Abstract
Poly(L-lysine) (PLL) dendrimer are amino acid based macromolecules and can be used as drug delivery agents. Their branched structure allows them to be functionalized by various groups to encapsulate drug agents into their structure. In this work, at first, an attempt was made on all-atom simulation of PLL dendrimer of different generations. Based on all-atom results, a course-grained model of this dendrimer was designed and its parameters were determined, to be used for simulation of three generations of PLL dendrimer, at two pHs. Similar to the all-atom, the coarse-grained results indicated that by increasing the generation, the dendrimer becomes more spherical. At pH 7, the dendrimer had larger size, whereas at pH 12, due to back folding of branching chains, they had the tendency to penetrate into the inner layers. The calculated radial probability and radial distribution functions confirm that at pH 7, the PLL dendrimer has more cavities and as a result it can encapsulate more water molecules into its inner structure. By calculating the moment of inertia and the aspect ratio, the formation of spherical structure for PLL dendrimer was confirmed.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Dendrimers are a family of branched chain polymers with regular repeating units which are joined together at a core. Due to their unique structural characteristics, dendrimers demonstrate significant applications [1, 2]. Based on various cores and branching chain units, dendrimers form a large family in which poly(L-lysine) (PLL), poly(amidoamine) (PAMAM), poly(propyleneine) (PPI) are the most common and well known family members [3]. PLL is a peptide dendrimer with dendritic structure and with peptide bonds. In peptide dendrimers, there are amino acid groups in the core and in the repeating units of their chain branches. PLL dendrimers are biocompatible, biodegradable, water soluble, flexible, and asymmetric. These characteristics which arise from their molecular structure, make PLL dendrimers desirable for biological application [4, 5]. For instance, they can be used for delivery of anticancer, antimicrobial, and antiviral drugs [6–9]. Recently, many studies have been conducted on PLL dendrimers and it has been shown that PLL with cationic NH2 in their terminal group can easily penetrate into the cell membrane [10]. In addition, Choi et al. [11] examined poly(ethyleneglycole)-block-poly(L-lysine) dendrimer interactions with DNA in water as the solvent and determined that these interactions create a self-assembly structure which increases the stability of DNA in solvents. Byrne et al. [12], studied complex formation of PLL with DNA and found that the star-shape structure of the complex can be utilized in gene delivery, and compared with linear-shape carriers, the size of spherically complex is very small. Yevlampieva et al. [13], examined the hydrodynamic behavior of PLL dendrimer in water and dimethylformamide DMF) as the solvents, and showed that PLL dendrimer has smaller size with flexible structure in comparison with other cationic dendrimers, such as PAMM. The PLL dendrimers can be modified to become suitable for a specific application. PEGylation (grafting polyethylene glycol chain to the terminal) is an example of these modifications. Okuda et al. [14] indicated that the PEGylation of PLL dendrimers can reduce their accumulation in kidneys. Kaminskas et al. [15] studied the effect of increasing PEG chain length on the functional groups of PLL dendrimers and reported that while PEGylation significantly reduces the PLL dendrimer cytotoxcity, the PEG chain length increases the half-life of the PLL-PEG complex in blood circulation. In another work, Okuda et al. [16] examined grafting of sixth generation of PLL-dendrimers with histidine and ararginine and they found that binding ability of histidine grafted PLL-dendrimer to DNA decreased, in comparison with the ungrafted one, but for the arginine grafted PLL dendrimer, the binding ability to DNA increased. Kaminskas et al. [17] and Fox et al. [5] studied the PLL dendrimer as an anticancer therapeutic agent. Rossi et al. [18] considered PLL dendrimer for drug delivery by complete grafting of PLL to different functional groups such as hydrophobic amino acids (alanine, valine, leucine), dicaboxylic acids (succinic, aspartic acid), guanidyl, and galacto-saccharide and partially grafted to lactose.
