Abstract
The bacterial diversity of two soil samples collected from the periphery of the Roopkund glacial lake and one soil sample from the surface of the Roopkund Glacier in the Himalayan ranges was determined by constructing three 16S rRNA gene clone libraries. The three clone libraries yielded a total of 798 clones belonging to 25 classes. Actinobacteria was the most predominant class (>10% of the clones) in the three libraries. In the library from the glacial soil, class Betaproteobacteria (24.2%) was the most predominant. The rarefaction analysis indicated coverage of 43.4 and 41.2% in the samples collected from the periphery of the lake thus indicating a limited bacterial diversity covered; at the same time, the coverage of 98.4% in the glacier sample indicated most of the diversity was covered. Further, the bacterial diversity in the Roopkund glacier soil was low, but was comparable with the bacterial diversity of a few other glaciers. The results of principal component analysis based on the 16S rRNA gene clone library data, percentages of OTUs and biogeochemical data revealed that the lake soil samples were different from the glacier soil sample and the biogeochemical properties affected the diversity of microbial communities in the soil samples.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Studies on bacterial diversity, based on the culture-independent 16S rRNA approach of water, soil, ice etc., of cold habitats such as Arctic (Bottos et al. 2008; Perreault et al. 2007; Kirchman et al. 2003), Antarctica (Yergeau et al. 2007; Niederberger et al. 2008; Stackebrandt et al. 2004; Tindall 2004; Bowman and McCuaig 2003; Nealson 1999; López-García et al. 2001; Brambilla et al. 2001; Bowman et al. 2000; Shivaji et al. 2004; Karr et al. 2003, 2006; Smith et al. 2006; Li et al. 2006; Prabagaran et al. 2007; Mikucki and Priscu 2007; Sjöling and Cowan 2003) and Himalayan glaciers (Xiang et al. 2005; Liu et al. 2006, 2009; Zhang et al. 2006, 2008; Gangwar et al. 2009) have established that the culture-independent approach reveals a greater degree of microbial diversity compared to the culture-dependent approach. The 16S rRNA gene clone libraries of cold regions like tundra soils, sub-glacial samples and permafrost samples revealed many sequences that were related to the Proteobacteria, Acidobacteria, Actinobacteria, Firmicutes, Cytophagae–Flavobacteriae–Bacteroides (CFB group), Planctomyces and Gemmatimonadetes (Cheng and Foght 2007; Neufeld et al. 2004; Steven et al. 2007; Carpenter et al. 2000; Amato et al. 2007; Gangwar et al. 2009). The Himalayas also has several distinct habitats such as glacial ice, permafrost, tundra wetlands, tundra soil, sub-glacial soil, peri-glacial soil and peri-glacial lakes. Earlier studies have reported bacterial diversity from sub-glacial samples and surface soil samples from the Himalayas (Liu et al. 2006, 2009), but the bacterial diversity of lakes formed due to glacial retreat water at high altitudes is lacking. In the current study, culture-independent approach was used to establish the bacterial diversity of soil samples collected from the Roopkund glacier and periphery of Roopkund glacial lake located in the Himalayan mountain ranges, India.
Materials and methods
Sampling site and sample collection
Roopkund glacier is located near the base of two Himalayan peaks namely Trisul (7120 m) and Nandghungti (6310 m), respectively, at an altitude of 5029 m. The glacial melt water pours into a shallow lake, the ‘Roopkund lake’ or the ‘Skeleton lake’, which is about 2 m deep. About 5 g of ice-free soil samples (RKS1 and RKS6) were collected in the month of September 2005 from about 1 m from the periphery of the Roopkund glacial lake (Fig. 1). In addition, another soil sample, RKS7, was also collected at the same time from the Roopkund glacier. Prior to collection, 1 cm of the surface soil was removed with a sterile spatula and using another sterile spatula the soil was collected, transferred into sterile polythene bags, transported to the laboratory under sterile conditions and stored at −20°C until use. The air temperature was 2°C at the time of sample collection.
Soil analysis
The physical and chemical characteristics of RKS1, RKS6 and RKS7 were analysed. Soil samples were thawed, dried, ground and allowed to pass through a 2-mm sieve. Soil pH (Rhoades 1982), water content (Blakemore et al. 1987) and the chemical characteristics (Jackson 1967) were determined using standard methods. Organic matter was determined in terms of organic carbon according to the method of Walkley and Black (1934). Further, the available/mineralizable Nitrogen (N) was determined by the method of Subbiah and Asija (1956), and the total element content was determined in soil after acid digestion. Total Phosphorous (P) was estimated spectrophotometrically by the ammonium vanadomolabdate method of Bray and Kurtz (1945). Macronutrients (Na, K, Ca and Mg) and micronutrients (Zn, Cu, Fe and Mn) were determined using an atomic absorption spectrophotometer (Perkin Elmer Aanalyst 100 model). Each sample was analysed in duplicate. Sulphate in the soil was determined by extracting soil with conc. H2SO4 and conc. HNO3 followed by analysis on ICP OR AAS (APHA 1995).
Bacterial count
Total bacterial count in the sediment samples was determined by epifluorescence using BacLight™ Bacterial Viability Kit (Invitrogen, Oregon, USA) as per the instructions given in the kit. Bacteria were counted in a Petroff-Hausser Counter using a fluorescent microscope (Axioplan 2, Zeiss, Germany).
Extraction of total DNA from soil and PCR amplification of the 16S rRNA gene
Total DNA was isolated from the soil samples essentially according to the methods described earlier (Shivaji et al. 2004; Tsai and Olson 1991, 1992). Primers 16S3 (5′-TCC TAC GGG AGG CAG CAG-3′) and 16S4 (5′-GGC GGT GTG TAC AAG GCC C-3′) corresponding to positions 339–356 and 1402–1384, respectively, of the Escherichia coli 16S rRNA gene (Lane, 1991) were used to amplify about 1.0 kb fragment of the total 1.5 kb 16S rRNA gene. Amplification was done as described earlier (Reddy et al. 2000; Shivaji et al. 2004). The PCR amplicon was electrophoresed on a 1.0% agarose gel and visualized following staining with ethidium bromide (1 μg/ml). The PCR product was purified with the Quiaquick PCR purification kit (Qiagen Inc, Chatsworth, USA) according to the instructions provided.
Cloning and library construction of soil 16S rRNA gene sequences
The purified PCR product obtained earlier was cloned into pMOS Blue Blunt End vector system (Amersham Biosciences, New Jersey, USA) following the instructions of the manual. Transformants were selected on a LB agar plate containing 20 μg/ml X-gal and 60-μg/ml ampicillin and incubated at 37°C overnight. Clones were maintained on LB agar plates containing X-gal and ampicillin.
