Abstract
The thermo-acidophilic archaeon, Sulfolobus tokodaii, was utilized for the production of Pd(0) bionanoparticles from acidic Pd(II) solution. Use of active cells was essential to form well-dispersed Pd(0) nanoparticles located on the cell surface. The particle size could be manipulated by modifying the concentration of formate (as electron donor; e-donor) and by addition of enzymatic inhibitor (Cu2+) in the range of 14–63 nm mean size. Since robust Pd(II) reduction progressed in pre-grown S. tokodaii cells even in the presence of up to 500 mM Cl−, it was possible to conversely utilize the effect of Cl− to produce even finer and denser particles in the range of 8.7–15 nm mean size. This effect likely resulted from the increasing stability of anionic Pd(II)–chloride complex at elevated Cl− concentrations, eventually allowing involvement of greater number of initial Pd(0) crystal nucleation sites (enzymatic sites). The catalytic activity [evaluated based on Cr(VI) reduction reaction] of Pd(0) bionanoparticles of varying particle size formed under different conditions were compared. The finest Pd(0) bionanoparticles obtained at 50 mM Cl− (mean 8.7 nm; median 5.6 nm) exhibited the greatest specific Cr(VI) reduction rate, with four times higher catalytic activity compared to commercial Pd/C. The potential applicability of S. tokodaii cells in the recovery of highly catalytic Pd(0) nanoparticles from actual acidic chloride leachate was, thus, suggested.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Palladium (Pd), one of the platinum group metals (PGMs), is regarded as one of the most important industrial catalysts. To secure a stable world supply of PGMs and other precious metals, recycling of secondary metal resources (e.g., spent catalysts and e-wastes) is considered increasingly important. For Pd recycling from such secondary resources, strong leaching lixiviants such as aqua regia, HCl, HNO3, and H2SO4 are used together with an oxidizing agent. To lower environmental impacts, cleaner alternatives, such as using diluted HCl with H2O2, are also investigated by different groups (e.g., Barakat et al. 2006). Since Pd catalysts today are mostly used as nanoparticles due to their greater specific surface area with higher reactivity (De Corte et al. 2012), developing recycling techniques of the metal in the nanoparticle form would be beneficial.
Microbiological production of precious metal nanoparticles is gaining increasing attention as a simple and clean technology which proceeds under ambient conditions without the use of hazardous chemicals (Zhang et al. 2011). Upon utilization of microbial cells, reduction of aqueous Pd(II) ions is triggered by enzymatic activity by an expense of externally added e-donor (or intracellular electron carriers such as NADH accumulated during pre-growth; Okibe et al. 2017). They are then deposited as solid Pd(0) nanoparticles at different cellular locations as a scaffold (on the cell wall, within the periplasmic space and inside cytoplasm), depending on the microbial species and conditions used (De Windt et al. 2006; Okibe et al. 2017). The enzymes proposed to be responsible for Pd(II) reduction include [NiFe]-hydrogenases in D. fructosivorans (Mikheenko et al. 2008) and E. coli (Deplanche et al. 2010), three [NiFe]-hydrogenases (Hyd-1, Hyd-2, and Hyd-3), and two formate dehydrogenase molybdoenzymes (FDH-N and FDH-H) in anaerobically grown E. coli (Foulkes et al. 2016), and molybdoenzyme (FDH-O) which is expressed both aerobically and anaerobically (Foulkes et al. 2016). These Pd(0) bionanoparticles have been effectively utilized as catalyst in reactions such as reduction of highly toxic Cr(VI) to Cr(III) (Humphries et al. 2007; Mabbett et al. 2004), dehalogenation of polychlorinated biphenyls (PCBs) (De Windt et al. 2006), and dechlorination of lindane (Mertens et al. 2007).
So far, the majority of Pd(0) bionanoparticles studies employed a variety of neutrophilic bacteria such as Desulfovibrio spp. (Mikheenko et al. 2008; Yong et al. 2002), Shewanella oneidensis (De Windt et al. 2006), Geobacter sulfurreducens (Yates et al. 2013), Bacillus sphaericus (Creamer et al. 2007), Escherichia coli, Serratia sp., Micrococcus luteus, Arthrobacter oxydans (Deplanche et al. 2014), Cupriavidus necator, Pseudomonas putida, and Paracoccus denitrificans (Bunge et al. 2010). In general, these Pd(0) bionanoparticles were observed with the particle size of a few tens of nanometers.