Despite various experimental studies on synthesis and application of PLL dendrimers, and the need to understand their structural behavior to achieve a meaningful insight into their application, theoretical studies on their structures still lag behind. Therefore it is necessary to recourse to possible theoretical approaches such as molecular simulations to cast light on this important matter. Molecular simulations as effective, accurate, and fast methods with low operating cost, are extending and improving their applications in recent years, and not only provide useful information on mechanism and theoretical aspects of experimental studies, but even, in some cases can replace the expensive and time consuming limitations of the experimental procedures. Molecular dynamics (MD) is a deterministic molecular modeling method that has produced very promising results in various fields of research, especially on biological systems. MD simulations have also been used to study the structural behavior of dendrimers [19–22]. Robert et al. [23], employed fully atomistic MD simulation to investigate the structural change of PLL dendrimers from generation 1 (G1) to generation 6 (G6) and indicated that G1 and G6 generations of the dendrimer complexes are spherical and regular in shape and have a highly grooved surface with a dense core structure. Neelov et al. [24], used atomistic MD simulation to investigate properties of different PLL dendrimers’ generations and reported that, their characteristics do not depend on temperature but their internal groups’ mobility depends on their generation.
The aim of this work is to study the structural characteristics of PLL dendrimers and their behavior in the solutions at two pHs. To facilitate and speed up the computational calculations in the MD simulations of the studied dendrimers, which have a large number of atoms, a course-grained (CG) approach is employed. The GC approach not only reduces the calculation time significantly, but larger step size and simulation box can be used which makes the simulations considerably faster to obtain the desired results. Lee et al. [25] obtained the bonded interaction parameters for the coarse-grained PAMAM dendrimer with the terminal groups of hystidine and arginine, at pH 5 and 7. They indicated that, with increasing the number of histidine in the dendrimer terminal groups, the size of the complex formed becomes larger. Kavyani et al. [26] by using the GC –MD approach, indicated that the length of PAMAM dendrimer core and its nature affect the dendrimer size and its encapsulation capacity. In another work, Lee et al. [27] studied the effect of PLL and PAMAM dendrimers on DMPC bilayer by using GC-MD simulation.
Methods
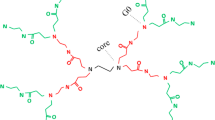
The chemical structure as obtained for PLL dendrimer by Kaminska et al. [15] and by Robert et al. [23] have been used in the present work. The benzyhydrylamine core and 1-lysine repeating units in the PLL dendrimer branches are shown in Fig. 1. Preliminary all-atom simulations of G3 and G5 of PLL dendrimer were performed to evaluate bonded interaction parameters, needed for the CG simulation. The obtained CG model was used for CG simulation of G3 and G5 at pH 7 and pH 12, to examine the structural changes of PLL dendrimer at these two protonated states.
All-atom simulation
The all atom (AA) simulation of the G3 and G5 PLL dendrimer was performed by employing GROMACS 4.5.4 software package [28, 29] along with GROMOS 53-a6 force field where the topological parameters were obtained from ABT website, as given in reference [30]. All atom simulations were performed for 30 ns with the time step of 2 fs. The simulations were carried out in a cubic box with the size of 6×6×6 nm3 where a single dendrimer was dissolved in 7000 water molecules. The simple point charge (SPC) model was used for the water molecules. The periodic boundary condition and a cutoff distance of 1.2 nm were applied in x, y and z directions. The temperature was adjusted at 305 K by using the modified Berendsen thermostat V-rescale method [31]. The pressure was set to 1 bar by utilizing Parrinello-Rahman barostat method [32]. The particle mesh Ewald (PME) summation was applied for electrostatic interactions [33]. The cutoff distance of 1.2 nm was used for both electrostatic and van der Waals interactions. The SHAKE constraints algorithm [33] was applied to all bonds.
