular mass (34 kDa). Recombinant Thxyn11A demonstrated a strong pH and temperature tolerance with a maximum activity at pH 9.0 and 70 °C. Xylotriose, the end-product of xylan hydrolysis by Thxyn11A, serves as a catalyst for hemicellulose pretreatment in industrial applications and can also function as a food source or supplement for enterobacteria. Due to its attractive biochemical properties, Thxyn11A may have potential value in many commercial applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The largest renewable carbon source on earth is the cell wall of plants, which consist primarily of cellulose, hemicellulose, pectin, and lignin [27]. Among these components, hemicellulose is the second-most abundant fraction after cellulose [2]. With the increasing awareness of the environmental issues caused by fossil fuel use and depletion, hemicellulose has been suggested to be a promising source of renewable energy. However, capturing this vast resource for energy, material, and chemicals presents a formidable challenge, due to the recalcitrant structure of hemicellulose. Xylan, the main component of hemicellulose, is a heterogeneous polysaccharide composed of a β-1,4-linked xylopyranose backbone with arabinofuranosyl, acetyl, or methylglucuronosyl side chains. Because of this complex structure, the complete degradation of xylan requires the synergistic actions of several enzymes [3].
Among the various xylanolytic enzymes, xylanases (1,4-β-d-xylan, EC 3.2.1.8) play a pivotal role in depolymerizing the xylan backbone. Based on the amino acid sequence similarities of their catalytic domains, xylanases are mainly classified into two glycoside hydrolases groups: family 10 (GH10) and family 11 (GH11) [7, 29]. Xylanases in the GH10 group have high molecular masses (>30 kDa) and low isoelectric points (pI), whereas GH11 xylanases have low molecular masses (<30 kDa) and high pIs. The major enzymes comprising the GH10 family are endo-1,4-β-xylanases and a small number of endo-1,3-β-xylanases (EC 3.2.1.32). By contrast, the GH11 family consists solely of endo-1,4-β-xylanases and usually gives larger end-products than the GH10 family members [5]. As a food source, larger xylo-oligosaccharides not only increase the populations of probiotics in the gut but also suppress the activity of enteric putrefactive bacteria, prevent the proliferation of pathogenic intestinal bacteria, facilitate digestion, and aid in the absorption of nutrients [28].
Many xylanases belonging to the GH11 family have been obtained from actinomycetes; however, few xylanases have been reported to be active and stable at an alkaline pH and elevated temperatures [24]. With the current methods of manufacturing cellulosic feedstocks, which depend on alkali and heat pretreatment, xylanases that are stably active at both high temperatures and under alkaline conditions are of particular value [10]. The Thermobifida halotolerans strain YIM 90462T is an aerobic, thermophilic, and halotolerant actinomycete found in the Yunnan Province of southwest China [31], which contains an alkaline thermostable GH9 endoglucanase and a thermostable xylanase that have been described previously [32, 33]. However, the xylanase is purified from fermentation broth of the native stain, and it is an acid-stable xylanase. This study describes the cloning, heterologous expression, and characterization of the alkaline thermostable xylanase (Thxyn11A) from this bacterial strain. Based on its distinctive features, Thxyn11A may be of potential use in biofuel production and other commercial applications.
Materials and methods
Bacterial strains, growth conditions, and genomic DNA isolation
Thermobifida halotolerans YIM 90462T was isolated from a salt mine sample during a previous study [31]. The bacteria were cultured for 1 week in Luria–Bertani medium at 45 °C. DNA was subsequently isolated from the mycelia using the method described by Li et al. [15].