PCR amplification of the 16S rRNA gene, sequencing and phylogenetic analysis
The 16S rRNA gene was amplified from the white colonies by colony PCR using the vector-targeted M13 forward (5′-GTAAAACGACGGCCAGT-3′) and M13 reverse (5′-GGAAACAGCTATGACCATG-3′) primers, respectively (Amersham Biosciences, New Jersey, USA). The amplified product was checked on 1% agarose gel for quality and then subjected to treatment with 1 μl of ExoSAP-IT (USB Corporation, USA) at 37°C for 15 min followed by incubation at 80°C. The ExoSAP-IT treated amplicon was sequenced using the primers M13 forward, M13 reverse, pD (5′-CAGCAGCCGCGGTAATAC-3′) and pF* (5′-ACGAGCTGACGACAGCCATG-3′) (Reddy et al. 2000). The sequences obtained were trimmed for vector and chimeric sequences using Gene Tool version 2 (http://www.biotools.com) and then aligned using the Autoassembler software (Macintosh software package from PE-Applied Bio-systems, Foster City, CA, USA). Approximately 1050-bp-long sequence obtained for all the clones was subjected to BLAST sequence similarity search (Altschul et al. 1990) and EzTaxon (Chun et al. 2007) to identify the nearest taxa. All the sequences were aligned with sequences, belonging to the nearest taxa, downloaded from the database (http://www.ncbi.nlm.nih.gov) using CLUSTAL W (Thompson et al. 1997). Pair-wise evolutionary distances were computed using the DNADIST programme with the Kimura 2-parameter model as developed by Kimura (1980). Phylogenetic and molecular evolutionary analyses were conducted using Neighbour-Joining option of MEGA version 4 software package (Tamura et al. 2007). Bootstrap analysis was performed employing 1000 replicate data sets in order to assess stability among the clades recovered in the phylogenetic tree.
Statistical analyses
In order to compare the bacterial diversity within the three samples, 16S rRNA gene sequences showing ≥97% sequence similarity were grouped into the same OTU (phylotype). Shannon–Wiener Diversity Index (http://www.changbioscience.com/genetics/shannon.html) was used to calculate Shannon index (H′), Evenness (J′) (Lefauconnier et al. 1994) and the Simpson’s index (D) (Magurran 1996). Rarefaction analysis was done using the site Online Calculation (http://biome.sdsu.edu/fastgroup/cal_tools.htm). Coverage of 16S rRNA gene clone libraries was calculated as described previously (Good 1954). Rarefaction curves were generated to compare the relative diversity and coverage of each.
The three 16S rRNA gene clone libraries (RUGL1, RUGL6 and RUPGL7) were compared using a JAVA based software Comm Cluster (Hur and Chun 2004) using three different cut-off values (97, 90 and 80%) and an ordination analysis was performed using principal component analysis (PCA). PCA was performed using the SPSS statistical computing package (version 16.0; SPSS, Inc., Chicago, IL, USA) and was employed to group or separate samples based on the biogeochemical parameters [altitude, total bacterial count, water content, pH value, organic carbon, organic matter, mineralisable Nitrogen (N), NO3 −, SO4 2−, total Phosphorous (P), Mg, Zn, Mn, Fe and Ca] and the percentages of OTUs in each sample. PCA analysis was also performed to compare the present 16S rRNA gene clone libraries with the earlier published data of clone libraries from different glaciers (refer to Table 4).
Nucleotide sequence accession numbers
All the sequences of the 16S rRNA gene clone library were deposited in GenBank with accession numbers GQ420851–GQ421153, GQ366384–GQ366653, GQ366655–GQ366699, EU852102–EU852155, EU852157–EU852260, EU852262–EU852266, EU852268–EU852274 and EU852276–EU852289.
Results
Viable bacterial count and the physico-chemical characteristics of the soil samples from Roopkund lake and Roopkund glacier
The total bacterial count in the soil samples (RKS1, RKS6 and RKS7) from the Roopkund lake and the glacier varied from 0.9 to 4.2 × 107 bacteria g−1 of soil. The pH and water content of the three soil samples were similar (Table 1). But, the organic carbon levels varied with very low content in the glacial soil sample RKS7 (0.29%) and high in the RKS1 (1.74%) and RKS6 (1.26%). The other chemical characteristics like mineralisable N, NO3 −, SO4 2−, total P, Mg, Zn, Mn and Fe content in the glacial lake samples (RKS1 and RKS6) were similar, but differed from RKS7, the glacial soil sample (Table 1).
Bacterial diversity from Roopkund lake and Roopkund glacier
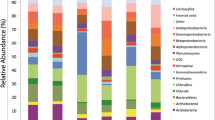
In the present study, two soil samples collected from the vicinity of the Roopkund Glacial Lake (RKS1 and RKS6) and one from the glacier (RKS7) were analysed for bacterial diversity. The soil samples yielded about 30–80 μg DNA g−1 of soil. About 200 ng of the DNA was used for constructing a 16S rRNA gene library. The libraries constructed from RKS1, RKS6 and RKS7 were designated RUGL1, RUGL6 and RUPGL7, respectively, and consisted of 302, 311 and 185 clones, respectively, with an insert size of approximately 1 kb. BLAST sequence similarity analysis of the clone libraries RUGL1 and RUGL6 indicated that clones belonging to the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Chlorobi, Chloroflexi, Firmicutes, Gemmatimonadetes, Nitrospira, Proteobacteria, Verrucomicrobia and TM7_s TM7a, respectively, were present in both the libraries (Table 2, Supplementary Table 1). In contrast to these two libraries, the 186 clones in RUPGL7 could be assigned to only two higher taxa namely Actinobacteria and Firmicutes (Table 2, Supplementary Table 1).
Frequency of distribution of the clones indicated that in the two libraries (RUGL1 and RUGL6) derived from the Roopkund glacial lake soil, clones belonging to phyla Acidobacteria (17.2 and 13.0%), Actinobacteria (10.6 and 32.8%), CFB group (10.6 and 6.4%), Chloroflexi (8.6 and 12.2%) and Proteobacteria (42.7 and 23.2%) were the most predominant (representing >10%) of the total clones. In contrast, in RUPGL7 clones assigned to Actinobacteria (86.5%) and Firmicutes (13.5%) were predominant (Tables 2, 4, Supplementary Table 1). The affiliation of each and every clone to the nearest phylogenetic neighbour based on 16S rRNA gene sequence is also presented in Supplementary Table 1.
Proteobacteria
In the 16S rRNA gene clone libraries of Roopkund glacial lake soils (RUGL1 and RUGL6) (Table 2, Supplementary Table 1) clones affiliated to all the five classes of the phylum Proteobacteria namely Alpha-, Beta-, Gama-, Delta- and Epsilonproteobaceria (Epsilonproteobaceria was absent in RUGL6 library) were present (Table 2, Supplementary Table 1). Phylogenetic analysis also confirmed the affiliation of the clone sequences to the five classes of phylum Proteobacteria (Figs. 2a, 3a). All the clones clustered with their related sequences (Figs. 2a, 3a) except RUGL1-52, RUGL1-298, RUGL1-415 and RUGL1-612 clones showed >90% pairwise sequence similarity based on BLAST with Geobacter lovleyi SZT (Deltaproteobacteria), Desulfuromonas thiophila DSM 8987T (Deltaproteobacteria), Thiobacillus sp. DDR2W1u61 (Betaproteobacteria) and Geoalkalibacter ferrihydriticus Z-0531T (Deltaproteobacteria) but did not cluster with their corresponding phylogenetic neighbours (Fig. 2a). Clones affiliated to Proteobacteria were absent in RUPGL7 library (Fig. 4).
