Abstract
Rare copy number variations by the nonrecurrent rearrangements involving PMP22 have been recently suggested to be associated with CMT1A peripheral neuropathy. As a mechanism of the nonrecurrent rearrangement, replication-based fork stalling template switching (FoSTeS) by microhomology-mediated break-induced replication (MMBIR) has been proposed. We found three Korean CMT1A families with putative nonrecurrent duplication. The duplications were identified by microsatellite typing and applying a CGH microarray. The breakpoint sequences in two families suggested an Alu–Alu-mediated rearrangement with the FoSTeS by the MMBIR, and a two-step rearrangement of the replication-based FoSTeS/MMBIR and meiosis-based recombination. The two-step mechanism has still not been reported. Segregation analysis of 17p12 microsatellite markers and breakpoint junction analysis suggested that the nonrecurrent rearrangements are stably inherited without alteration of junction sequence; however, they may allow some alteration of the genomic contents in duplication across generations by recombination event. It might be the first study on the pedigree analysis of the large CMT1A families with nonrecurrent rearrangements. It seems that the exact mechanism of the nonrecurrent rearrangements in the CMT1A may have a far more complex process than has been expected.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Charcot–Marie–Tooth disease (CMT) is a genetically and clinically heterogeneous hereditary motor and sensory neuropathy with an estimated prevalence of 1/2,500. Based on the electrophysiological criteria, CMT is frequently classified into two forms, the demyelinating form (CMT1) and the axonal form (CMT2) [1]. Up to date, more than 50 genes or loci have been reported as the underlying cause of CMT at the Inherited Peripheral Neuropathies Mutation Database (http://www.molgen.ua.ac.be/CMTMutations/Mutations/). Of them, the nonallelic homologous recombination (NAHR) between proximal and distal homologous repeat elements (CMT1A d-REP and p-REPs) on chromosome 17p12 is the most frequent genetic cause of the demyelinating CMT, particularly CMT1A (MIM# 118220) [2–4]. Reciprocal deletion of the same region by the unequal crossover is associated with the hereditary neuropathy with liability to pressure palsies (HNPP, MIM# 162500) [5, 6]. The two flanking CMT1A–REPs share 98.7% sequence identity, and the duplication comprises approximately 1.4-Mb genomic region which includes the dosage-sensitive myelin gene PMP22 [7]. Duplication is found up to approximately 70% of patients with CMT1 [8].
Rearrangements of the human genome can be grouped into two types according to breakpoint sequences, recurrent rearrangements, and nonrecurrent rearrangements. The major mechanism of recurrent rearrangements is NAHR which is associated with several genomic repeated sequences, e.g., low copy repeats (LCRs), long interspersed repetitive sequences (LINEs), and transposons. Because significant regions of homology are required for recombination, short interspersed repetitive sequences (SINEs), such as Alu, are not usually substrates for the NAHR [9, 10]. The molecular mechanism of the nonrecurrent rearrangements by nonhomologous end joining (NHEJ) has been little understood. Recently, a replication-based mechanism of fork stalling template switching (FoSTeS) has been proposed to explain the nonrecurrent rearrangements of the NHEJ [11–15]. According to the FoSTeS DNA replication model, the active replication fork can stall and switch templates using complementary template microhomology to anneal and prime DNA replication (microhomology-mediated break-induced replication, MMBIR). Chen et al. suggested a serial replication slippage as a possible cause of smaller genomic complex rearrangements [16]. Template switching during replication may be exacerbated by the presence of cruciform or other non-B DNA structures [12, 17].
Although the genomic disorders of CMT1A duplication and HNPP deletion are prevalently associated with recurrent rearrangements caused by NAHR events between two LCRs, recently, rare copy number variations (CNVs) on the chromosome 17p12 by the nonrecurrent rearrangements have also been suggested as the underlying cause of the CMT1A and HNPP [15, 18, 19]. Breakpoint sequence analyses have also revealed that various molecular mechanisms are involved in generating the nonrecurrent 17p12 rearrangements, such as NHEJ, Alu-Alu-mediated rearrangement, and FoSTeS and/or MMBIR mechanisms.