Cloning the full sequence of the thxyn11a gene
By comparing ten amino-acid sequences of GH11 xylanases from actinomycetes, two degenerate primers (DP1 and DP2) (Table 1) were designed using the CODEHOP method (http://bioinformatics.weizmann.ac.il/blocks/codehop.html). The polymerase chain reaction (PCR) was performed using the following parameters: one cycle at 94 °C for 5 min, 30 cycles at 94 °C for 45 s, 64 °C for 45 s, and 72 °C for 1 min, and then a final extension at 72 °C for 10 min. Similarly, a conserved gene fragment of inositol monophosphatase, which is located downstream of the xylanase gene, was amplified using two additional degenerate primers (IMFP and IMRP) (Table 1) employing the method described above. The amplified fragments were purified and ligated into the pEASY-T1 vector (TransGen, Beijing; China) for sequencing and BLAST analysis. After amplification, sequencing; and BLAST analysis, a 417-bp DNA fragment of the xylanase gene and a 589-bp DNA fragment of the inositol monophosphatase gene were amplified using the aforementioned primers. Based on these two DNA fragments, a pair of specific primers (SPRP and SPFP) (Table 1) were designed to amplify the C-terminal encoding sequence of the xylanase gene. In order to amplify the N-terminal encoding sequence of xylanase, the SiteFinding-PCR method [25] was implemented using three nested primers (SP1U, SP2U; and SP3U) (Table 1). Both amplified fragments were purified and ligated into the pEASY-T1 vector for sequencing and BLAST analysis, as described above.
Nucleotide sequence analysis and accession number assignment
The sequenced DNAs were compared to available sequences from GenBank using the BLASTX program (http://blast.ncbi.nlm.nih.gov/Blast/), and all gene fragments were assembled using DNAstar software (DNAStar, Madison, WI, USA). The primary structure of the amino acid sequence was deduced and analyzed using EXPASY tools (http://expasy.org/). The signal peptide sequence of the protein was predicted using SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/). The phylogenetic tree was drawn with Mega 4.0, and a conserved domain analysis was conducted using Pfam (http://pfam.wustl.edu/hmmsearch.shtml). The sequence of the thxyn11A gene was submitted to GenBank and assigned the accession number JN016522.
Expression vector construction
After the complete thxyn11A gene sequence was obtained, two specific primers (Thxyn11A-FP and Thxyn11A-RP; NdeI and XhoI sites shown in italics and bold) (Table 1) were designed to amplify the gene. The PCR mixture (50 μl) consisted of 2× PCR buffer, 0.4 mM of each dNTP, 1.0 unit of KOD FX DNA polymerase (TOYOBO, Osaka, Japan), 15 pmol of each primer, and 50 ng of the template DNA. The PCR cycle conditions consisted of an initial step of 5 min at 94 °C followed by 30 cycles of 10 s at 98 °C and 1 min at 68 °C with a final extension at 68 °C for 10 min. The PCR products were gel-purified, digested with NdeI and XhoI (Fermentas, Maryland, USA), and cloned into the pET28a vector (Novagen, Darmstadt, Germany) to generate the recombinant plasmid pET28a-thxyn11A. The construct was subsequently transformed into Top10 competent cells (Invitrogen, Shanghai, China) for sequencing and BLAST analysis.
Xylanase gene expression and purification
The pET28a-thxyn11A plasmid was extracted from positive Top10 cells according to the manufacturer’s protocol (Tiangen, Beijing, China) and transformed into the E.coli BL21 (DE3) strain for protein expression. The transformants were grown in LB medium with 50 μg/ml kanamycin (Sigma, St. Louis, MO, USA) at 37 °C overnight. Three milliliters of a saturated culture were inoculated into 300 ml of Terrific broth [23] and incubated at 37 °C with shaking until the cell density reached an absorbance of 0.6 at 600 nm. To induce protein expression, 300 μl IPTG (100 mM) was added into the culture and incubated for approximately 24 h at 37 °C on a rotary shaker (200 rpm). The culture supernatant, which contained the recombinant Thxyn11A, was obtained by centrifugation at 12,000× g for 20 min at 4 °C. Next, the supernatant was loaded onto a Ni–NTA column (Merck, Darmstadt, Germany). After allowing binding to proceed for 30 min, the resin was washed with five column volumes of buffer A (50 mM Tris pH 8.0, 300 mM NaCl, and 20 mM imidazole) and eluted with five column volumes of buffer B (50 mM Tris pH 8.0, 300 mM NaCl, and 500 mM of imidazole). The eluted protein was concentrated at 4 °C with an Amicon centrifugal filter unit (MWCO 10,000, Millipore, Massachusetts, USA). The homogeneity of 10 μl concentrated protein was monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using a 12 % (w/v) acrylamide gel. The proteins were visualized by Coomassie Brilliant Blue R-250 or Congo Red staining [17].