On the other hand, considering metallurgical processes for the metal extraction reaction being often highly acidic, extremely acidophilic microorganisms are worth investigating for their potential in the precious metal bionanoparticles production. However, compared with the number of Pd(0) bionanoparticles studies done on neutrophilic microorganisms, limited studies on extreme acidophiles are so far available. Enzymatic activity of neutrophiles may be more susceptible to acidic leachates. In fact, pre-palladized neutrophilic cells were prepared prior to exposure to highly acidic leachates, to promote autocatalytic chemical Pd(II) reduction to Pd(0) (Creamer et al. 2006; Mabbett et al. 2006). In addition, H2 gas (instead of formate) was used as e-donor to promote Pd(II) reduction using neutrophilic D. desulfuricans at pH 2.0 (Yong et al. 2002).
As for acidophilic bacteria, Acidocella aromatica PFBC and Acidiphilium cryptum SJH were utilized to produce Pd(0) bionanoparticles from acidic Pd(II) solutions as well as from Pd(II)-containing spent catalyst leachate (Okibe et al. 2017). The former strain was also utilized for size-controlled production of Au(0) nanoparticles (Rizki and Okibe 2018). While, regarding acidophilic archaea, Au(0) and Ag(0) nanoparticles were formed using Sulfolobus islandicus (Kalabegishvili et al.2014, 2015). To our knowledge, studies on Pd(0) bionanoparticles formation using extremely acidophilic archaea are not yet available.
Hence, this study focused on the extremely acidophilic archaea to reveal the utility of this third domain of life in the production of Pd(0) nanoparticles. From the genome sequence, the presence of the putative formate dehydrogenase enzyme (FDH) (encoded by the ST0348 gene; http://www.uniprot.org/) was indicated with the thermophilic, extremely acidophilic archaeon Sulfolobus tokodaii (optimal growth conditions 80 °C and pH 2.5–3; Suzuki et al. 2002). This archaeon was also shown to have the Fe3+-reducing ability under both anaerobic and micro-aerobic conditions (Masaki et al. 2018). The study, therefore, investigated the capability of S. tokodaii in Pd(0) bionanoparticles production from acidic Pd(II) solutions. The effect of chloride ions (Cl−) was also studied to gain fundamental knowledge on its potential utility in actual acidic industrial leachates.
Materials and methods
Microorganism
The thermo-acidophilic archaeon, Sulfolobus tokodaii 7T (NBRC 100140), was routinely sub-cultured and pre-grown aerobically in 300 mL Erlenmeyer flasks containing 100 mL heterotrophic basal salts (HBS) medium (per L; 450 mg (NH4)2SO4, 50 mg KCl, 50 mg KH2PO4, 500 mg MgSO4 7H2O, 14 mg Ca(NO3)2 4H2O, 142 mg Na2SO4; pH 2.0 with H2SO4) with 10 mM glucose and 0.025% (w/v) yeast extract. Flasks were incubated at 70 °C, shaken at 120 rpm.
Pd(II) reduction test by S. tokodaii cell suspension
Sulfolobus tokodaii cells were pre-grown aerobically, harvested at the late-exponential phase, and washed and resuspended in 50 mL HBS medium in 70 mL vials (1.0 × 109 cells/mL; pH 2.0). Pd(II) was added (as Na2PdCl4) at 50 mg/L. As e-donor, sodium formate (HCOONa) was added at 0, 5 or 10 mM. It should be noted that HCOONa (pKa = 3.75) added to the media hereafter was likely present predominantly as formic acid (HCOOH) under the acidic conditions used in this study. For comparison, active cells (± 5 mM Cu2+; as CuSO4·7H2O), heat-killed cells (autoclaved at 120 °C for 20 min), and cell-free controls were prepared. All solutions were prepared aerobically, but vials were sealed with butyl-rubber stoppers and aluminum crimps to establish the micro-aerobic condition and incubated unshaken at 70 °C. Liquid samples were regularly withdrawn using syringe needles and analyzed for Pd(II) concentration spectrophotometrically using the PAR method (Mizuno and Miyatani 1976). Following Pd(II) reduction, cells were harvested, washed, and freeze-dried overnight for XRD analysis (Rigaku UltimaIV; CuKα 40 mA, 40 kV). All of the experiments were conducted in duplicate.
Ultra-thin section transmission electron microscopy (TEM) observation
After completion of Pd(II) reduction, S. tokodaii cells were fixed with 2% (v/v) paraformaldehyde/2.5% (v/v) glutaraldehyde mixture in 0.1 M phosphate buffer solution (PBS) (4 °C, 30 min), washed twice with 0.1 M PBS (pH 7.2), post-fixed with 1% OsO4 in 0.1 M PBS (4 °C, 1–2 h), and washed twice again. Cells were then dehydrated using ascending series of ethanol concentration (70, 80, 90, and 99.5% for 5 min each, followed by 100% for 10 min twice), washed twice with propylene oxide (5 min each), and finally embedded in epoxy resin (polymerized at 70 °C, 48 h). Ultra-thin sectioning (70 nm) was performed using an ultramicrotome (Leica EM UC7). Sections were placed onto copper grids, stained with EM stainer (Nisshin EM) and lead acetate, and sputter-coated with carbon, prior to observation with TEM (Tecnai G-20; accelerating voltage 200 kV).