In the present study, we identified three Korean CMT1A families who are suggested to be associated with the nonrecurrent rearrangements by the replication-based FoSTeS. Analysis of the breakpoint junctions in each of the unique rearrangements suggested involvement of several mechanisms, such as Alu–Alu mediation, two-step process combining replication error and meiotic recombination, and complex rearrangement of two discrete duplications. The haplotype and breakpoint junction analyses also suggested that the nonrecurrent rearrangements may allow a slight modification of genomic contents by recombination event within duplication regions across generations.
Materials and methods
Patients
This study included three Korean CMT1A families (FC85, FC116, and FC388) with 34 individuals (19 affected and 15 unaffected individuals) (Fig. 1). This study also included four healthy controls for the CNV test. As the gain and loss controls, each two CMT1A duplication and HNPP deletion patients with the common 1.4 Mb recurrent recombination on 17p12 was involved. For the examination of clinical phenotypes, 149 CMT1A patients were also involved. All participants in this study provided written informed consent according to the protocol approved by the Institutional Review Board for Ewha Womans University Hospital. Total DNA was isolated from peripheral blood by using a QIAamp Blood DNA purification kit (Qiagen, Hilden, Germany).
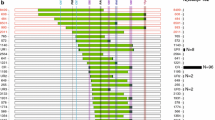
CMT1A pedigrees with nonrecurrent duplication. Haplotypes (indicated below each member) were obtained by genotyping of nine microsatellites, but the genotypes in parentheses were deduced from the pedigree analysis. Asterisks (*) indicate individuals whose DNA were used for this analysis. The gray boxes on the haplotypes indicate the duplicated region, and open boxes indicate chromosomal regions before de novo event, respectively. Arrows (↗) indicate probands (open squares and circles unaffected individuals, filled squares and circles affected individuals). a FC116 family. Except for a putative crossover (III-11 to IV-17) on the duplication region (black left-pointing pointer) (a-1), the duplication was stably inherited without alteration (a-2). b FC388 family. Putative de novo rearrangement occurred from I-1 to II-2. c FC85 family. A de novo discrete rearrangement was observed (I-1 to II-2)
Determination of duplication by microsatellite typing and real-time PCR
The nine microsatellites within the 17p12 CMT1A duplication region were amplified using the hexaplex PCR system that consisted of D17S921, D17S9A, D17S918, D17S4A, D17S122, and D17S2230, or single PCR method (D17S1296, D17S1357, and D17S9B) with fluorescent-labeled primers [20]. The PCR products were resolved with the automatic genetic analyzer ABI3100, and genotypes were determined by the Genotyper software (Applied Biosystems, Foster City, CA, USA). Duplication of PMP22 was also determined by real-time quantitative PCR. The real-time PCR was performed on the RotorGene real-time PCR machine (Corbett Research, Australia) using the QuantiTech SYBR Green PCR kit (Qiagen). GAPDH was co-amplified for the control gene.
CNV determination by comparative genomic hybridization microarray
A high resolution of the rearrangement map and narrowed range of breakpoint junctions were obtained by determining the CNVs with a customer-designed high-density comparative genomic hybridization (CGH) microarray (HG18 CGH 2X 135K; Roche-NimbleGen, Madison, WI, USA). The mean probe size and spacing length of the array were 60-mer and 215 bp, respectively. The CGH microarray data were analyzed with NimbleScan (ver. 2.4) and SignalMap (ver. 1.9) software. The gain and loss threshold used in this study were log2 ratio >0.3 and <−0.3, respectively. Healthy individuals were used as normal control, and recurrent CMT1A duplication and HNPP deletion patients were used as gain and loss controls, respectively.