Enzyme and protein assays
The purified, concentrated protein fraction containing 3 μg Thxyn11A was used for enzyme characterization. The protein concentration was determined by the Sangon protein assay kit (Sangon, Shanghai, China) using bovine serum albumin as a standard. The xylanase activity was determined by measuring the amount of reducing sugars released from birch wood xylan using the 3,5-dinitrosalicylic acid (DNS) reagent according to the method described by Miller [20]. The reaction mixture, which contained 80 μl of 1.0 % (w/v) birch wood xylan and 20 μl of suitably diluted enzyme, was incubated in 0.05 M Glycine-NaOH buffer (pH 9.0) at 70 °C for 10 min. The reaction was terminated by the addition of 150 μl of 1.0 % (w/v) DNS. The mixture was subsequently boiled for 5 min and cooled, and the optical density was measured at 540 nm. All experiments were performed in triplicate, and the statistical analyses were performed using SigmaPlot 12.0.
The effect of pH and the pH stability of purified Thxyn11A
The effect of pH on the enzymatic activity of Thxyn11A was assessed by measuring the relative activity using 20 μl of a xylanase solution incubated with 80 μl of 1 % birch wood xylan in buffers of varying pH [0.05 M McIlvaine buffer (pH 4.0–7.5), 0.05 M Tris–HCl buffer (pH 7.5–9.0) and 0.05 M Glycine-NaOH buffer (pH 9.0–10.5)]. All of the experiments were performed at 70 °C for 10 min, and the maximum activity was set as 100 %. To determine the pH stability, the enzyme was exposed to four buffers with different pH values, 6.0, 7.0, 8.0, and 9.0, and incubated at 70 °C for 240 min. An aliquot of xylanase was removed and assayed every 30 min for enzymatic activity, as described above.
The effect of temperature and the thermostability of purified Thxyn11A
The effect of temperature on the enzymatic activity of Thxyn11A was determined by measuring the relative activity using the reaction system described above with temperatures ranging between 30 and 90 °C. All experiments were performed in 0.05 M Glycine-NaOH buffer (pH 9.0) for 10 min. The thermal stability of the enzyme was measured as the residual enzyme activity after the incubation of the enzyme at 70, 80, or 90 °C for 240 min. An aliquot of xylanase was removed every 30 min to measure the enzymatic activity, as described above.
The effect of different metal ions and other compounds on Thxyn11A activity
Purified Thxyn11A was incubated with 1 % birch wood xylan in 0.05 M Glycine-NaOH buffer (pH 9.0) containing 1 mM MgCl2, PbAc2, BaCl2, LiCl, KCl, CaCl2, NaCl, CuCl2, MnCl2, AlCl3, FeCl3, NiSO4, ZnCl2, CoCl2, BiCl2, CdSO4, EDTA, DTT, PMSF, or 1 % SDS for 10 min at 70 °C. The relativity activity of the enzyme was measured, and the activity of the enzyme in buffer alone was defined as 100 %.
Analysis of the hydrolysis products
To obtain the hydrolysis products, 20 μl of purified xylanase (15.6 μg/ml) was incubated with 2 % birch wood xylan in 0.05 M Glycine-NaOH buffer (pH 9.0) at 70 °C. An aliquot of the reaction mixture was withdrawn after 12 h, and the reaction was stopped by boiling the solution for 5 min. The reaction mixture was subsequently centrifuged, and the supernatant was subjected to thin layer chromatography (TLC) using TLC plates (silica gel 60 F254, Jingdao, China). Xylobiose, xylotriose, xylotetraose, and xylopentaose were used as standards (Megazyme, Wicklow, Ireland). The TLC plates were developed with chloroform–acetic acid–H2O (6:7:1, v/v/v), sprayed with a methanol–sulfuric acid mixture (95:5, v/v), and heated at 150 °C in an oven until spots appeared.