Coarse-grained simulation
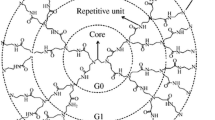
The coarse-grained simulation of CG of G3 and G5 PLL dendrimer were performed at two pHs (7 and 12), by using MARTINI force field (MFF) package [34, 35]. By placing the atoms of the PLL dendrimer in the defined virtual atomic groups (or beads) of the CG model, the bonded parameters were evaluated from the average distribution functions of all bonds and angles from the center of mass (COM) of the atom groups. The bead types of lysine were obtained from Marrink et al. [34]. The GC model of benzhydrylamine in the PLL dendrimer core, was obtained from Monticelli et al. [35] as illustrated in Fig. 2. In this figure, the terminal bead types of 1-lysine are indicated as P3, C3, and P1 and as it shows, the interior lysines have the protonation state as represented by P5-C3-P1 bead type. At pH 7, the protonated bead types of the surface lysines are changed to Qd-C3-Qd but at pH 12 where they are nonprotonated, the bead types are again represented as P3-C3-P1. The bond parameters for CG simulations, as calculated from bond and angle distribution function are reported in Tables 1 and 2. These tables indicate that, the obtained bond lengths and angles for lysine bead type (P5-C3-P1) in the dendrimer framework are different from those reported by Marrink et al. [34]. The reason for this difference is explained by considering the fact that, their results is for the presence of a single lysine in the dendrimer, where no lysine-lysine interactions are involved. However, in our case study, there are lysine-lysine and lysine-core interactions and therefore, as expected, these interactions have changed the bond angles of lysine from 180 to 130 dgrees. To simulate the interactions of PLL dendrimer with water beads, the MARTINI water model was used where four water molecules are represented as a single P4 bead. The negative Cl− ions were added to the simulation boxes to neutralize the system. A 1.2 nm cutoff distance was used for the van der Waals interactions for shifting the potential function to vanish smoothly in the range of 0.9 to 1 nm. The electrostatic interactions were modeled by utilizing a combination of short range electrostatic interactions with a cutoff distance of 1.2 nm and by employing the particle mesh Ewald summation (PME) for long range interactions [33]. The simulation time for the CG simulation was 600 ns with a time step of 20 fs. The temperature was set at 305 K and the pressure at 1 bar by applying Berendsen coupling method in the NPT ensemble [36]. All the simulations and the corresponding analysis were performed by GROMACS4.5.4 simulation package and the visualization of the simulation results were obtained by employing the Visual Molecular Dynamics (VMD) software [37]. The simulation results indicated that, after 200 ns the system attained the equilibrium state and the calculated properties remained constant.
Results and discussion
After observing that the system has attained the equilibrium state, the simulation results were obtained, in the last 200 ns of the simulation time. The characteristics of the simulation systems are listed in Table 3, where they are indicated by appropriate abbreviations, for example, CG-G4-12 means a coarse-grained generation 4 PLL dendrimer at pH 12. Furthermore, the size of simulation box and the number of solvent molecules (H2O) in each system are also presented in this table. To confirm that, the PLL dendrimers structure achieved equilibrium state, the radius of gyration (R g ) of randomly selected coarse grained PLL dendrimers were calculated and are plotted in Fig. 3, where the plot indicates that after 200 ns, the R g remains constant, and the systems have attained the equilibrium state. Also the R g values evaluated for both CG and AA simulations along with those reported by Flcovich et al. [38], are presented in Table 4. The R g values as expected, show that, higher PLL dendrimer generations have larger size and the calculated values by CG simulations are in good agreement with those of AA simulations. Additionally, our results show a dramatic decrease in the PLL dedrimer size at pH 12, whereas Liu et al. [19] in their molecular simulation study obtained an insignificant, less than 5 %, change in the PAMAM dendrimer size at different pHs. However, their other results in the same work, indicate an almost significant decrease in the internal volume and surface of PAMAM dendrimer, upon pH decrease. However, there are other reports that confirm and lend support to our simulation results. For example, Hong et al. [21] by using NMR spectroscopy measurements observed a significant change in the hydrodynamic radius of PAMAM dendrimer, due to molecular protonation increase. Also considering the R g values reported by Falkovich et al. [38], it becomes evident that, their results