Neighbour Joining phylogenetic tree of 16S rRNA gene clones from RUGL1, a library constructed using RKS1, a soil sample from the vicinity of Roopkund glacier lake, showing the phylogenetic relationship of clones affiliated to Proteobacteria (a) and clones affiliated to Acidobacteria, Actinobacteria, Bacilli, CFB, Chlorobea, Chloroflexi, Clostridia, Elusimicrobia, Gemmatimondetes, Holophagae, Nitrospira, Solibacteres, TM7_s TM7a and Verrucomicrobiae (b). In a the following clones have been compressed: 9, 56, 300, 321, 425, 559 and 567 as Clade-1; 3, 170, 333, 467, 471, 518 and 580 as Clade-2; 303, 334, 442, 507, 530, 537, 569, 577 and 605 as Clade-3; 57, 174, 178, 247, 296, 305 and 591 as Clade-4 and 253, 271, 275 and 297 as Clade-5. In b the following clones have been compressed 1, 29, 83, 95, 164, 276, 403, 416, 434, 461, 464 and 514 as Clade-1; 37, 251, 318, 344, 440, 468, 557 and 626 as Clade-2; 12, 114, 223, 339, 350, 382, 391, 410, 449, 493, 511 and 589 as Clade-3; 6, 59, 67, 167, 197, 201, 209, 385, 398, 413, 554 and 595 as Clade-4; 11, 28, 81, 397, 470 and 604 as Clade-5; 230, 363, 366, 380 and 572 as Clade-6; and 71, 196, 248, 483, 617 as Clade-7. Phylogenetic trees were constructed by Neighbour-Joining method. Numbers at the nodes are bootstrap values. The bar represents 0.05 and 0.02 substitutions per alignment position in a and b, respectively
Neighbour-Joining phylogenetic tree of 16S rRNA gene clones from RUGL6, a library constructed using RKS6, a soil sample from the vicinity of Roopkund glacier lake, showing the phylogenetic relationship of clones affiliated to Proteobacteria (a) and clones affiliated to Acidobacteria, Actinobacteria, CFB, Chloroflexi, Clostridia, Gemmatimondetes, Nitrospira, Planctomycetes, Solibacteres, TM7_s TM7a and Verrucomicrobiae (b). In b the following clones have been compressed 28, 54, 62, 72, 87, 117, 128, 182, 203, 247, 278, 372, 395, 399, 411, 412 and 472 as Clade-1; 34, 126, 222, 306, 373, 463, 466, 474 and 485 as Clade-2; 36, 68, 113, 120, 131, 141, 150, 152, 168, 175, 197, 265, 327, 360, 382, 394, 410, 413, 468, 470 and AM292622 as Clade-3; 97, 111, 112, 135, 195 and 489 as Clade-4; 1, 20, 92, 101, 102, 107, 109, 115, 205, 261, 264, 316, 351, 404, 414, 432, and 441 as Clade-5; 91, 291, 329, 348, 364 and 467 as Clade-6; 17, 29, 183, 225, 235, 244, 289, 303, 420 and AY571791 as Clade-7; 5, 8, 24, 40, 138, 153, 158, 174, 177, 184, 186, 188, 193, 215, 228, 241, 277, 319, 328, 330, 343, 384, 421, 436, 442, 444, 455, 457, 471 and EU192969 as Clade-8; 23, 25, 82, 83, 123, 196, 242, 255, 294, 314, 324 and 422 as Clade-9; 56, 78, 118, 132, 172, 226, 266, 283, 288, 302, 333, 337, 374, 428, 439, 453 and AM902609 as Clade-10; 98, 151, 176, 237, 243, 271, 313, 354, 371, 379, 401, 407 and 426 as Clade-11; 66, 116, 169, 202, 240, 254, 257, 349, 391, 469, 475 and Prolixibacter as Clade-12. Numbers at nodes are bootstrap values. The bar represents 0.02 substitutions per alignment position
Neighbour Joining phylogenetic tree of 16S rRNA gene clones from RUPGL7, a library constructed using RKS7, a soil sample from the Roopkund glacier) showing the phylogenetic relationship of clones with their nearest phylogenetic relatives. In this library consisting of 185 clones a majority of the clones (160) were phylogenetically close to Cryobacterium roopkundense (93–100%), and the remaining (25 clones) were close to Bacillus firmus (95–99%) and Bacillus foraminis (96.9%). The tree was constructed using 15, 16, 19, 88, 135, 219, 351, 24, 208, 216, 266, 381 and 327 clones of Cryobacterium roopkundense, Bacillus firmus and Bacillus foraminis, respectively. Numbers at nodes are bootstrap values. The bar represents 0.02 substitutions per alignment position
Actinobacteria
Clones affiliated to Actinobacteria were present in libraries RUGL1, RUGL6 and RUPGL7 (32, 102 and 160 clones), and they shared a 16S rRNA gene sequence similarity of 83–100% with the nearest phylogenetic neighbour (Table 2, Supplementary Table 1). In RUGL1 library, all the clones related to Actinobacteria formed two major clades (Fig. 2b) along with the other reported strains of Actinobacteria. In RUGL6 library also all the clone sequences related to Actinobacteria clustered with the Actinobacterial clade except one sequence RUGL6-301 which clustered with Solobacteres (Fig. 3b). In RUPGL7 library all the clones phylogenetically related to Cryobacterium roopkundense RuGl_7T (which was earlier isolated from the same habitat (Reddy et al. 2009) clustered with Cryobacterium roopkundense (In Fig. 4 only 7 sequences out of 160 clone sequences were taken for the tree construction) (Fig. 4).
Acidobacteria
In RUGL1 phylum Acidobacteria was represented by 17.2% of the total clones in the library and were affiliated to four genera Acidobacterium, Edaphobacter, Geothrix and Solibacter. These clones related to the class Acidobacteria formed three branches and clustered with the branch of the class Halopahagae (Fig. 2b). Further, the clones related to Solibacter formed two branches along with Solibacter usitatus Ellin6076 and clustered with the branch of Clostridia (Fig. 2b). In RUGL6, 13% of the clones belonged to phylum Acidobacteria, and they formed two clades (10 and 11 clones) and branched from Verrucomicrobiae and Planctomycetacia, respectively (Fig. 3b). It is interesting that two clones related to Solibacteres clustered with uncultured Solibacter sp. AMIB8 (Fig. 3b). Clones related to Acidobacteria were not present in the RUPGL7 library.
Bacteroidetes
Clones similar to the phylum Bacteroidetes constituted 10.6 and 6.4% of the total clones in RUGL1 and RUGL6 libraries, respectively (Table 2, Supplementary Table 1) and shared a 16S rRNA gene sequence similarity of 85–99 and 83–99%, respectively, with the nearest phylogenetic neighbour (Supplementary Table 1). All clone sequences related to Bacteroidetes clustered within the Bacteroidetes clade which appeared to be polyphyletic (Figs. 2b, 3b). Clones related to Bacteroidetes were not present in the RUPGL7 library.
Chloroflexi
Phylum Chloroflexi was represented by 9 and 12.1% of the total clones in RUGL1 and RUGL6 libraries (Table 2, Supplementary Table 1), respectively, and formed a single clade in both RUGL1 and RUGL6 libraries (Figs. 2b, 3b). Clones related to Chloroflexi were not present in the RUPGL7 library.