The substrate specificity and kinetic parameters of Thxyn11A
Xylanase was incubated with 1 % (w/v) Avicel, beech wood xylan, birch wood xylan, Carboxyl Methyl Cellulose (CMC), β-glucan, Lichenan, or oat spelt xylan (Sigma, St. Louis, Missouri, USA) in 0.05 M Glycine-NaOH buffer (pH 9.0) at 70 °C for 10 min to test for substrate specificity. The reaction was stopped, and the relative activity of the enzyme was measured and compared to the enzymatic activity using a standard substrate, which was defined as 100 %. The K m and V max values for the purified recombinant enzyme were determined using the standard reaction conditions with 2–12 mg/ml birch wood xylan as a substrate. The data were plotted according to the Lineweaver–Burk method [16].
Results and discussion
Gene cloning
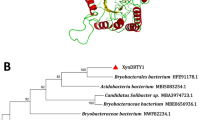
A 417-bp gene fragment was amplified by the CODEHOP method. The nucleotide sequence of this fragment shared 87 % identity with the xylanase gene from Streptomyces sp. S9, indicating that it was a partial xylanase gene. After two PCR products from the 5′ and 3′ flanking regions were isolated, sequenced, and assembled with the core gene region, the resulting DNA sequence was 1,185 bp, which contained a 1,008-bp ORF. The resulting protein sequence showed a high degree of similarity to several known xylanases including Nocardiopsis dassonvillei subsp. dassonvillei DSM 43111 xylanase (80 %; gi|9246561), T. fusca YX (78 %; gi|3580704), S. sp. S9 (75 %; gb|ACF57947.1), and S. viridosporus (74 %; gb|AAF09501.1) (Fig. 1). To date, many xylanase genes have been cloned from varying microorganisms, including Aspergillus versicolor MKU3 [12], Nesterenkonia xinjiangensis CCTCC AA001025 [9], Actinomadura sp. S14 [26], Fusarium oxysporum [6], Phanerochaete chrysosporium [8], Chaetomium thermophilum [1], Paenibacillus sp. 12–11 [34] and S. sp. S27 [14]. However, this study is the first to clone a xylanase gene from T. halotolerans YIM 90462T.
Sequence analysis
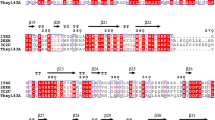
A conserved domain search using Pfam confirmed the presence of a GH11 xylanase catalytic domain and two putative catalytic glutamate residues (E128 and E217). Therefore, the cloned xylanase was designated Thxyn11A. Using the neural networks of SignalP, a potential signal peptide was predicted within amino acids 1–12. The mature protein consists of 323 amino acids with a calculated molecular weight of 34 kDa and an isoelectric point of 9.1. As a typical GH11 xylanase, Thxyn11A contains an N-terminal GH11 catalytic domain and two highly conserved Glu residues, which are important for the hydrolytic activity. However, the molecular weight of Thxyn11A is larger than the majority of xylanases, which usually are less than 30 kDa, as mentioned above [5]. Nonetheless, the molecular weight of Thxyn11A is notably similar to those of Cfl Xyn11A [18] and XynB119 [35]. The reason of this phenomenon is an extra cellulose-binding module (CBM) appended the C-terminals of them, and this accessory structure can potentiate the activity of enzymes that attack the plant cell wall by proximity effects [4]. In addition, there is a report that C-terminal region plays an important role in thermostability of GH11 Xylanase from S. lividans [30].
Gene expression and protein purification
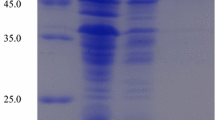
The intact coding region of the thxyn11A gene within the pET28a-thxyn11A vector was introduced into E. coli BL21 (DE3) cells. After inducing the cells with 1 mM IPTG, the C-terminal His6-tagged xylanase was produced intracellularly and was also partially secreted into the culture supernatant. No xylanase was detected in the non-induced cells harboring the pET28a-thxyn11A construct (Fig. 2). In order to rapidly purify the protein, only the xylanase from the supernatant was collected and purified by Ni2+ affinity chromatography (Fig. 2). SDS-PAGE analysis of the eluted fraction revealed a band of approximately 34 kDa (Fig. 2, lane 4, 5), which corresponded to the theoretical molecular weight of the mature xylanase protein (34,112 Da). A similar-weight xylanase, XynB119, has also been shown to be secreted when recombinantly expressed [35]; however, the signal peptide of XynB119 was artificially introduced from the pET-22b (+) vector. In contrast, Thxyn11A has a secretion signal peptide inherently within the gene and therefore is more likely to keep its native structure and activity upon secretion [13].