belong to PLL dendrimers with different cores, from those used in the present work. Therefore, the difference between R g values of this work and those of Falkovich [38] et al., is quite expected and is justifiable, although the trend of variations is the same. It is reported that at higher generations the dendrimers, have spherical shape [23, 38]. The calculated values for principle moments of inertia I x , I y , and I z in x, y, and z directions and their ratios, namely the aspect ratios, (I x / Iy ), (I x /I z ), and (I y /I z ) are reported in Table 5. The aspect ratio can be used as is a proper indication of the shape of the simulated PLL dendrimer generations. The results in Table 5 are in agreement with the power law expression R g =M β as proposed by Robert et al. [23], where M is the molecular weight and β is approximately 0.33, for spherical shape molecules. Falcovich et al. [38], presented a similar expression for spherical shape of branching chain dendrimers in the following form: R g =(MG 2 ) β, where M is the molecular weight, G is the dendrimer generation, and β is approximately 0.19. In Fig. 4, we have plotted log (R g ) versus log (MG 2 ) which shows a linear fit with the slope of β = 0.19. In Fig. 5, the radial density probabilities of water beads are plotted versus the distance from the central beads, located at the center of mass (COM) of the PLL dendrimer, as defined by the Gromacs software. The results in Fig. 5 indicate that at pH 7 and for the studied generations, the water beads have penetrated into the PLL dendrimer. According to this plot at pH 7 more water beads have penetrated deeper inside the dendrimer, compared with those at pH 12. This means that, more internal cavities have been created inside the structural layers of the dendrimer, at pH 7. The reason for this behavior can be explained by considering the interactions of the chain branching terminals in the PLL dendrimer. At pH 7 the terminal groups have a net positive partial charge, which causes repulsion between the terminal groups and as a result, the branching chains tend to extend toward the outer surface of the spherical PLL dendrimer, to become separated from each other and increase their distance, to reduce their interaction energy arisen from repulsion forces between the terminals. Therefore, path ways are created for free movement and penetration of water molecules, into the inner cavities of the dendrimer. However, at pH 12, the terminal groups are not positively charged, and they remain close to each other, and then to reduce the surface energy of the PLL dendrimer, they will back fold toward the PLL dendrimer core and as a result fewer water molecules can penetrate into the inner layers compared with those at pH 7. In Fig. 6, the radial distribution function (RDF) for the PLL dendrimer terminal beads, are plotted versus the distance from the central beads (as explained earlier) located at COM. This figure shows that, for the three studied generations (G3, G4, and G5), at pH 12, the first peak has the highest intensity which means that the terminal groups have been back folded toward the core. These results are similar to those presented by the probability distribution curves, in Fig. 5, and as explained earlier, are due to neutralization of the PLL dendrimer at this pH. These effect may also be explained by the charged hydrophobic terminal groups [4, 12], where their absence at higher pH, leads to collapse of PLL dendrimer and this can be facilitated by semi rigidity of the pLL dendrimer structure. The layer number in the dendrimer structure for each generation is denoted by Ln. The RDF of the PLL dendrimer layers (L0, L1, L2,….,Ln) versus their distance from the central beads at COM, are shown in Figs. 7 and 8, where the RDF peaks at pH 7 have lower intensity. These results can be attributed to branching chain extension as explained earlier. On comparing the RDF plots in Figs. 7 and 8, for G4 and G5 at pH 7, it is seen that the RDF peaks of G5, have higher intensity than those of G4. This difference in the intensity can be explained by the fact that, in G5, more terminals are involved in the interaction with the core, and then the probability of back folding of G5 layers toward the PLL dedrimer core is higher than G4. For a more clear representation of the obtained results, the water beads which were outside of the hydrodynamic radius of PLL dendrimer were eliminated, and the calculated water beads which penetrated into the PLL dendrimer were calculated. The results are presented in Table 6, which demonstrates that, the number of water beads penetrated into the inner layers, for G5 is more than G4, which means that, the G5 generation has more and larger cavities to encapsulate water beads. Therefore, G5 can be considered as a more suitable choice to be used in drug delivery applications.