Firmicutes and Elusimicrobia
Clones affiliated to phylum Firmicutes constituted only 1.7 and 1.3%, respectively, in RUGL1 and RUGL6 (Tables 2, 4, Supplementary Table 1). All the Firmicutes sequences were related to Clostridia and in the phylogenetic trees they formed two and three branches, respectively, in RUGL1 and RUGL6 libraries (Figs. 2b, 3b). One clone RUGL1-448 showing 83% pairwise sequence similarity with Thermacetogenium phaeum PBT belonging to the class Bacilli clustered with RUGL1-632 which is related to Elusimicrobium minutum Pei191 (Fig. 2b). In RUPGL7 13.5% of all clones (Tables 2, 4, Supplementary Table 1) are phylogenetically close to Bacillus firmus AU9 (Fig. 4). (In Fig. 4 only five sequences out of 24 clone sequences were taken for the tree construction). One clone sequence clustered with Bacillus foraminis CV53T which is its phylogenetic relative (Fig. 4).
Chlorobi, Gemmatimonadetes, Nitrospira, Planctomycetes and Verrucomicrobia
BLAST analysis indicated that clones affiliated to phylum Chlorobi (5 and 1 clones in RUGL1 and RUGL6 libraries), Gemmatimonadetes (8 and 5 clones in RUGL1 and 6 libraries), Nitrospira (6 and 8 clones in RUGL1 and 6 libraries), Planctomycetes (only two clones in the RUGL6 library), Verrucomicrobia (4 and 3 clones in RUGL1 and 6 libraries) (Table 2, Supplementary Table 1) shared a 16S rRNA gene sequence similarity with the nearest phylogenetic neighbours (Supplementary Table 1). Further, all the clone sequences clustered with the respective phylogenetic neighbour except RUGL1-266 which branched from the Solibacteres, RUGL1-400 which clustered with Chloroflexi and RUGL1-404, which clustered with RUGL1-448 (related to Bacilli) (Figs. 2b, 3b). Clone sequences related to Chlorobi branched from the CFB group in both libraries and clone sequences related to the phyla Gemmatimonadetes and Nitrospira formed separate branches and clustered together (Figs. 2b, 3b). Clones related to above five phyla were not present in the RUPGL7 library.
TM7_s TM7a
Few of the clones were related to the candidate phylum TM7_s TM7a (2 and 16 clones in RUGL1 and 6 libraries, respectively) (Table 2, Supplementary Table 1) and they shared a 16S rRNA gene sequence similarity of 80–93% and clustered together (Figs. 2b, 3b). Clones related to phylum TM7_s TM7a were not present in the RUPGL7 library.
In the present study, unculturable bacteria were also identified in various phyla in RUGL1 and RUGL6 libraries (Supplementary Table 1), and the maximum number was seen in phylum Acidobacteria (9.9 and 12% in RUGL1 and 6 libraries, respectively).
Statistical analysis
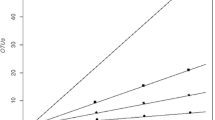
The three libraries from the three soil samples (RKS1, RKS6 and RKS7) are represented by 302, 311 and 185 clones, respectively, of 16S rRNA gene. These clones based on 16S rRNA gene sequence similarity criteria of >97% could be categorised into 171, 183 and 3 phylotypes (Fig. 5; Table 3, Supplementary Table 1). The most common phylotype was Cryobacterium psychrophilum, which was shared by all the three libraries (Supplementary Table 1). Species richness varied from 3 to 183 and the diversity coverage ranged from 41.2 to 98.4% (Table 3). The rarefaction curves indicated that the bacterial populations in both the samples from Roopkund lake did not plateau whereas the soil sample from Roopkund glacier, with diversity coverage of 98.4% plateaued (Fig. 5). The rarefaction curve analysis implied that these are likely to be minimal estimates of diversity. These observations are supported by bacterial diversity parameters, such as Shannon index, Simpson’s index, Coverage, Chao1 and Evenness (Table 3).
Rarefaction curves of observed OTUs in the two soil samples collected in the vicinity of Roopkund glacier (RKS1 and RKS6) and a soil sample collected from the Roopkund glacier (RKS7). The symbols represent the rarefaction curves generated for the clone libraries from the three samples RKS1 (filled circle), RKS6 (filled triangle) and RKS7 (minus) respectively
The sampling sites RKS1 and RKS6 which were close to each other and located on the vicinity of the Roopkund glacial lake were found to be closely associated in the ordination diagram based on the first two principal components (Fig. 6). It was also evident from the clone library sequences that samples from the vicinity of the Roopkund lake RKS1 and RKS6 clustered together at the cut-off values of (97, 90 and 80%) (Fig. 6a, b, c), whereas the sample collected from the Roopkund glacier (RKS7) differed from the lake samples.
Ordination diagrams based on principal component analysis using frequency tables obtained from the three 16S rDNA clone libraries (RUGL1, RUGL6 and RUPGL7) defined at different cutoff similarity values of 97% (a), 90% (b) and 80% (c) using Comm Cluster Software. PC 1 principal component analysis factor 1, PC 2 principal component analysis factor 2
The result of PCA based on percentage of a specific OTU (Phyla) in the three libraries (RUGL1, RUGL6 and RUPGL7) is shown in Fig. 7a, and it appears that the principal component factors 1 and 2 (PC1-74.084% and PC2-25.216%) explain 99.3% of the total variances. From the PCA plot considerable differences in the distribution of phyla in the three soil samples has been observed. The phyla Actinobacteria and Firmicutes which are dominant in RUPGL7 library grouped closely and were separated from the other phyla present only in RUGL1 and RUGL6 libraries. The result of PCA based on the biogeochemical properties is shown in Fig. 7b and the principal component factors 1 and 2 (PC1-87.107% and PC2-12.893%) explained 100% of the total variances. The PCA plot indicates that altitude and sulphate content which are higher in the glacier sample (RKS7) grouped closely. Further, when all the other parameters were checked it was observed that except for water content, nitrate and calcium the remaining parameters appeared to be associated. Thus, it may imply that altitude, sulphate, nitrate, calcium and water content are the key parameters that influence the observed differences in the percentage of specific OTUs in the three 16S rRNA gene clone libraries.
a Principal component analysis based on percentage of specific OTUs from three 16S rRNA gene clone libraries (PC1-74.084% and PC2-25.216%). b PCA plot based on biogeochemical properties (altitude, total bacterial count, water content, pH value, organic carbon, organic matter, N, NO3−, SO42−, total P, Mg, Zn, Mn, Fe and Ca) of three soil samples (PC1-87.107% and PC2-12.893%). c PCA plot based on percentage of specific OTUs from 11 16S rRNA gene clone libraries (PC1-72.962% and PC2-16.544%). The data for eight of the libraries were taken from published literature (See footnote to Table 4). PC 1 principal component analysis factor 1; PC 2 principal component analysis factor 2; TPG Tibetan Plateau Glaciers, MLME Moraine lakes, Mount Everest; GMWME Glacial meltwater, Mount Everest; JEGC John Evans glacier, Canada; BGA Bench Glacier, Alaska; SOA Schirmacher Oasis, Antarctica; ST Siberian tundra; SIS Samoylov Island, Siberia
Principal component analysis based on percentage of specific OTUs (Phyla) in three samples from the present study and eight samples from the published literature (RKS1; RKS6; RKS7; Tibetan Plateau Glaciers, moraine lakes, Mount Everest; glacial meltwater, Mount Everest; John Evans glacier, Canada; Bench Glacier, Alaska; Schirmacher Oasis, Antarctica; Siberian tundra; Samoylov Island, Siberia) are shown in Fig. 7c. The principal component factors 1 and 2 (PC1-72.962% and PC2-16.544%) explain 89.506% of the total variances. The PCA plot indicated that RKS7 from the Roopkund glacier clustered separately from all the other samples; RKS6 and Samoylov Island sample clustered together and all the other samples including RKS1 grouped closely. This analysis also suggested that the bacterial diversity in the Roopkund glacier is different from all the other samples studied.