Particle-size analysis using Image-J software
TEM images of S. tokodaii cells were analyzed using Image-J software (National Institute of Health, USA). First, the images were calibrated and thresholded by selecting the ROI (region of interest) and removing the background noise (based on the contrast between nanoparticles and cell components). Pd(0) bionanoparticles were then analyzed with the “Analyze Particles” function, which calculates the projected area of an individual particle. The diameter of each particle was calculated from its projected area, assuming that the particle is spherical. For each condition, 4–5 cells displaying a total of around 180–1400 particles were analyzed to calculate the mean and median particle sizes.
In the case of cell-free Pd(0) particles, scanning electron microscope (SEM; Keyence VE9800) images were adjusted with the “Bandpass filter” function to create a better contrast between each particle. The adjusted images were then thresholded and analyzed using the same protocol as described above.
Functional group analysis
Fourier transform infrared spectroscopy (FTIR) analysis was performed to study the overtime change in the functional groups of S. tokodaii biomass upon exposure to Pd(II) ions. After addition of 50 mg/L Pd(II) and 5 mM formate to S. tokodaii active cell suspensions (at 109 cells/mL in 50 mL HBS medium; in 70 mL vials under micro-aerobic condition, 70 °C, pH 2.0), the cells were collected at 0, 3, 5 or 10 h [Pd(II) reduction profile corresponding to the active cells experiment, as shown in Fig. 1] by centrifugation, washed thoroughly using deionized water, freeze-dried overnight, and quantitatively mixed with KBr (FTIR grade). The infrared spectra were measured in the range of 400–4000 cm−1 at a resolution of 2 cm−1 with 128 scan times (JASCO FTIR-670 Plus).
Pd(II) reduction to Pd(0) in S. tokodaii cell suspensions (filled circle active cells, filled up-pointing triangle active cells + 5 mM Cu2+, unfilled circle heat-killed cells; all at 1.0 × 109 cell/mL) or in cell-free controls (multiplication sign), under the micro-aerobic condition at 70 °C, pH 2.0, in the presence of 5 mM formate as e-donor
Zeta-potential measurement
Sulfolobus tokodaii cells were pre-grown as described above, harvested by centrifugation, washed twice, and resuspended in 10 mL of 1 mM KCl (pH 3.0, 4.0 or, 5.0 with KOH or HCl) at 108 cells/mL. After addition of 50 mg/L Pd(II) (as Na2PdCl4), cells were left for 30 min prior to zeta-potential measurement (Malvern ZETASIZER Nano series). The measurement was conducted in duplicate.
Effect of Cl− on Pd(0) bionanoparticles formation
As an alternative to highly corrosive aqua regia leaching, chloride-peroxide leaching was purposed for Pd extraction from spent catalysts (Barakat et al. 2006). To study fundamental aspects of the effect of Cl− on Pd(0) bionanoparticles formation, Cl− was added (as NaCl) at 10, 50, 100, and 1000 mM to S. tokodaii active cell suspensions, based on the methodology described in the previous section. Formate was added as e-donor at 10 mM.
Thermogravimetry analysis
The Pd(0) load on Pd(0) bionanoparticles was estimated by thermogravimetry analysis (TG–DTA 2000SA, Bruker AXS), by heating 5 mg of a Pd(0) bionanoparticles sample in the Pt-sample-pan (from room temperature (25 °C) to 1200 °C at 10 °C/min) under N2 atmosphere to prevent formation of PdO. Pd(0) bionanoparticles samples were regularly collected following the completion of reduction of 50 mg/L Pd(II) (in 109 cells/mL cell suspensions).
Evaluation of the catalytic activity of Pd(0) bionanoparticles
Catalytic activities of the Pd(0) bionanoparticles produced under different conditions as well as cell-free Pd(0) particles were compared with that of commercial palladium on carbon catalyst [Pd/C; 10% w/w Pd(0) loading; Sigma-Aldrich], based on Cr(VI) reduction reaction.
After completion of Pd(0) bionanoparticle formation by S. tokodaii from 50 mg/L Pd(II) using 5 or 10 mM formate (as described in previous sections), precipitates were collected by centrifugation and freeze-dried. An aliquot of the precipitate [equivalent to 0.5 mg net-Pd(0)] was used as a catalyst for the following Cr(VI) reduction reaction in 20 mL deionized water containing 10 mg/L Cr(VI) (as Na2CrO4·4H2O) and 10 mM sodium formate as e-donor (pH 2.0 with H2SO4; 25 mL vials). Solutions used in the Cr(VI) reduction reaction were prepared aerobically, but vials were sealed with butyl-rubber stoppers and aluminum crimps to establish the micro-aerobic condition and incubated unshaken at 30 °C. Samples were withdrawn with syringe needles to analyze the Cr(VI) concentration using the diphenylcarbazide method (Noroozifar and Khorasani-Motlagh 2003).