Long-range template PCR and determination of breakpoint sequence
Long-range template PCR was attempted to amplify the breakpoint junctions. PCR primers were designed as approximate intervals of 2 kb within about 10-kb ranges with either side of estimated breakpoints by the CGH analysis (Supplemental Table 1). The PCR was performed using the long-range template PCR kit (Roche, Mannheim, Germany). PCR products were sequenced on the automatic genetic analyzer ABI3100 using the BigDye terminator cycle sequencing kit (Applied Biosystems). DNA sequences were compared to reference sequences, UCSC hg18 at the UCSC Genome Browser (http://genome.ucsc.edu/) or Build 36.1 at the NCBI website (http://blast.ncbi.nlm.nih.gov/) by using the SeqScape (ver. 2.1, Applied Biosystems) and Chromas software (ver. 2.33; Technelysium, Australia).
Clinical assessment
Clinical information was obtained in a standardized manner, including motor and sensory impairments, and deep tendon reflexes. Histopathological examination of the affected individuals included the light and electron microscopic analyses of a sural nerve. Magnetic resonance imaging study was performed on the lumbar spine and lower legs of the patients using a supine position using a 1.5-T system (Siemens Vision, Erlangen, Germany). Physical disability was determined by two scales, the functional disability scale (FDS) [21] and the CMT neuropathy score (CMTNS) [22].
Results
Identification of partial duplication of chromosome 17p12 in CMT1A families
From the genotyping of nine microsatellites on 17p12 in subjects with CMT1A families, we found three families with unusual partial duplication. CMT1A patients with common duplication showed 1.4 Mb duplication containing all the markers from D17S921 to D17S2230; however, FC116 and FC388 revealed duplication from D17S1296 to D17S2230 (Fig. 1a) and from D17S1296 to D17S122 (Fig. 1b), respectively. Particularly, FC85 showed a complex rearrangement with two discrete regions, D17S921, and D17S1357 to D17S2230 (Fig. 1c). The duplication regions included the PMP22 gene in all three families, which was also confirmed by amplification of the PMP22 using the real-time PCR. De novo rearrangements were suggested in two families with both paternal origins: father (I-1)-to-daughter (II-2) in FC388 and father (I-1)-to-son (II-2) in FC85. Although the proband’s father (I-1) in FC388 was not directly tested in this study, we considered that a de novo rearrangement occurred during transmission because he was determined to be an unaffected individual by history taking.
The exact duplication regions were further narrowed into 14.84–15.44 Mb in FC116 and 14.59–15.31 Mb in FC388 by applying the CGH microarray (Fig. 2a). The complex duplication of FC85 included 14.00–14.46 Mb and 14.98–15.43 Mb. Figure 2b explains the genomic structure of the identified rare rearrangements.
Characterization of the rare nonrecurrent rearrangements in CMT1A patients. a Log2 ratio plots obtained by the high-density CGH microarray (NimbleGen). Compared to healthy control (CTL), common CMT1A and HNPP patients showed duplication or deletion of 1.4 Mb on the 17p12 region. However, three families revealed unique partial duplications different from each other. b Diagram of genomic structure on 17p12 and rearrangements identified in CMT1A patients with partial duplication. All CNVs included the PMP22 duplication. Map distances were from Map build UCSC hg18 (http://genome.ucsc.edu/). c Sequences of the break junctions in FC116 and d FC388. The proximal and distal reference sequences are given by black and blue letters, respectively. The sequences with microhomology are indicated by red letters with gray boxes (– deletion)
Nonrecurrent rearrangements by Alu–Alu and microhomology mediation
The breakpoint junctions were successfully amplified by long-range template PCR from the patients in FC116 and FC388 (Supplementary Fig. S1). The range of distal breakpoints in FC116 and FC388 could be narrowed within 14,839,425–14,841,529 bp and 14,582,503–14,584,531 bp, respectively (hg18). However, we failed to obtain the PCR products from FC85 even though we tried to amplify the breakpoint junction by using various primer pairs.