SDS-PAGE analysis of the expression and purification of recombinant Thxyn11A. Lane 1 contains protein markers. Lane 2 contains the total cell extract of E. coli BL21 (DE) expressing the pET28a-thxyn11A plasmid induced by IPTG. Lane 3 contains the total cell extract of E. coli BL21 (DE) expressing the pET28a-thxyn11A plasmid without induction. Lane 4 contains Ni–NTA purified recombinant Thxyn11A. Lane 5 contains a zymogram of the purified Thxyn11A
Temperature and pH optimization and stability
The purified xylanase exhibited a high activity at temperatures ranging between 40 and 90 °C (Fig. 3a) with a maximum activity at 70 °C. More than 90 % of its maximal activity was retained at 70 °C for 30 min at pH 9.0 (Fig. 3b). Although Thxyn11A cannot maintain its maximal activity for longer than 30 min at 80 or 90 °C, it displayed greater than 50 % of its maximal activity at 70 °C for up to 90 min, thereby indicating that it is a thermostable enzyme. The influence of pH on the xylan hydrolytic activity of the recombinant xylanase is presented in Fig. 3c. We found the enzyme displayed more than 70 % of its maximal activity between pH 6.0 and 10.0 with an optimal activity at pH 9.0. Although the purified Thxyn11A retained more than 70 % of its initial activity after incubation at pH 7.0 for 150 min, its alkaline stability dramatically decreased after incubation at pH 8.0 for 120 min or pH 9.0 for 30 min (Fig. 3d). Interestingly, Thxyn11A could maintain its activity at pH 6.0 longer than at pH 7.0 (data not shown). To our knowledge, three GH11 xylanases have been cloned from the genus Thermobifida [26]. The optimum catalysis temperature for the Thermobifida sp. xylanase is the highest known of the xylanases (80 °C), but it is an acid-stable xylanase. The xylanase from Thermobifida fusca NTU22 has the same optimum catalysis temperature as Thxyn11A but exhibits its maximal activity at a neutral pH, which limits its application for hemicellulose bioprocessing.
The effect of pH and temperature on the activity and stability of the recombinant xylanase. a The effect of temperature on xylanase activity. b The thermostability of the xylanase. c The effect of pH on xylanase activity. d The pH stability of the enzyme. The data shown in this figure represent the mean ± SD of three experimental replicates
The effect of various chemicals on Thxyn11A activity
The xylanase activity of Thxyn11A in the presence of different metal ions or chemical reagents was determined with CMC as a substrate. Among the metal ions tested, 1 mM Co2+or Mn2+ enhanced enzymatic activity approximately 1.2-fold, while Fe3+ and Pb2+ inhibited the xylanase activity. Incubation with other cations only induced a partial stimulation or inhibition of the enzyme (80 %< activity remaining <110%). Although one fungal GH11 xylanase has been shown to be inhibited by Co2+ [21], other reports have demonstrated that Co2+ can increase the activity of xylanase, even in 5 mM to 10 mM Co2+ [13, 19].
Interestingly, as a heavy-metal ion, the inhibition of Thxyn11A by Pb2+ was not as high as expected (only approximately 20 %), which implies that Thxyn11A could have great potential for wastewater-treatment applications. Among the chemical reagent tested, the activity of Thxyn11A was enhanced to 118 % by DTT, inhibited by EDTA (59 % remaining) and SDS (60 %), and caused no measurable effect by PMSF treatment. The enhanced activity in the presence of DTT suggests a potentially reactive thiol group may be found in the enzyme [19]. Contrarily, the lack of change with PMSF treatment suggests the absence of a potentially cleavable serine group in the enzyme active site. Additionally, the decreased activity of the xylanase in the presence of EDTA indicates that a metal-ion-binding site may be found within the enzyme active site, and the inhibition of xylanase in the presence of SDS may be due to the denaturation of the enzyme from its native conformation.