Results and discussion
Pd(II) reduction and Pd(0) bionanoparticle production by S. tokodaii
Without the addition of e-donor, an only negligible amount of Pd(II) (3–5 mg/L in 16 h) was reduced in all cases (data not shown). Use of 5 mM formate as e-donor resulted in complete Pd(II) reduction (under micro-aerobic conditions) regardless of the presence/condition of the cells, but with different speed; the first “induction phase” before the initiation of rapid Pd(II) reduction was generally shortened by the presence of cell biomass, especially with active cells (without Cu2+) (Fig. 1). This first “induction phase” corresponds to the period of Pd(0) crystal nucleation, which takes place most effectively on the active cell surface due to the intact enzymatic activity. After which, the autocatalytic property of Pd(0) nuclei accelerates the speed of Pd(II) reduction [Pd(0) crystal growth phase] (Okibe et al. 2017). Upon completion of Pd(II) reduction under each condition, black cell-Pd(0) precipitates (confirmed by XRD; Fig. 2) were recovered for ultra-thin section TEM observation and the following particle-size analysis (Fig. 3). Pd(0) bionanoparticles of relatively homogeneous size were found localized mostly on the cell surface when active cells were used (Fig. 3a, b). In contrast, Pd(0) particles of diverse size were formed randomly (extracellularly and intracellularly) with heat-killed cells (Fig. 3c). Partially deactivating the enzymatic activity by Cu2+ decreased the number of Pd(0) nucleation sites, but instead increased the size of individual Pd(0) bionanoparticle (Fig. 3c). Heat-kill treatment of the cells completely disrupted enzymatic activities and selective cell permeability: Pd(II) ions were able to freely diffuse into the cells and deformed cell components with enlarged surface area, as the scaffold for Pd(0) nucleation. As a result, the Pd(II) reduction speed in heat-killed cells was even somewhat higher than that in active cells with Cu2+ (Fig. 1). Nonetheless, the lack of intact cell functions produced highly heterogeneous Pd(0) particles (Fig. 3c). In cell-free controls, the resultant Pd(0) particles were highly aggregated, as shown in Supplemental Fig. 1.
TEM cross-sectional images of Pd(0) bionanoparticles formed on S. tokodaii cells at 5 mM formate, and their particle-size distributions: a active cells (n = 750 in 5 cells), b active cells + 5 mM Cu2+ (n = 185 in 5 cells), and c heat-killed cells (n = 370 in 4 cells). Arrows point extracellular Pd(0) particles (c)
The presence of putative formate dehydrogenase enzyme (FDH) (encoded by the ST0348 gene; http://www.uniprot.org/) may be responsible for the initial Pd(0) crystal nucleation in S. tokodaii, by facilitating the production of H2 from formate (Sinha et al. 2015) on the active cell surface. To clarify the mechanism, however, further biochemistry studies are needed for the archaeal cells.
Contribution of functional groups on the S. tokodaii cell surface for Pd(II) biosorption and the following Pd(0) bionanoparticles formation
Overtime changes in FTIR spectra were analyzed during microbial Pd(II) reduction and Pd(0) bionanoparticle formation by S. tokodaii cells (Fig. 4). It was shown from zeta-potential measurement that the weakly positive cell surface charge shifted towards negative upon exposure to Pd(II) (from +3.3 to +2.4 mV at pH 2.0; Fig. 5). This implies that biosorption of negatively charged Pd(II) ions precedes microbial Pd(II) reduction. Based on the spectrum of original cells (0 h; Fig. 4a) the bands at 1650 and 1535 cm−1 resulted mainly from v C = O and δ N–H, respectively, of amides from proteins (Goormaghtigh et al. 1994). The band at 1454 cm−1 is attributed to δ C–H of CH3 and CH2 groups, and those at 1398 and 1235 cm−1 are assigned to v C–O from carboxylate groups and ʋ P = O of phosphodiester groups in nucleic acids and phospholipids, respectively (Giordano et al. 2001). The spectral region between 900 and 1200 cm−1 is assigned to ʋ C–O–C of diverse polysaccharides groups, which also derive from EPS (Extracellular polymeric substances) as well as surface layers (S layers) from Sulfolobus spp. (Koerdt et al. 2011; Zhang et al. 2015).