The exact DNA sequences at the breakpoints and their surrounding regions were determined from the long-range template PCR products (Table 1). In FC116, the breakpoint junction was found within two different Alu sequence families (distal end, AluY; proximal end, AluSc) with 34-bp exact microhomology (Fig. 2c). In FC388, each proximal and distal breakpoint was within a long terminal repeat (LTR, family MALR) sequence and an intron of CDRT4 gene with 3-bp “TCA” microhomology. The breakpoint junction of FC388 also showed an 11-bp deletion, 7 bp after the microhomology (Fig. 2d). The 11-bp deletion was not observed in either non-rearranged regions corresponding to distal breakpoint end (presumptive ancestral sequence before de novo event) in proband (II-2) or same chromosomal regions of her mother (I-2) and unaffected siblings (II-3, 4, 5) (Supplementary Fig. S2). We also could not find the variation of 11-bp deletion from the dbSNP (http://www.ncbi.nlm.nih.gov/projects/SNP) and BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi?). Therefore, we considered that the 11-bp deletion is due to a complex nonrecurrent rearrangement during the de novo event. All the affected members in FC116 (n = 15) and FC388 (n = 3) showed exactly the same breakpoint DNA sequences in each pedigree.
When the microsatellite haplotypes were analyzed in FC116, the deduced two alleles by duplication (gray boxes) were always the same in six corresponding markers as shown in Fig. 1a-2 (4-4/7-7/6-6/4-4/5-5/6-6). Therefore, it seems that the rearrangement might be resulted from the FoSTeS by the MMBIR via 34-bp microhomology. The haplotypes of the duplicated region were the same in all the affected members except for IV-17 (Fig. 1a-1). The different haplotype of IV-17 might be due to a meiosis-based recombination event between D17S1357 and D17S918 during transmission of rearrangement region from mother (III-11) to son (IV-17) (Fig. 3a). The haplotype analysis of FC388 suggested a more complex two-step rearrangement process (Figs. 1b and 3b). First, a nonrecurrent rearrangement between D17S1296 and D17S122 might occur by the FoSTeS via 3-bp microhomology during DNA replication, which produced a presumptive intermediate haplotype, “4-4/8-8/8-8/1-1/7-7”. Second, the crossover event between D17S1357 and D17S918 during meiosis might produce final rearrangement with the haplotype of “4-4/8-8/8-9/1-1/4-7”.
Putative crossover and two-step process of a rearrangement (blue X crossover). a Crossover on the duplication region in FC116. A crossover event might occur between D17S1357 and D17S918 during transmission from III-11 to IV-17 (Fig. 1a-1). b De novo rearrangement by putative two-step process in FC388. The FoSTeS/MMBIR event might be preceded during DNA replication (red horizontal arrows), and then crossover occurred during meiosis between D17S1357 and D17S918 during transmission from II-2 to III-1 (Fig. 1b). The regions indicated by single asterisk and double asterisk were used for sequencing analysis to determine the 11-bp deletion
Clinical manifestations
The clinical phenotypes of 19 patients with the rare duplication showed no significant different features from the common CMT1A patients with 1.4-Mb recurrent duplications (Table 2). However, FC116 family revealed broad intrafamilial phenotypic variations, e.g., age at onset: 4 to 60 years, CMTNS: 2/30 to 27/30, FDS: 0/8 to 5/8, and median nerve conduction velocities: 0 to 30.7 m/s. The histological examination of a sural nerve biopsy in a 52-year-old woman (FC116—IV-25) showed severe loss of myelinated fibers of all calibers and revealed diverse features of onion bulb and pseudo-onion bulb formations as the characteristics of common CMT1A patients (data not shown).