Conclusions
In this study all-atom and coarse-grained simulations were performed on the poly(L-lysine) PLL dendrimers and the obtained results indicated that due to polar group interactions of the chain branching terminals and their protonation at the pHs 7 and 12, the generations of the PLL dendrimer indicate different behaviors. The reason for this was explained by chain branch back folding and was confirmed by radial distribution function calculations. The calculated aspect ratios showed that their structural shape becomes more spherical at higher generations and at higher pH. At pH 7, the higher generations can encapsulate more water molecules and then they are preferred for drug delivery applications.
References
Baoukina S, Monticelli L, Tieleman DP (2013) Interaction of pristine and functionalizedcarbon nanotubes with lipid membranes. J Phys Chem B 117:12113–12123
Menjoge AR, Kannan RM, Tomalia DA (2010) Dendrimer-based drug and imaging conjugates: design considerations for nanomedical applications. Drug Discov Today 15:171–185
Yonetake K, Masuko T (1999) Poly(propyleneimine) dendrimers peripherally modified with mesogens. Macromolecular 32:6578–6586
Zhang X, Oulad-abdelghani M, Zelkin AN, Wang Y, Haı Y, Benkirane-jessel N, Mainard D, Voegel J, Caruso F (2010) Poly(L-lysine) nanostructured particles for gene delivery and hormone stimulation. Biomaterials 31:1699–1706
Fox ME, Guillaudeu S, Fre JMJ, Jerger K, Macaraeg N, Szoka FC (2009) Synthesis and in Vivo antitumor efficacy of PEGylated Poly(L-lysine) dendrimer-camptothecin conjugates. Mol Pharm 6:1562–1572
Gillies ER, Frechet JM (2005) Dendrimers and dendritic polymers in drug delivery. Drug Discov Today 10:35–43
Stiriba S, Frey H, Haag R (2002) Dendritic polymers in biomedical applications: From potential to clinical use in diagnostics and therapy. Angew Chem Int Ed 41:1329–1334
Niedrhafner P, Šebestik J, Ježek J (2005) Peptide dendrimers. J Peptide Sci 11:757–788
Boas U, Heegaard PM (2004) Dendrimers in drug research. Chem Soc Rev 33:43–63
Perumal OP, Inapagolla R, Kannan S, Kannan RM (2008) The effect of surface functionality on cellular trafficking of dendrimers. Biomaterials 29:3469–347611
Choi JS, Lee EJ, Choi YH, Jeong YJ, Park JS (1999) Poly(ethylene glycol)-block-poly(L-lysine) dendrimer: novel linear polymer/dendrimer block copolymer forming a spherical water-soluble polyionic complex with DNA. Bioconj Chem 10:62–65
Byrne M, Victory D, Hibbitts A, Lanigan M, Heise A, Cryan S (2013) Molecular weight and architectural dependence of well-defined star-shaped poly(lysine) as a gene delivery vector. Biomater Sci 1:1223–1234
Yevlampieva N, Dobrodumov A, Nazarova O, Okatova O (2012) Hydrodynamic behavior of dendrigraft polylysines in water and dimethylformamide. Poly 4:20–31
Okuda T, Kawakami S, Maeie T, Niidome T (2006) Biodistribution characteristics of amino acid dendrimers and their PEGylated derivatives after intravenous administration. J Contrl Rel 114:69–77
Kaminskas LM, Boyd BJ, Karellas P, Krippner GY, Lessene R, Kelly B, Porter CJH (2008) The impact of molecular weight and PEG chain length on the systemic pharmacokinetics of PEGylated poly L-Lysine dendrimers. Mol Pharm 5:449–463
Okuda T, Sugiyama A, Niidome T, Aoyagi H (2004) Characters of dendritic poly(l-lysine) analogues with the terminal lysines replaced with arginines and histidines as gene carriers in vitro. Biomater 25:537–544
Kaminskas LM, Kelly BD, Mcleod VM, Boyd BJ, Krippner GY, Williams ED, Porter CJH (2009) Pharmacokinetics and tumor disposition of PEGylated, methotrexate conjugated Poly-L-lysine dendrimers. Mol Pharm 6:1190–1204