Upon exposure to Pd(II) ions, relatively major shifts were observed with the bands at 1650, 1535, and 1235 cm−1 towards lower energy (Fig. 4). Changes were also observed in the region between 900 and 1200 cm−1 (Fig. 4). This suggests that amide groups, which are positively charged at acidic pH, were responsible for sorption of major anionic Pd(II) species under this condition, PdCl3− (Colombo et al. 2008). Phosphate and polysaccharides groups may have electrically attracted minor PdCl+ species (Colombo et al. 2008).
Effect of Cl− on Pd(0) bionanoparticles production
Since the use of 5 mM formate as e-donor (as in the previous test) was shown insufficient in the presence of additional Cl−, its concentration was increased to 10 mM this time (Fig. 6). Without extra Cl− addition, increasing the formate concentration in active cells from 5 mM (Fig. 3a) to 10 mM (Fig. 7a) upsized the Pd(0) bionanoparticles roughly by twofold, but instead decreased the particle density. Addition of elevating concentrations of Cl− (0–1000 mM) increasingly prolonged the apparent “induction phase” before the initiation of rapid Pd(II) reduction (Fig. 6). This likely resulted from the form of major Pd(II) species increasingly shifting from PdCl3− towards more stable PdCl42− at higher Cl− concentrations (Colombo et al. 2008), causing the initiation of Pd(0) crystal nucleation more time-consuming. Nonetheless, the following rapid Pd(II) reduction was triggered in all the cases, except at the highest Cl− concentration of 1000 mM (Fig. 6). TEM images revealed that this prolonged “induction phase” due to the presence of Cl− likely allowed involvement of a sufficient number of crystal nucleation site (enzymatic sites), leading to the formation of finer and denser Pd(0) bionanoparticles (Fig. 7). The smallest and densest particles (mean size = 8.7 nm; 354/cell) were recovered at 50 mM Cl− (Fig. 8c), followed by those (mean size = 12 nm; 330/cell) at 100 mM Cl− (Fig. 8d). The results suggest that the presence of Cl− in the range of up to 50–100 mM does not deactivate enzymatic sites for Pd(0) nucleation (Fig. 8c, d), unlike the case with Cu2+ (Fig. 3b). At 500–1000 mM formate, cells started to disintegrate, and no evidence of Pd(0) deposition was found at 1000 mM. Overall, the enzymatic Pd(0) nucleation activity of S. tokodaii at the expense of formate seems to be more robust than that of neutrophilic Desulfovibrio desulfuricans in the presence of high-concentration Cl− (Yong et al. 2002). Therefore, using S. tokodaii cells, Pd(0) bionanoparticles could be recovered from acidic chloride leachates of secondary Pd sources (e.g., spent catalyst; Barakat et al. 2006) upon appropriate dilution. In abiotic studies by other groups, Cl− was utilized to shape-control Pd(0) nanoparticles (Nalajala et al. 2016) or size-control Au(0) nanoparticles in the citrate reduction system (the particle size becomes larger at higher Cl− concentrations due to the decrease of the surface charge; Zhao et al. 2012). The results in this study suggest that the effect of Cl− can also be utilized in biological systems to size-control Pd(0) nanoparticles by selecting favorable microbial strains.
Pd(II) reduction to Pd(0) in S. tokodaii active cell suspensions (1.0 × 109 cell/mL), under the micro-aerobic condition at 70 °C, pH 2.0, in the presence of 10 mM formate as e-donor. NaCl was added at 0 mM (filled circle), 10 mM (filled square), 50 mM (filled up-pointing triangle), 100 mM (filled down-pointing triangle), 500 mM (filled diamond), or 1000 mM (multiplication sign)
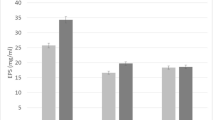