Discussion
Human genomic disorders are caused by an alteration of the genome architectures which are mostly resulted from recurrent genomic rearrangements between region-specific LCRs. However, replication-based nonrecurrent rearrangement has been also proposed to be one of the important underlying causes of genomic disorders. Several CMT1A patients have been recently reported to be associated with the nonrecurrent rearrangements [15, 18, 19, 23]. This study also identified three CMT1A families with the rare nonrecurrent rearrangements. It might be the first haplotype analysis of large CMT1A families with the NHEJ. The exact breakpoint junctions were determined by the long-range template PCR after analysis of the CGH microarray in FC116 and FC388. However, we failed to obtain PCR product from FC85 with two discrete duplicated regions. The rearrangement of FC85 may contain more complex events such as inversion or microduplication/deletion. Two paternal originated de novo rearrangements were observed (FC388 and FC85). This is comparable to the predominant paternal origin of general CMT1A de novo duplications by the unequal crossover even though the rearrangement mechanisms might be considerably different between the two events [24–26].
A careful examination of clinical phenotypes for all the affected individuals showed no significant difference compared with the common CMT1A patients, which is consistent with the idea that the copy number of the PMP22 gene may contribute to the phenotype. However, the broad intrafamilial phenotype variation in FC116 (e.g., broad range of age at onset from 4 to 60 years) might suggest an existence of a genetic modifier or environmental factor(s) which influence on the expressivity of the clinical symptoms rather than incomplete penetrance of the phenotype. Kleopa et al. suggested that mutations in the PMP22 gene are relevant with the broad phenotypic spectrum [27].
The Alu–Alu-mediated rearrangement in the FC116 could be explained by either of two mechanisms, meiosis-based nonallelic homologous recombination (unequal crossover) or replication-based FoSTeS by MMBIR [15, 28, 29]. However, Alu sequences usually do not act as the preferring substrates for the homologous recombination because significant regions of homology are required for recombination [9]. When we performed the segregation analysis of 17p12 markers, both duplicated alleles were always the same. This segregation analysis indicated that the rearrangement might result from the FoSTeS by the MMBIR via 34-bp microhomology. If the rearrangement has occurred by NAHR mechanism via the unequal crossover between two homologous chromosomes, two duplicated alleles would be different. However, the evidence of the identical alleles could not completely exclude the potential involvement of NAHR in this rearrangement because an unequal crossover event between sister chromatids might generate identical alleles on the duplication region [30, 31]. The involvement of the FoSTeS mechanism via decades-bp microhomology for the Alu–Alu-mediated rearrangement was also suggested by Zhang et al. [15].
Although the rare rearrangement in the FC388 was regarded as by the FoSTeS/MMBIR via 3-bp microhomology, the haplotype analysis suggested that a more complex mechanism might be involved in the rearrangement. The de novo duplication from I-1 to II-2 might be achieved by two different events, FoSTeS/MMBIR during DNA replication and recombination during meiosis (Fig. 3b). This two-step rearrangement mechanism has still not been reported. The “serial replication slippage” suggested by Chen et al. might involve in the nonrecurrent rearrangement of FC388 since the breakpoint sequence also revealed an 11-bp deletion [16].
Little has still been understood whether the nonrecurrent rearrangements are stably transmitted to the next generation. As expected, this study revealed that the breakpoint junctions are stably inherited with no sequence modification since all the 15 affected members revealed the same breakpoint sequence in the FC116 family including five generations. Three affected members in the FC388 also showed the same breakpoint sequence. But the other concern is whether the duplicated genomic contents of nonrecurrent rearrangements are stably transmitted across generations. One of the interesting findings of this study is the occurrence of meiotic recombination within the duplication. The recombinations may exchange the contents between duplicated allele and wild-type allele in genomic duplications. Moreover, this reminds us that the duplicated PMP22 copies might be different in the same family (like FC116), although the copy number of PMP22 is the same. It may also potentially contribute to variability in gene expression that could partially explain some of the broad phenotypic spectrum in the same CMT1A family.
Considering the combined replication error and meiotic recombination events, it seems that the exact mechanisms of the rare partial duplications may have far more complex and delicate processes than has been expected. It also seems that the genomic contents in duplication may be unstable across generations to some extent, although the breakpoint junction is stable. We believe that this study provides a clue to discover the molecular mechanisms of the nonrecurrent rearrangements in many other human genomic disorders with rare CNVs.