Analysis of the hydrolysis products
Birchwood xylan was hydrolyzed with purified Thxyn11A, and the resulting products were analyzed by TLC (Fig. 4). The reaction produced xylotriose as the end-product when the reaction was allowed to proceed for 12 h. In addition, larger xylooligomers, like xylotetroase and xylopentaose, were also detected. The observed product profile demonstrates Thxyn11A is a xylan endo-acting enzyme that belongs to the GH11 family. Although this hydrolysis pattern is consistent with a GH11 xylanase from Bacillus licheniformis [13], other GH11 xylanases produce a mixture of xylose and xylooligosaccharide from the hydrolysis of xylan [14, 20, 22]. The reason Thxyn11A fails to produce smaller sugar units remains unclear; however, this enzymatic property will make Thxyn11A more appealing for use in bioconversion and the food industry, due to its larger end-products, which protect probiotics from pathogenic microorganisms.
Thin-layer chromatography showing the hydrolysis products obtained by the action of Thxyn11A on birch wood xylan. Lane 1 contains the standard enzyme reaction mix. Lane 2 contains the reaction mix without Thxyn11A. Lane 3 contains the reaction mix with Thxyn11A. All reactions were performed under standard conditions for 12 h
The substrate specificity and kinetic parameters of purified Thxyn11A
The purified xylanase activities for various substrates are assayed under a standard condition. The enzyme had a relatively narrow substrate preference, exhibiting 100 % relative activity for birch wood xylan, 92 % for beech wood xylan, and 89 % for oat spelt xylan, but only 6 % of its maximal activity was detected when Avicel was used as a substrate. Additionally, almost no activity (lower than 1 % remaining) was detected when CMC, barley glucan, or lichenan were used in the reaction. These data indicate that Thxyn11A shows high activity for the less branched and more homogeneous xylans (birch and beech wood), which consist primarily of xylose units (90 %). In contrast, Thxyn11A shows reduced activity for oat-spelt xylan, which contains 10 % arabinose units and 15% glucose units. Contrarily, a GH11 xylanase from A. fumigatus MKU1 exhibited its highest activity towards oat spelt xylan, but showed only 66 and 77 % of its relative activity when birch and beech wood xylans were used as the substrates, respectively [11]. Moreover, the low activity of Thxyn11A towards Avicel, CMC and barley glucan demonstrates the xylanase does not exhibit cellulase activity, which is in agreement with previous findings [35]. Using a Lineweaver–Burk plot, the K m, and V max values of Thxyn11A were calculated to be 3.5 mg/ml and 470.7 μmol mg−1 min−1, respectively, with birch wood xylan as the substrate. Although the V max of Thxyn11A was lower than that of a GH11 xylanase from another actinomycete, the K m value indicated that Thxyn11A has a higher affinity for birch wood xylan [26].
Conclusions
In this study, a new xylanase gene, thxyn11A, was cloned from the actinomycete strain T. halotolerans YIM 90462T. A sequence analysis of the gene showed that it belongs to the GH11 xylanase family and has an extra CBM. The recombinant xylanase demonstrated broad pH stability, a strong tolerance to high temperatures, and unusual hydrolysis products. These properties make Thxyn11A a promising enzyme for industrial applications in the food and feed industries, as well as for the pre-treatment of the lignocellulosic biomass required to improve the yields of fermentable sugars in bioethanol production.