Comparison of the catalytic activity (based on the specific Cr(VI) reduction rate) of Pd(0) bionanoparticles (a–h) and commercial Pd/C catalyst (i). Pd(0) bionanoparticles were produced by S. tokodaii in the presence of either 5 mM formate (a–d) or 10 mM formate (e–h), under different conditions: a active cells, b active cells + Cu2+, c heat-killed cells, d cell-free controls, e active cells, f active cells + 10 mM Cl−, g active cells + 50 mM Cl−, and h active cells + 100 mM Cl−. a–h Mean and median particle sizes of Pd(0) nanoparticles are indicated. h The mean particle size of Pd/C catalyst is cited from Yang et al. (2018). The specific Cr(IV) reduction rate was calculated for the time interval of 0–5 h (a), 0–30 h (b), 0–30 h (c), 0–48 h (d), 0–10 h (e), 0–9 h (f), 0–1 h (g), 0–4 h (h), and 0–4 h (i) (Supplemental Fig. 3). As e-donor for Cr(VI) reduction, 10 mM formate was used in all cases
Catalytic activity of Pd(0) bionanoparticles
Pd(0) bionanoparticles produced under different conditions were compared for their catalytic activity based on the Cr(VI) reduction reaction. Based on the thermogravimetric analysis [50% (w/w) Pd(0) loaded on dry-cell weight; Supplemental Fig. 3], reaction mixtures were prepared to contain an equivalent amount of Pd(0). Neither Pd(0) bionanoparticles only nor formate only as e-donor reduced Cr(VI) to Cr(III) (data not shown). Therefore, formate decomposition via Pd(0) catalyst to produce H2 represents the effectiveness of Cr(VI) reduction (Okibe et al. 2017). The catalytic activity of Pd(0) bionanoparticles highly depended on the particle size; those of smaller size exhibited higher specific Cr(VI) reduction rate (Fig. 8). The finest Pd(0) bionanoparticles produced by active cells at 50 mM Cl− possessed the highest catalytic activity which was approximately four times greater compared to commercial Pd/C (Fig. 8; Supplemental Fig. 3). The results here suggested that the size of Pd(0) bionanoparticles can be manipulated by modifying the concentration of formate as e-donor and by the use of Cu2+ as enzyme inhibitor (as was observed with Au(0) bionanoparticles; Rizki and Okibe 2018), as well as by taking advantage of the effect of Cl−. This has the important implication that this approach using S. tokodaii cells could be effectively applied to actual Pd–chloride leachates even to produce Pd(0) nanoparticles of higher catalytic activity.
Most prokaryotes (bacteria and archaea) possess S layers (a monomolecular planar array of proteinaceous subunits as the outermost component of the cell envelope). Gram-negative archaea including Sulfolobus spp. possess S layers as the only cell-wall component external to the plasma membrane. While, in Gram-positive bacteria and Gram-positive archaea, S layers are found on the surface of the rigid wall matrix (mainly peptidoglycan and pseudopeptidoglycan, respectively). In Gram-negative bacteria, cell envelope is composed of a thin peptidoglycan wall and an outer membrane which S layer is attached to (Sleytr and Beveridge, 1999). In addition to the particle size, such differences in the cell envelope structure could affect the accessibility of the Pd(0) bionanoparticle catalyst to reaction substrates. Simpler and thinner archaeal cell surface structure may be advantageous in this regard. More studies on metal bionanoparticles using archaeal cells would further clarify the importance of the third domain of life in nanobiotechnology.
Conclusion
Pd(0) bionanoparticles were effectively produced from acidic Pd(II) solution using the thermophilic, extremely acidophilic archaeon, S. tokodaii. Use of enzymatically active S. tokodaii cells was essential to produce well-dispersed Pd(0) bionanoparticles deposited on the cell surface. The particle size could be manipulated (14–63 nm mean size) by means of modification of the formate concentration and addition of enzymatic inhibitor (Cu2+). The effect of additional Cl− was conversely utilized to produce even finer and denser Pd(0) bionanoparticles (8.7–15 nm mean size). The finest Pd(0) bionanoparticles produced at 50 mM Cl− (mean 8.7 nm; median 5.6 nm) exhibited the highest catalytic activity (four times higher compared to commercial Pd/C). Therefore, the potential applicability of S. tokodaii cells in the recovery of highly catalytic Pd(0) nanoparticles from actual acidic chloride leachate was suggested. Further studies on extreme archaea for metal bionanoparticles production may benefit the development of nanobiotechnology.