References
Harding AE, Thomas PK (1980) The clinical features of hereditary motor and sensory neuropathy types I and II. Brain 103:259–280
Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao L, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI (1991) DNA duplication associated with Charcot–Marie–Tooth disease type 1A. Cell 66:219–232
Lupski JR, Wise CA, Kuwano A, Pentao L, Parke JT, Glaze DG, Ledbetter DH, Greenberg F, Patel PI (1992) Gene dosage is a mechanism for Charcot–Marie–Tooth disease type 1A. Nat Genet 1:29–33
Pentao L, Wise CA, Chinault AC, Patel PI, Lupski JR (1992) Charcot–Marie–Tooth type 1A duplication appears to arise from recombination at repeat sequences flanking the 1.5 Mb monomer unit. Nat Genet 2:292–300
Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, Odelberg SJ, Disteche CM, Bird TD (1993) DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 72:143–151
Mariman EC, Gabreels-Festen AA, van Beersum SE, Valentijn LJ, Baas F, Bolhuis PA, Jongen PJ, Ropers HH, Gabreels FJ (1994) Prevalence of the 1.5-Mb 17p deletion in families with hereditary neuropathy with liability to pressure palsies. Ann Neurol 36:650–655
Reiter LT, Murakami T, Koeuth T, Gibbs RA, Lupski JR (1997) The human COX10 gene is disrupted during homologous recombination between the 24 kb proximal and distal CMT1A–REPs. Hum Mol Genet 6:1595–1603
Szigeti K, Garcia CA, Lupski JR (2006) Charcot–Marie–Tooth disease and related hereditary polyneuropathies: molecular diagnostics determine aspects of medical management. Genet Med 8:86–92
Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422
Stankiewicz P, Lupski JR (2002) Genome architecture, rearrangements and genomic disorders. Trends Genet 18:74–82
Lieber MR (2008) The mechanism of human nonhomologous DNA end joining. J Biol Chem 283:1–5
Lee JA, Carvalho CM, Lupski JR (2007) A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131:1235–1247
Gu W, Zhang F, Lupski JR (2008) Mechanisms for human genomic rearrangements. Pathogenetics 1:4
Hastings PJ, Ira G, Lupski JR (2009) A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet 5:e1000327
Zhang F, Khajavi M, Connolly AM, Towne CF, Batish SD, Lupski JR (2009) The DNA replication FoSTeS/MMBIR mechanism can generate genomic, genic and exonic complex rearrangements in humans. Nat Genet 41:849–853
Chen JM, Chuzhanova N, Stenson PD, Ferec C, Cooper DN (2005) Complex gene rearrangements caused by serial replication slippage. Hum Mutat 26:125–134
Kosmider B, Wells RD (2007) Fragile X repeats are potent inducers of complex, multiple site rearrangements in flanking sequences in Escherichia coli. DNA Repair (Amst) 6:1580–1863
Zhang F, Seeman P, Liu P, Weterman MA, Gonzaga-Jauregui C, Towne CF, Batish SD, De Vriendt E, De Jonghe P, Rautenstrauss B, Krause KH, Khajavi M, Posadka J, Vandenberghe A, Palau F, Van Maldergem L, Baas F, Timmerman V, Lupski JR (2010) Mechanisms for nonrecurrent genomic rearrangements associated with CMT1A or HNPP: rare CNVs as a cause for missing heritability. Am J Hum Genet 86:892–903
Huang J, Wu X, Montenegro G, Price J, Wang G, Vance JM, Shy ME, Züchner S (2010) Copy number variations are a rare cause of non-CMT1A Charcot–Marie–Tooth disease. J Neurol 257:735–741
Choi BO, Kim J, Lee KL, Yu JS, Hwang JH, Chung KW (2007) Rapid diagnosis of CMT1A duplications and HNPP deletions by multiplex microsatellite PCR. Mol Cells 23:39–48