Rossi J, Boiteau L, Collet H, Tsamba BM, Larcher N (2012) Functionalisation of free amino groups of lysine dendrigraft (DGL) polymers. Tetrahedron Lett 53:2976–2979
Liu Y, Bryantsev VS, Diallo MS, Goddard WA III (2009) PAMAM dendrimers undergo pH responsive conformational changes without swelling. J Am Chem Soc 131:2798–2799
Markelov DA, Falkovich SG, Neelov IM, Ilyash MY, Matveev VV, Lähderanta E, Ingman P, Darinskii AA (2015) Molecular dynamics simulation of spin–lattice NMR relaxation in poly- l -lysine dendrimers: manifestation of the semiflexibility effect. Phys Chem Chem Phys 17:3214–3226
Hong K, Liu Y, Porcar L, Liu D, Gao CY, Smith GS, Herwig KW, Cai S, Li X, Wu B, Chen WR, Liu L (2012) Structural response of polyelectrolyte dendrimer towards molecular protonation: the inconsistency revealed by SANS and NMR. J Phys Condens Matter 24:064116
Amjad-Iranagh GK, Modarress H (2014) Molecular simulation study of PAMAM dendrimer composite membranes. J Mol Model 20:2119–2124
Roberts BP, Scanlon MJ, Krippner GY, Chalmers DK (2009) Molecular dynamics of poly(L-lysine) dendrimers with naphthalene disulfonate Caps. Macromol 42:2775–2783
Neelov IM, Markelov DA, Falkovich SG, Okrugin BM, Darinskii AA (2013) Mathematical simulation of lysine dendrimers: temperature dependences. Poly Sci 55:154–161
Lee H, Choi JS, Larson RG (2011) Molecular dynamics studies of the size and internal structure of the PAMAM dendrimer grafted with arginine and histidine. Macromol 44:8681–8686
Kavyani S, Amjad-iranagh S, Modarress H (2014) Aqueous poly(amidoamine) dendrimer G3 and G4 generations with several interior cores at pHs 5 and 7: a molecular dynamics simulation study. J Phys Chem B 118:3257–3266
Lee H, Larson R (2008) Lipid bilayer curvature and pore formation induced by charged linear polymers and dendrimers: the effect of molecular shape. J Phys Chem B 112:12279–12285
Hess B, Kutzner C (2008) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC (2005) GROMACS: fast, flexible, and free. J Comput Chem 26:1701–1718
Malde AK, Zuo L, Breeze M, Stroet M, Poger D, Nair PC, Oostenbrink C, Mark AE (2011) An automated force field topology builder (ATB) and repository: version 1.0. J Chem Theory Comput 7:4026–4037
Bussi G, Donadio D, Parrinello M (2007) Canonical sampling through velocity rescaling. J Chem Phys 126:14101–14107
Parrinello M, Rahman A (1981) Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys 52:7182–7190
Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG (1995) A smooth particle mesh Ewald method. J Chem Phys 103:8577–8593
Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH (2007) The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B 111:7812–7824
Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ (2008) The MARTINI coarse-grained force field: extension to proteins. J Chem Theory and Comput 4:819–834
Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular Ddnamics. J Mol Graph 14:33–38
Falkovich S, Markelov D, Neelov I, Darinskii A (2013) Are structural properties of dendrimers sensitive to the symmetry of branching? Computer simulation of lysine dendrimers. J Chem Phys 139:64903–64913
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rahimi, A., Amjad-Iranagh, S. & Modarress, H. Molecular dynamics simulation of coarse-grained poly(L-lysine) dendrimers. J Mol Model 22, 59 (2016). https://doi.org/10.1007/s00894-016-2925-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00894-016-2925-0