References
Barakat MA, Mahmoud MHH, Mahrous YS (2006) Recovery and separation of palladium from spent catalyst. Appl Catal A 301:182–186
Bunge M, Sobjerg LS, Rotaru AE, Gauthier D, Lindhardt AT, Hause G, Finster K, Kingshott P, Skrydstrup T, Meyer RL (2010) Formation of palladium(0) nanoparticles at microbial surfaces. Biotechnol Bioeng 107:206–215
Colombo C, Oates CJ, Monhemius AJ, Plant JA (2008) Complexation of platinum, palladium and rhodium with inorganic ligands in the environment. Geochem-Explor Env A 8:91–101
Creamer NJ, Baxter-Plant VS, Henderson J, Potter M, Macaskie LE (2006) Palladium and gold removal and recovery from precious metal solutions and electronic scrap leachates by Desulfovibrio desulfuricans. Biotechnol Lett 28:1475–1484
Creamer NJ, Mikheenko IP, Yong P, Deplanche K, Sanyahumbi D, Wood J, Pollmann K, Merroun M, Selenska-Pobell S, Macaskie LE (2007) Novel supported Pd hydrogenation bionanocatalyst for hybrid homogeneous/heterogeneous catalysis. Catal Today 128:80–87
De Corte S, Hennebel T, De Gusseme B, Verstraete W, Boon N (2012) Bio-palladium: from metal recovery to catalytic applications. Microb Biotechnol 5:5–17
De Windt W, Boon N, Van den Bulcke J, Rubberecht L, Prata F, Mast J, Hennebel T, Verstraete W (2006) Biological control of the size and reactivity of catalytic Pd(0) produced by Shewanella oneidensis. Antonie Van Leeuwenhoek 90:377–389
Deplanche K, Caldelari I, Mikheenko IP, Sargent F, Macaskie LE (2010) Involvement of hydrogenases in the formation of highly catalytic Pd(0) nanoparticles by bioreduction of Pd(II) using Escherichia coli mutant strains. Microbiol SGM 156:2630–2640
Deplanche K, Bennett JA, Mikheenko IP, Omajali J, Wells AS, Meadows RE, Wood J, Macaskie LE (2014) Catalytic activity of biomass-supported Pd nanoparticles: influence of the biological component in catalytic efficacy and potential application in ‘green’ synthesis of fine chemicals and pharmaceuticals. Appl Catal B 147:651–665
Foulkes JM, Deplanche K, Sargent F, Macaskie LE, Lloyd JR (2016) A novel aerobic mechanism for reductive palladium biomineralization and recovery by Escherichia coli. Geomicrobiol J 33:230–236
Giordano M, Kansiz M, Heraud P, Beardall J, Wood B, McNaughton D (2001) Fourier transform infrared spectroscopy as a novel tool to investigate changes in intracellular macromolecular pools in the marine microalga Chaetoceros muellerii (bacillariophyceae). J Phycol 37:271–279
Goormaghtigh E, Cabiaux V, Ruysschaert J (1994) Determination of soluble and membrane protein structure by Fourier transform infrared spectroscopy: I. Assignments and model compounds. In: Hilderson HJ, Ralston GB (eds) Subcellular biochemistry. Physicochemical methods in the study of biomembranes. Springer, Boston, pp 329–362
Humphries AC, Penfold DW, Macaskie LE (2007) Cr(VI) reduction by bio and bioinorganic catalysis via use of bio-H2: a sustainable approach for remediation of wastes. J Chem Technol Biotechnol 82:182–189
Kalabegishvili TL, Murusidze IG, Prangishvili DA, Kvachadze LI, Kirkesali EI, Rcheulishvili AN, Ginturi EN, Janjalia MB, Tsertsvadze GI, Gabunia VM, Frontasyeva MV, Zinicovscaia I, Pavlov SS (2014) Gold nanoparticles in Sulfolobus islandicus biomass for technological applications. Adv Sci Eng Med 6:1–7
Kalabegishvili TL, Murusidze IG, Prangishvili DA, Kvachadze LI, Kirkesali EI, Rcheulishvili AN, Ginturi EN, Janjalia MB, Tsertsvadze GI, Gabunia VM, Frontasyeva MV, Zinicovscaia I, Pavlov SS (2015) Silver nanoparticles in Sulfolobus islandicus biomass for technological applications. Adv Sci Eng Med 7:1–8
Koerdt A, Orell A, Pham TK, Mukherjee J, Wlodkowski A, Karunakaran E, Biggs CA, Wright PC, Albers S (2011) Macromolecular fingerprinting of Sulfolobus species in biofilm: a transcriptomic and proteomic approach combined with spectroscopic analysis. J Proteome Res 10:4105–4119
Mabbett AN, Yong P, Farr JP, Macaskie LE (2004) Reduction of Cr(VI) by “palladized” biomass of Desulfovibrio desulfuricans ATCC 29577. Biotechnol Bioeng 87:104–109
Mabbett AN, Sanyahumbi D, Yong P, Macaskie LE (2006) Biorecovered precious metals from industrial wastes: single-step conversion of a mixed metal liquid waste to a bioinorganic catalyst with environmental application. Environ Sci Technol 40:1015–1021
Masaki Y, Tsutsumi K, Okibe N (2018) Iron redox transformation by the thermo-acidophilic archaea from the genus Sulfolobus. Geomicrobiol J 35:757–767