Discussion
In the present study, two soil samples collected from the vicinity of the Roopkund Glacial Lake (RKS1 and RKS6) and one from the glacier (RKS7) were analysed for bacterial diversity. The results indicated a low bacterial count and decreased species richness in the glacial soil (RKS7) compared to the glacial lake soils (RKS1 and RKS6) and this may be due to the decreased organic carbon %, organic matter %, mineralisable N and total P in glacial soil samples compared to the lake soil samples (Table 1). This observation is in accordance with earlier studies which had demonstrated that factors such as organic carbon content (LaMontagne et al. 2003; Zhou et al. 2002; Fierer et al. 2007) and nutrient availability (Fierer et al. 2003), influence microbial community composition and diversity. The high carbon in the lake soils might also be due to the decomposition of human carcasses indicated by the presence of skeletons present near the lake and also the availability of water for the microbial metabolic activities (Bill 1994).
The results obtained by PCA based on the 16S rRNA gene clone library data, percentages of OTUs and biogeochemical properties indicated that the three samples (RKS1, RKS6 and RKS7) and their microbial communities were different, but the samples from the lake (RKS1 and RKS6) are similar compared to the sample from glacier (RKS7). The results of PCA also suggested that biogeochemical properties such as altitude, sulphate, nitrate, calcium and water content may influence the heterogeneity observed in the three samples with respect to the specific OTUs. High altitude, sulphate content and soil texture might influence the decreased diversity observed in the glacial soil sample. However, in an earlier study Zhang et al. (2008) demonstrated that the bacterial diversity was not related to Ca2+ concentration but factors such as pH (Eichorst et al. 2007), and water content (Treves et al. 2003) influence microbial community composition and diversity. In the present study also water content appears to be a key parameter that influences the observed differences in the percentage of specific OTUs in the glacial soil compared to the lake soil 16S rRNA gene clone libraries. Earlier studies have indicated that coarse sediments are less favourable than fine sediment as a microbial substrate (Certini et al. 2004; Kastovská et al. 2005), and this indeed may be the reason for the decreased diversity observed in the glacial soil compared to the lake soil since the texture of the soil from the glacier was coarse (sandy), moist, yellowish-brown and had small stones whereas the soil from the lake was fine, moist, blackish, silty, but also had small stones. An overall comparison indicated that the physical and chemical properties of the glacial soil sample and lake soil samples determined in this study are related to other glacial habitats (Liu et al. 2006; Mindl et al. 2007; Foght et al. 2004; Zhang et al. 2008). For instance, the pH, the water content (Foght et al. 2004), organic carbon content (Liu et al. 2006; Mindl et al. 2007; Foght et al. 2004), nitrogen content (Foght et al. 2004), phosphorous (Mindl et al. 2007) and bacterial count (Mindl et al. 2007) were comparable with the earlier studies from cold habitats.
BLAST sequence similarity analysis of the clone libraries RUGL1 and RUGL6 constructed using the soil samples from the lakes indicated that clones belonging to the phyla Acidobacteria, Actinobacteria, Bacteroidetes, Chlorobi, Chloroflexi, Firmicutes, Gemmatimonadetes, Nitrospira, Proteobacteria, Verrucomicrobia and TM7_s TM7a were present in both the libraries (Table 2, Supplementary Table 1) except clones belonging to phylum Planctomycetes and Elusimicrobia which were present only in one of the libraries and constituted a very small fraction of the total library (<1%), indicating that the variation may due to variation in recovery of the specific DNA in the two samples. It is not surprising that the two libraries are very similar with respect to the various taxa present since both the soil samples are from the vicinity of the same lake and the physico-chemical characteristics are also very similar (Table 1). In contrast to the aforementioned two libraries, library RUPGL7 (consisting of 185 clones) from the glacial soil, which is a harsh habitat compared to the soil samples collected from the periphery of the glacial lake could be assigned to only two higher taxa, namely Actinobacteria and Firmicutes (Table 2, Supplementary Table 1). Xiang et al. (2004) reported only two phyla Proteobacteria and Cytophaga–Flavobacterium–Bacteroides from Malan ice core drilled from the Tibetan Plateau, which is a cold desert like the present site.
Frequency of distribution of the clones indicated that in the two libraries (RUGL1 and RUGL6) derived from the Roopkund glacial lake soil, clones belonging to phyla Acidobacteria (17.2 and 13.0%), Actinobacteria (10.6 and 32.8%), CFB group (10.6 and 6.4%), Chloroflexi (8.6 and 12.2%) and Proteobacteria (42.7 and 23.2%) were the most predominant (representing >10%) of the total clones (Supplementary Table 1). All these phyla have been reported earlier in different cold habitats of John Evans glacier, Canada (Cheng and Foght 2007); Tibetan plateau glacier (Liu et al. 2009); Samoylov island, Siberia (Wagner et al. 2009) and Schirmacher oasis soil, Antarctica (Shivaji et al. 2004). However, in the Bench glacier, Alaska, only clones belonging to phyla Acidobacteria (1.1%), CFB group (1.1%), Proteobacteria (96.8%) and Spirochaetes (1.1%) were present (Skidmore et al. 2005) (Table 4). In contrast, in RUPGL7 clones assigned to Actinobacteria (86.5%) and Firmicutes (13.5%) were predominant (Tables 2, 4, Supplementary Table 1), thus indicating a decrease in bacterial diversity in the glacial soil.