References
Abdul G, Sher AK, Zahid M, Muhammad IR, Farooq L (2011) Heterologous expression of a gene for thermostable xylanase from Chaetomium thermophilum in Pichia pastoris GS115. Mol Biol Rep 38:3227–3233
André RLD, Tony MS, Fausto BRA, Fábio MS, Daniela AR, Adriana FP, Fernando S, Rolf AP, João AJ, Hector FT, Maria LTMP (2011) Heterologous expression of an Aspergillus niveus xylanase GH11 in Aspergillus nidulans and its characterization and application. Process Biochem 46:1236–1242
Biely P (1985) Microbial xylanolytic systems. Trends Biotechnol 3:286–290
Bolam DN, Ciruela A, McQueen-Mason S, Simpson P, Williamson MP, Rixon JE, Boraston A, Hazlewood GP, Gilbert HJ (1998) Pseudomonas cellulose-binding domains mediate their effects by increasing enzyme substrate proximity. Biochem J 331:775–781
Collins T, Gerday C, Feller G (2005) Xylanases, xylanases families and extremophilic xylanases. FEMS Microbiol Rev 29:3–23
George EA, Anastasia Z, Panagiota MS, Kosmas H, George D, Amalia DK, Dimitris GH (2011) Homologous overexpression of xylanase in Fusarium oxysporum increases ethanol productivity during consolidated bioprocessing (CBP) of lignocellulosics. J Biotechnol 152:16–23
Henrissat B (1991) A classification of glycosyl hydrolasess based on amino acids similarities sequences. Biochem J 280:309–316
Huy ND, Kim SW, Park SM (2011) Heterologous expression of endo-1,4-beta-xylanase C from Phanerochaete chrysosporium in Pichia pastoris. J Biosci Bioeng 111:654–657
Kui H, Luo H, Shi P, Bai Y, Yuan T, Wang Y, Yang P, Dong S, Yao B (2010) Gene cloning, expression, and characterization of a thermostable xylanase from Nesterenkonia xinjiangensis CCTCC AA001025. Appl Biochem Biotechnol 162:953–965
Kulkarni N, Shendye A, Rao M (1999) Molecular and biotechnological aspects of xylanases. FEMS Microbiol Rev 23:411–456
Jeya M, Thiagarajan S, Lee JK, Gunasekaran P (2009) Cloning and expression of GH11 xylanase gene from Aspergillus fumigatus MKU1 in Pichia pastoris. J Biosci Bioeng 108:24–29
Jeya M, Thiagarajan S, Lee JK, Gunasekaran P (2009) Identification of new GH 10 and GH 11 xylanase genes from Aspergillus versicolor MKU3 by genome-walking PCR. Biotechnol Bioproc Eng 14:13–19
Lee CC, Kibblewhite-Accinelli RE, Smith MR, Wagschal K, Orts WJ, Wong DWS (2008) Cloning of Bacillus licheniformis xylanase gene and characterization of recombinant enzyme. Curr Microbiol 57:301–305
Li N, Shi P, Yang P, Wang Y, Luo H, Bai Y, Zhou Z, Yao B (2009) Cloning, expression, and characterization of a new Streptomyces sp. S27 xylanase for which xylobiose is the main hydrolysis product. Appl Biochem Biotechnol 159:521–531
Li WJ, Xu P, Schumann P, Zhang YQ, Pukall R, Xu LH, Stackebrandt E, Jiang CL (2007) Georgenia ruanii sp. nov., a novel actinobacterium isolated from forest soil in Yunnan (China), and emended description of the genus Georgenia. Int J Syst Evol Microbiol 57:1424–1428
Lineweaver H, Burk D (1934) The determination of enzyme dissociation constants. J Am Chem Soc 56:658–666
Liu SY, Shibu MA, Jhan HJ, Lo CT, Peng KC (2010) Purification and characterization of novel glucanases from Trichoderma harzianum ETS 323. J Agric Food Chem 58:10309–10314
Lorena AD, Teresa MC, Alejandro SH, Jesús VE, Farrés GSA, Beatriz XC, Roberto RM, María MH, María EHL (2010) Cloning and expression of a novel, moderately thermostable xylanase-encoding gene (Cfl xyn11A) from Cellulomonas flavigena. Bioresour Technol 101:5539–5545
Luo H, Wang Y, Li J, Wang H, Yang J, Yang Y, Huang H, Fan Y, Yao B (2009) Cloning, expression and characterization of a novel acidic xylanase, XYL11B, from the acidophilic fungus Bispora sp. MEY-1. Enzyme Microbial Technol 45:126–133