Mertens B, Blothe C, Windey K, De Windt W, Verstraete W (2007) Biocatalytic dechlorination of lindane by nano-scale particles of Pd(0) deposited on Shewanella oneidensis. Chemosphere 66:99–105
Mikheenko IP, Rousset M, Dementin S, Macaskie LE (2008) Bioaccumulation of palladium by Desulfovibrio fructosivorans wild-type and hydrogenase-deficient strains. Appl Environ Microbiol 74:6144–6146
Mizuno K, Miyatani G (1976) Successive spectrophotometric determination of palladium and platinum. Bull Chem Soc Jpn 49:2479–2480
Nalajala N, Chakraborty A, Bera B, Neergat M (2016) Chloride (Cl−) ion-mediated shape control of palladium nanoparticles. Nanotechnology 27:065603
Noroozifar M, Khorasani-Motlagh M (2003) Specific extraction of chromium as tetrabutylammonium-chromate and spectrophotometric determination by diphenylcarbazide: speciation of chromium in effluent streams. Anal Sci 19:705–708
Okibe N, Nakayama D, Matsumoto T (2017) Palladium bionanoparticles production from acidic Pd(II) solutions and spent catalyst leachate using acidophilic Fe(III)-reducing bacteria. Extremophiles 21:1091–1100
Rizki IN, Okibe N (2018) Size-controlled production of gold bionanoparticles using the extremely acidophilic fe(iii)-reducing bacterium. Acidocella aromatica. Miner 8:3
Sinha P, Roy S, Das D (2015) Role of formate hydrogen lyase complex in hydrogen production in facultative anaerobes. Int J Hydrog Energy 40:8806–8815
Sleytr UB, Beveridge TJ (1999) Bacterial S-layers. Trends Microbiol 7:253–260
Suzuki T, Iwasaki T, Uzawa T, Hara K, Nemoto N, Kon T, Ueki T, Yamagishi A, Oshima T (2002) Sulfolobus tokodaii sp. nov. (f. Sulfolobus sp. strain 7), a new member of the genus Sulfolobus isolated from Beppu Hot Springs, Japan. Extremophiles 6:39–44
Yang G, Bauer TJ, Haller GL, Baráth E (2018) H-transfer reactions of internal alkenes with tertiary amines as H-donors on carbon supported noble metals. Org Biomol Chem 16:1172–1177
Yates MD, Cusick RD, Logan BE (2013) Extracellular palladium nanoparticle production using Geobacter sulfurreducens. Acs Sustain Chem Eng 1:1165–1171
Yong P, Rowson NA, Farr JP, Harris IR, Macaskie LE (2002) Bioreduction and biocrystallization of palladium by Desulfovibrio desulfuricans NCIMB 8307. Biotechnol Bioeng 80:369–379
Zhang XL, Yan S, Tyagi RD, Surampalli RY (2011) Synthesis of nanoparticles by microorganisms and their application in enhancing microbiological reaction rates. Chemosphere 82:489–494
Zhang R, Neu TR, Zhang Y, Bellenberg S, Kuhlicke U, Li Q, Sand W, Vera M (2015) Visualization and analysis of EPS glycoconjugates of the thermoacidophilic archaeon Sulfolobus metallicus. Appl Microbiol Biotechnol 99:7343–7356
Zhao L, Jiang D, Cai Y, Ji X, Xie R, Yang W (2012) Tuning the size of gold nanoparticles in the citrate reduction by chloride ions. Nanoscale 4:5071
Acknowledgement
This work was partly supported by a grant from the Japan Society for the Promotion of Science (JSPS Kakenhi No. 26820394). We are grateful to Dr Yumi Fukunaga at the Ultramicroscopy Research Center, Kyushu University, for her support in TEM analysis. S.K. is grateful for financial assistance provided by the Kyushu University Advanced Graduated Program in Global Strategy for Green Asia.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by G. Antranikian.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (PDF 1379 kb). SEM image of Pd(0) precipitates formed in cell-free controls
792_2019_1106_MOESM3_ESM.pdf
Supplementary material 3 (PDF 247 kb). Comparison of the catalytic activity (based on the specific Cr(VI) reduction rate) of Pd(0) bionanoparticles (a–h) and commercial Pd/C catalyst (i). Pd(0) bionanoparticles were produced by S. tokodaii in the presence of either 5 mM formate (a–d) or 10 mM formate (e–h), under different conditions: (a) active cells, (b) active cells + Cu2+, (c) heat-killed cells, (d) cell-free controls, (e) active cells, (f) active cells + 10 mM Cl−, (g) active cells + 50 mM Cl−, and (h) active cells + 100 mM Cl−. The specific Cr(IV) reduction rates in Fig. 8 were calculated for the time interval of 0–5 h (a), 0–30 h (b), 0–30 h (c), 0–48 h (d), 0–10 h (e), 0–9 h (f), 0–1 h (g), 0–4 h (h), and 0–4 h (i). As e-donor for Cr(VI) reduction, 10 mM formate was used in all cases
Rights and permissions
About this article
Cite this article
Kitjanukit, S., Sasaki, K. & Okibe, N. Production of highly catalytic, archaeal Pd(0) bionanoparticles using Sulfolobus tokodaii. Extremophiles 23, 549–556 (2019). https://doi.org/10.1007/s00792-019-01106-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-019-01106-7