Phylum Proteobacteria is the most dominating community in the RUGL1 and RUGL6 libraries (129 and 72 clones) constructed using Roopkund glacial lake soil (Supplementary Table 1). Earlier studies on the bacterial diversity of glaciers have also indicated predominance of Proteobacteria under cold conditions (Cheng and Foght 2007; Liu et al. 2006, 2009; Skidmore et al. 2005; Shivaji et al. 2004; Wagner et al. 2009; Zhou et al. 1997). As compared to Phylum Proteobacteria which was represented only in libraries RUGL1 and RUGL6 phylum Actinobacteria appears to be a dominating community in RUGL1, RUGL6 and RUPGL7. Majority of the Actinobacteria clones of RUGL1 and RUGL6 libraries (26 and 78) are related to different known strains which belong to 36 different genera (Supplementary Table 1). But, in RUPGL7 library, 160 clones are related to Cryobacterium psychrophilum (Suzuki et al. 1997) of the class Actinobacteria. Earlier studies on the bacterial diversity of glaciers have also indicated predominance of Actinobacteria (Burkert et al. 2003; Glöckner et al. 2000; Cheng and Foght 2007; Mosier et al. 2007; Liu et al. 2006; 2009; Wagner et al. 2009; Zhou et al. 1997). Clones with similarity to Phylum Acidobacteria were present in libraries of RUGL1 and RUGL6. In fact, in this phylum vast majority of the clones were similar to uncultured Acidobacterium (12%). Earlier studies on the bacterial diversity of glaciers have also indicated predominance of Acidobacteria (Cheng and Foght 2007; Skidmore et al. 2005; Shivaji et al. 2004; Liu et al. 2009; Wagner et al. 2009; Zhou et al. 1997).
In the phylum Bacteroidetes, the class Sphingobacteria represents highest number of clones (19 and 9) in RUGL1 and RUGL6 libraries (Supplementary Table 1), respectively, and the remaining clones belong to classes Bacteroidia and Flavobacteria, thus confirming earlier studies (Cheng and Foght 2007; Skidmore et al. 2005; Shivaji et al. 2004; Liu et al. 2006, 2009; Wagner et al. 2009; Zhou et al. 1997).
An interesting feature is that most of the clones in the phylum Chloroflexi identified with uncultured bacteria. This is a clear indication that in these cold habitats clones belonging to the phylum Chloroflexi are diverse and attempts are needed to culture these organisms. The presence of Chloroflexi in other cold habitats (Costello and Schmidt 2006; Li et al. 2008; Lysnes et al. 2004; Reed et al. 2006; Shivaji et al. 2004; Liu et al. 2006; 2009; Wagner et al. 2009) is also documented.
A phylogenetic analysis of the clones affiliated to phylum Firmicutes indicated a distinct difference between the clones in the soil from the glacial lake soil (RKS1 and RKS6) compared to glacial soil (RKS7). In the lake soil, though the frequency of clones belonging to Firmicutes was low the diversity was significantly higher with the clones showing similarity to genera Caldalkalibacillus, Clostridium, Desulfosporosinus and Syntrophobotulus, whereas all the clones in the glacial soil were affiliated only to the genus Bacillus (Supplementary Table 1). As of now, only one new species of Bacillus has been described from a Himalayan glacier (Reddy et al. 2008). The presence of Firmicutes in other cold habitats (Liu et al. 2009; Wagner et al. 2009; Zhou et al. 1997) is also documented.
Clones belonging to many other phyla such as Chlorobi, Verrucomicrobia, Planctomycetes, Nitrospira and Gemmatimonadetes were present in at least two of the three libraries, but the frequency of distribution was low (<3%). This observation is in agreement with earlier studies which had also indicated that clones affiliated to Chlorobi (Zhou et al. 1997), Verrucomicrobia (Cheng and Foght 2007; Liu et al. 2006, 2009; Wagner et al. 2009; Zhou et al. 1997), Planctomycetes (Cheng and Foght 2007; Liu et al. 2006, 2009; Zhou et al. 1997), Nitrospira (Shivaji et al. 2004) and Gemmatimonadetes (Shivaji et al. 2004; Steven et al. 2007) are present in other cold habitats (Table 4) at a low frequency. Phylum Elusimicrobia is represented only by a single clone in the RUGL1 library, and reports on the presence of these bacteria in cold habitats is lacking. Apart from all the above-known phyla, few clones were related to the candidate phylum TM7_s TM7a (Marcy et al. 2007). The presence of the candidate phylum TM7 was reported from Tibetan plateau glacier sample but at a very low frequency (0.1%). The presence of these clones in Roopkund lake soil may be justified by the presence of human skeletons at the edge of the skeleton lake (Bill 1994) since these were first reported from the human oral cavity (Marcy et al. 2007).
This study demonstrates that the diversity of bacteria in two soil samples obtained from the vicinity of the Roopkund glacial lake and a sample from the Roopkund glacier is comparable with that reported from other cold habitats such as glacial soil, oasis soil and tundra soil samples (Table 4). For example, members of Actinobacteria, Gammaproteobacteria, Betaproteobacteria and Acidobacteria have been reported from Tibetan plateau glacier, China; Mount Everest, Nepal; John Evans glacier, Canada; Bench Glacier, Alaska; Schirmacher Oasis soil, Antarctica and Siberian tundra soil samples (Table 4). The presence of Bacillus is reported for the first time from a glacier, though an isolate was also reported from the Pindari glacier of Himalayas earlier (Reddy et al. 2008). Bacillus and Cryobacterium were also found in Guliya and Vostok (Antarctica) ice core samples (Christner et al. 2001, 2003). The occurrence of related phylotypes in geographically diverse cold environments is indicative of the ability of microorganisms to adapt similar strategies to survive freezing and remain active at low temperatures (Abyzov et al. 1998; Priscu and Christner 2003). Noticeably, neither enteric nor pathogenic bacteria were detected in Roopkund glacier though enteric bacteria have been isolated from glacial ice on Ellesmere Island (Dancer et al. 1997) and surface snow of Tateyama Mountains in Japan (Segawa et al. 2005).
Conclusions
Bacterial diversity of Roopkund glacier and the glacial lake based on 16S rRNA gene sequence analysis led to the identification of Actinobacteria, Bacilli, Acidobacteria and Proteobacteria as the major groups in two soil samples collected near Roopkund glacier. But, molecular sequences representing Proteobacteria could not be detected in the glacial soil. PCA indicated that biogeochemical properties such as altitude, sulphate, nitrate, calcium and water content might influence the heterogeneity observed in the three samples with respect to the specific OTUs.