Birouk N, Gouider R, Le Guern E, Gugenheim M, Tardieu S, Maisonobe T, Le Forestier N, Agid Y, Brice A, Bouche P (1997) Charcot–Marie–Tooth disease type 1A with 17p11.2 duplication. Clinical and electrophysiological phenotype study and factors influencing disease severity in 119 cases. Brain 120:813–823
Shy ME, Blake J, Krajewski K, Fuerst DR, Laura M, Hahn AF, Li J, Lewis RA, Reilly M (2005) Reliability and validity of the CMT neuropathy score as a measure of disability. Neurology 64:1209–1214
Shchelochkov OA, Cheung SW, Lupski JR (2010) Genomic and clinical characteristics of microduplications in chromosome 17. Am J Med Genet A 152A:1101–1110
Palau F, Löfgren A, De Jonghe P, Bort S, Nelis E, Sevilla T, Martin JJ, Vilchez J, Prieto F, Van Broeckhoven C (1993) Origin of the de novo duplication in Charcot–Marie–Tooth disease type 1A: unequal nonsister chromatid exchange during spermatogenesis. Hum Mol Genet 2:2031–2035
Lopes J, Vandenberghe A, Tardieu S, Ionasescu V, Lévy N, Wood N, Tachi N, Bouche P, Latour P, Brice A, LeGuern E (1997) Sex-dependent rearrangements resulting in CMT1A and HNPP. Nat Genet 17:136–137
Lopes J, Ravisé N, Vandenberghe A, Palau F, Ionasescu V, Mayer M, Lévy N, Wood N, Tachi N, Bouche P, Latour P, Ruberg M, Brice A, LeGuern E (1998) Fine mapping of de novo CMT1A and HNPP rearrangements within CMT1A–REPs evidences two distinct sex-dependent mechanisms and candidate sequences involved in recombination. Hum Mol Genet 7:141–148
Kleopa KA, Georgiou DM, Nicolaou P, Koutsou P, Papathanasiou E, Kyriakides T, Christodoulou K (2004) A novel PMP22 mutation ser22phe in a family with hereditary neuropathy with liability to pressure palsies and CMT1A phenotypes. Neurogenetics 5:171–175
Casarin A, Martella M, Polli R, Leonardi E, Anesi L, Murgia A (2006) Molecular characterization of large deletions in the von Hippel–Lindau (VHL) gene by quantitative real-time PCR: the hypothesis of an alu-mediated mechanism underlying VHL gene rearrangements. Mol Diagn Ther 10:243–249
Franke G, Bausch B, Hoffmann MM, Cybulla M, Wilhelm C, Kohlhase J, Scherer G, Neumann HP (2009) Alu–Alu recombination underlies the vast majority of large VHL germline deletions: molecular characterization and genotype–phenotype correlations in VHL patients. Hum Mutat 30:776–786
Le Guern E, Sturtz F, Gugenheim M, Gouider R, Bonnebouche C, Ravise N, Gonnaud P-M, Tardieu S, Bouche P, Chazot G, Agid Y, Vandenberghe A, Brice A (1994) Detection of deletion within 17p11.2 in 7 French families with hereditary neuropathy with liability to pressure palsies (HNPP). Cytogenet Cell Genet 65:261–264
Lopes J, Tardieu S, Silander K, Blair I, Vandenberghe A, Palau F, Ruberg M, Brice A, LeGuern E (1999) Homologous DNA exchanges in humans can be explained by the yeast double-strand break repair model: a study of 17p11.2 rearrangements associated with CMT1A and HNPP. Hum Mol Genet 8:2285–2292
Acknowledgments
This study was supported by Mid-career Researcher Program through NRF grant funded by the MEST (R01-2008-000-20604-0 and KRF-2008-313-C00750), Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Additional information
B.-O. Choi and N.K. Kim contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 191kb)
Rights and permissions
About this article
Cite this article
Choi, BO., Kim, N.K., Park, S.W. et al. Inheritance of Charcot–Marie–Tooth disease 1A with rare nonrecurrent genomic rearrangement. Neurogenetics 12, 51–58 (2011). https://doi.org/10.1007/s10048-010-0272-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10048-010-0272-3