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428
Qu W, Shao W (2011) Cloning, expression and characterization of glycoside hydrolases family 11 endoxylanase from Bacillus pumilus ARA. Biotechnol Lett 33:1407–1416
Ricardo SST, Félix GS, Marcelo VS, Edivaldo XFF, Elba PSB (2010) Purification and characterization studies of a thermostable β-xylanase from Aspergillus awamori. J Ind Microbiol Biotechnol 37:1041–1051
Sambrook J, Russell DW (2001) Molecular cloing: a laboratory manual, 3rd edn. Cold Spring Harbor, New York, Appendix A2.4
Shrinivas D, Savitha G, Raviranjan K, Naik GR (2010) A highly thermostable alkaline cellulase-free xylanase from thermoalkalophilic Bacillus sp. JB 99 suitable for paper and pulp industry: purification and characterization. Appl Biochem Biotechnol 162:2049–2057
Tan G, Gao Y, Shi M, Zhang X, He S, Chen Z, An C (2005) SiteFinding-PCR: a simple and efficient PCR method for chromosome walking. Nucleic Acids Res 33:e122
Thayat S, Peechapack S, Kenji M, Fusako K, Kosum C (2011) Cloning of a thermostable xylanase from Actinomadura sp. S14 and its expression in Escherichia coli and Pichia pastoris. J Biosci Bioeng 111:528–536
Thomson JA (1993) Molecular biology of xylan degradation. FEMS Microbiol Lett 104:65–82
Vazquez MJ, Alonso JL, Dominguez H, Parajo JC (2000) Xylooligosaccharides: manufacture and applications. Trends Food Sci Technol 11:387–393
Wang G, Luo H, Meng K, Wang Y, Huang H, Shi P, Pan X, Yang P, Diao Q, Zhang H, Yao B (2011) High genetic diversity and different distributions of glycosyl hydrolase family 10 and 11 xylanases in the goat rumen. PLoS ONE 6:e16731
Wang Q, Xia T (2008) Importance of C-terminal region for thermostability of GH11 xylanase from Streptomyces lividans. Appl Biochem Biotechnol 144:273–282
Yang LL, Tang SK, Zhang YQ, Zhi XY, Wang D, Xu LH, Li WJ (2008) Thermobifida halotolerans sp. nov., isolated from a salt mine sample, and emended description of the genus Thermobifida. Int J Syst Evol Microbiol 58:1821–1825
Zhang F, Chen JJ, Ren WZ, Nie GX, Ming H, Tang SK, Li WJ (2011) Cloning, expression and characterization of an alkaline thermostable GH9 endoglucanase from Thermobifida halotolerans YIM 90462T. Bioresour Technol 102:10143–10146
Zhang F, Hu SN, Chen JJ, Lin LB, Wei YL, Tang SK, Xu LH, Li WJ (2012) Purification and partial characterisation of a thermostable xylanase from salt-tolerant Thermobifida halotolerans YIM 90462T. Process Biochem 47:225–228
Zhao Y, Meng K, Luo H, Yang P, Shi P, Huang H, Bai Y, Yao B (2011) Cloning, expression, and characterization of a new xylanase from alkalophilic Paenibacillus sp. 12–11. J Microbiol Biotechnol 21:861–868
Zhou J, Shi P, Zhang R, Huang H, Meng K, Yang P, Yao B (2011) Symbiotic Streptomyces sp. TN119 GH 11 xylanase: a new pH-stable, protease- and SDS-resistant xylanase. J Ind Microbiol Biotechnol 38:523–530
Acknowledgments
We are grateful to Dr. Chao Huang for technological guidance on TLC and to Dr. Guang-Yu Yang for many useful discussions. This research was supported by the National Basic Research Program of China (No. 2010CB833801), the National Natural Science Foundation of China (No. 31070007), and the International Cooperation Research Program of Yunnan Province (No. 2009AC017). W-J Li was also supported by ‘Hundred Talents Program' of the Chinese Academy of Sciences.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, F., Chen, JJ., Ren, WZ. et al. Cloning, expression, and characterization of an alkaline thermostable GH11 xylanase from Thermobifida halotolerans YIM 90462T . J Ind Microbiol Biotechnol 39, 1109–1116 (2012). https://doi.org/10.1007/s10295-012-1119-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10295-012-1119-8