References
Abyzov SS, Mitskevich IN, Poglazova MN (1998) Microflora of the deep glacier horizons of central Antarctica. Microbiology 67:66–73
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Amato P, Hennebelle R, Magand O, Sancelme M, Delort AM, Barbante C, Boutron C, Ferrari C (2007) Bacterial characterization of the snow cover at Spitzberg, Svalbard. FEMS Microbiol Ecol 59:255–264
APHA (1995) Standard methods for the examination of water and wastewater, 19th edn. American Public Health Association, Washington, DC
Bill A (1994) The Nanda Devi affair, Penguin Books India, New Delhi. ISBN 0-14-024045-4
Blakemore LC, Searle PL, Daly BK (1987) Methods for chemical analysis of soils. NZ Soil Bureau Scientific Report 80
Bottos EM, Vincent CW, Greer WF, Whyte LG (2008) Prokaryotic diversity of arctic ice shelf microbial mats. Environ Microbiol 10:950–966
Bowman JP, McCuaig RD (2003) Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol 69:2463–2483
Bowman JP, Rea SM, McCammon SA, McMeekin TA (2000) Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ Microbiol 2:227–237
Brambilla E, Hippe H, Hagelstein A, Tindall BJ, Stackebrandt E (2001) 16S rDNA diversity of cultured and uncultured prokaryotes of a mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23–33
Bray RH, Kurtz LT (1945) Determination of total organic and available forms of phosphorus in soils. Soil Sci 59:39–45
Burkert U, Warnecke F, Babenzien D, Zwirnmann E, Pernthaler J (2003) Members of a readily enriched beta-proteobacterial clade are common in surface waters of a humic lake. Appl Environ Microbiol 69:6550–6559
Carpenter EJ, Lin S, Capone DG (2000) Bacterial activity in South Pole snow. Appl Environ Microbiol 66:4514–4517
Certini G, Campbell CD, Edwards AC (2004) Rock fragments in soil support a different microbial community from the fine earth. Soil Biol Biochem 36:1119–1128
Cheng SM, Foght JM (2007) Cultivation-independent and -dependent characterization of bacteria resident beneath John Evans Glacier. FEMS Microbiol Ecol 59:318–330
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2001) Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ Microbiol 3:570–577
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2003) Bacterial recovery from ancient glacial ice. Environ Microbiol 5:433–436
Chun J, Lee J-H, Jung Y, Kim M, Kim S, Kim BK, Lim YW (2007) EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int J Syst Evol Microbiol 57:2259–2261
Costello EK, Schmidt SK (2006) Microbial diversity in alpine tundra wet meadow soil: novel Chloroflexi from a cold, water-saturated environment. Environ Microbiol 8:1471–1486
Dancer SJ, Shears P, Platt DJ (1997) Isolation and characterization of coliforms from glacial ice and water in Canada’s High Arctic. J Appl Microbiol 82:597–609
Eichorst SA, Breznak JA, Schmidt TM (2007) Isolation and characterization of soil bacteria that define Terriglobus gen. nov. in the phylum Acidobacteria. Appl Environ Microbiol 73:2708–2717
Fierer N, Schimel JP, Holden PA (2003) Controls on microbial CO2 production: a comparison of surface and subsurface soil horizons. Glob Chang Biol 9:1322–1332
Fierer N, Bradford MA, Jackson RB (2007) Toward an ecological classification of soil bacteria. Ecology 88:1354–1364
Foght J, Aislabie J, Turner S, Brown CE, Ryburn J, Saul DJ, Lawson W (2004) Culturable bacteria in subglacial sediments and ice from two southern hemisphere. Glaciers Microbiol Ecol 47:329–340
Gangwar P, Alam SI, Bansod S, Singh L (2009) Bacterial diversity of soil samples from the western Himalayas, India. Can J Microbiol 55:564–577
Glöckner FO, Zaichikov E, Belkova N, Denissova L, Pernthaler J, Pernthaler A, Amann R (2000) Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl Environ Microbiol 66:5053–5065
Good IJ (1954) The population frequencies of species and the estimation of population parameters. Biometrica 40:237–264
Hur I, Chun J (2004) A method for comparing multiple bacterial community structures from 16S rDNA clone library sequences. J Microbiol 42:9–13
Jackson ML (1967) Soil chemical analysis. Prentice-Hall of India, New Delhi
Karr EA, Sattley WM, Jung DO, Madigan MT, Achenbach LA (2003) Remarkable diversity of phototrophic purple bacteria in a permanently frozen Antarctic lake. Appl Environ Microbiol 69:4910–4914
Karr EA, Ng JM, Belchik SM, Sattley WM, Madigan MT, Achenbach LA (2006) Biodiversity of methanogenic and other archaea in the permanently frozen Lake Fryxell, Antarctica. Appl Environ Microbiol 72:1663–1666
Kastovská K, Elster J, Stibal M, Santrůcková H (2005) Microbial assemblages in soil microbial succession after glacial retreat in Svalbard (High Arctic). Microbiol Ecol 50:396–407
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kirchman DL, Yu L, Cottrell MT (2003) Diversity and abundance of uncultured cytophaga-like bacteria in the Delaware estuary. Appl Environ Microbiol 69:6587–6596
LaMontagne MG, Schimel JP, Holden PA (2003) Comparison of subsurface and surface soil bacterial communities in California grassland as assessed by terminal restriction fragment length polymorphisms of PCRamplified 16S rRNA genes. Microbiol Ecol 46:216–227
Lane DJ (1991) 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M (eds) Nucleic acid techniques in bacterial systematics. Wiley, New York, pp 115–147
Lefauconnier B, Hagen JO, Rudant JP (1994) Flow speed and calving rate of Kongsbreen glacier, Svalbard, using SPOT images. Polar Res 10:56–65
Li S, Xiao X, Yin X, Wang F (2006) Bacterial community along a historic lake sediment core of Ardley Island, west Antarctica. Extremophiles 10:461–467
Li Y, Li F, Zhang X, Qin S, Zeng Z, Dang H, Qin Y (2008) Vertical distribution of bacterial and archaeal communities along discrete layers of a deep-sea cold sediment sample at the East Pacific Rise (approximately 13 degrees N). Extremophiles 12:573–585
Liu Y, Yao T, Jiao N, Kang S, Zeng Y, Huang S (2006) Microbial community structure in moraine lakes and glacial meltwaters, Mount Everest. FEMS Microbiol Lett 265:98–105
Liu Y, Yao T, Jiao N, Kang S, Xu B, Zeng Y, Huang S, Liu X (2009) Bacterial diversity in the snow over Tibetan Plateau Glaciers. Extremophiles 13:411–423
López-García P, López-López A, Moreira D, Rodríguez-Valera F (2001) Diversity of free-living prokaryotes from a deep-sea site at the Antarctic Polar Front. FEMS Microbiol Ecol 36:193–202
Lysnes K, Thorseth IH, Steinsbu BO, Øvreås L, Torsvik T, Pedersen RB (2004) Microbial community diversity in seafloor basalt from the Arctic spreading ridges. FEMS Microbiol Ecol 50:213–230
Magurran AE (1996) Ecological diversity and its measurement. Chapman & Hall, London, pp 274–286
Marcy Y, Ouverney C, Bik EM, Losekann T, Ivanova N, Martin HG, Szeto E, Platt D, Hugenholtz P, Relman DA, Quake SR (2007) Dissecting biological ‘dark matter’ with single-cell genetic analysis of rare and uncultivated TM7 microbes from the human mouth. Proc Natl Acad Sci USA 104:11889–11894
Mikucki JA, Priscu JC (2007) Bacterial diversity associated with Blood Falls, a subglacial outflow from the Taylor Glacier, Antarctica. Appl Environ Microbiol 73:4029–4039
Mindl B, Anesio AM, Meirer K, Hodson AJ, Laybourn-Parry J, Sommaruga R, Sattler B (2007) Factors influencing bacterial dynamics along a transect from supraglacial runoff to proglacial lakes of a high Arctic glacier. FEMS Microbiol Ecol 59:307–317
Mosier AC, Murray AE, Fritsen CH (2007) Microbiota within the perennial ice cover of Lake Vida, Antarctica. FEMS Microbiol Ecol 59:274–288
Nealson KH (1999) Post-Viking microbiology: new approaches, new data, new insights. Orig Life Evol Biosph 29:73–93
Neufeld JD, Yu Z, Lam W, Mohn WW (2004) Serial analysis of ribosomal sequence tags (SARST): a high-throughput method for profiling complex microbial communities. Environ Microbiol 6:131–144
Niederberger TD, McDonald IR, Hacker AL, Soo RM, Barrett JE, Wall DH, Cary SC (2008) Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ Microbiol 10:1713–1724
Perreault NN, Andersen DT, Pollard WH, Greer CW, Whyte LG (2007) Characterization of the prokaryotic diversity in cold saline perennial springs of the Canadian high Arctic. Appl Environ Microbiol 73:1532–1543
Prabagaran SR, Manorama R, Delille D, Shivaji S (2007) Predominance of Roseobacter, Sulfitobacter, Glaciecola and Psychrobacter in seawater collected off Ushuaia, Argentina, Sub-Antarctica. FEMS Microbiol Ecol 59:342–355
Priscu JC, Christner BC (2003) Microbial diversity and bioprocessing. In: Bull AT (ed) Earth’s icy biosphere. ASM press, Washington DC, pp 130–145
Reddy GSN, Aggarwal RK, Matsumotto GI, Shivaji S (2000) Arthrobacter flavus sp. nov., a psychrophilic bacterium isolate from a pond in McMurdo Dry Valley, Antarctica. Int J Syst Evol Microbiol 50:1553–1561
Reddy GSN, Uttam A, Shivaji S (2008) Bacillus cecembensis sp. nov., isolated from the Pindari glacier of the Indian Himalayas. Int J Syst Evol Microbiol 58:2330–2335
Reddy GSN, Pradhan S, Manorama R, Shivaji S (2009) Cryobacterium roopkundense sp. nov., a psychrophilic bacterium from a Himalayan glacier. Int J Syst Evol Microbiol 60:866–870
Reed AJ, Lutz RA, Vetriani C (2006) Vertical distribution and diversity of bacteria and archaea in sulfide and methane-rich cold seep sediments located at the base of the Florida Escarpment. Extremophiles 10:199–211
Rhoades JD (1982) Soluble salts. In: Page AL (ed) Methods of soil analysis. Chemical and microbiological properties, 2nd edn. Soil Science Society of America, Madison, pp 167–179
Segawa T, Miyamoto K, Ushida K, Agata K, Okada N, Kohshima S (2005) Seasonal change in bacterial flora and biomass in mountain snow from the Tateyama Mountains, Japan, analyzed by 16S rRNA gene sequencing and real-time PCR. Appl Environ Microbiol 71:123–130
Shivaji S, Reddy GSN, Aduri RP, Kutty R, Ravenschlag K (2004) Bacterial diversity of a soil sample from Schirmacher Oasis, Antarctica. Cell Mol Biol 50:525–536
Sjöling S, Cowan DA (2003) High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 7:275–282
Skidmore M, Anderson SP, Sharp M, Foght J, Lanoil BD (2005) Comparison of microbial community composition in two subglacial environments reveals a possible role for microbes in chemical weathering processes. Appl Environ Microbiol 71:6986–6997
Smith JJ, Tow LA, Stafford W, Cary C, Cowan DA (2006) Bacterial diversity in three different Antarctic Cold Desert mineral soils. Microbiol Ecol 51:413–421
Stackebrandt E, Brambilla E, Cousin S, Dirks W, Pukall R (2004) Culture-independent analysis of bacterial species from an anaerobic mat from Lake Fryxell, Antarctica: prokaryotic diversity revisited. Cell Mol Biol 50:517–524
Steven B, Briggs G, McKay CP, Pollard WH, Greer CW, Whyte LG (2007) Characterization of the microbial diversity in a permafrost sample from the Canadian high Arctic using culturedependent and culture-independent methods. FEMS Microbiol Ecol 59:513–523
Subbiah BH, Asija GL (1956) A rapid procedure for determination of available nitrogen in soils. Curr Sci 25:259–260
Suzuki K, Sasaki J, Uramoto M, Nakase T, Komagata K (1997) Cryobacterium psychrophilum gen. nov., sp. nov., nom. rev., comb. nov., an obligately psychrophilic actinomycete to accommodate ‘‘Curtobacterium psychrophilum’’ Inoue and Komagata 1976. Int J Syst Bacteriol 47:474–478
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Thompson JD, Higgins DG, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tindall BJ (2004) Prokaryotic diversity in the Antarctic: the tip of the iceberg. Microb Ecol 47:271–283
Treves DS, Xia B, Tiedje JM (2003) A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microbiol Ecol 45:20–28
Tsai YL, Olson BH (1991) Rapid method for direct extraction of DNA from soil and sediments. Appl Environ Microbiol 57:1070–1074
Tsai YL, Olson BH (1992) Rapid method for seperation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl Environ Microbiol 58:2292–2295
Wagner D, Kobabe S, Liebner S (2009) Bacterial community structure and carbon turnover in permafrost-affected soils of the Lena Delta, northeastern Siberia. Can J Microbiol 55:77–83
Walkley A, Black CA (1934) An examination of Degtjareff methods for determining soil organic matter and a proposed modification of chromic acid titration method. Soil Sci 37:29–38
Xiang SR, Yao TD, An LZ, Xu BQ, Li Z, Wu GJ, Wang YQ, Ma S, Chen XR (2004) Bacterial diversity in Malan ice core from the Tibetan Plateau. Folia Microbiol 49:269–275
Xiang S, Yao T, An L, Xu B, Wang J (2005) 16S rRNA sequences and differences in bacteria isolated from the Muztag Ata glacier at increasing depths. Appl Environ Microbiol 71:4619–4627
Yergeau E, Newsham KK, Pearce DA, Kowalchuk GA (2007) Patterns of bacterial diversity across a range of Antarctic terrestrial habitats. Environ Microbiol 9:2670–2682
Zhang X, Tandong Y, Lizhe A, Lide T, Shijian X (2006) A study on the vertical profile of bacterial DNA structure in the Puruogangri (Tibetan Plateau) ice core using denaturing gradient gel electrophoresis. Ann Glaciol 43:160–166
Zhang X, Yao T, Tian L, Xu S, An L (2008) Phylogenetic and physiological diversity of bacteria isolated from Puruogangri ice core. Microbiol Ecol 55:476–488
Zhou J, Davey ME, Figueras JB, Rivkina E, Gilichinsky D, Tiedje JM (1997) Phylogenetic diversity of a bacterial community determined from Siberian tundra soil DNA. Microbiology 143:3913–3919
Zhou J, Xia B, Treves DS, Wu L-Y, Marsh TL, O’Neill RV, Palumbo AV, Tiedje JM (2002) Spatial and resource factors influencing high microbial diversity in soil. Appl Environ Microbiol 68:326–334
Acknowledgments
We would like to thank the Department of Biotechnology and Council of Scientific and Industrial Research, Government of India for financial support to Dr. S. Shivaji. TNRS acknowledges the CSIR, Government of India, for the award of Research Associateship. We would like to thank Dr. M. K. Reddy, Scientist and Head and Dr. M. Kalita, Scientist from National Environmental Engineering Research Institute, Zonal Laboratory, Hyderabad, and Dr. M. K. Singh, Scientist from National Centre for Antarctic Ocean Research, Goa, for providing the data of the soil chemical characteristics.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Oren.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pradhan, S., Srinivas, T.N.R., Pindi, P.K. et al. Bacterial biodiversity from Roopkund Glacier, Himalayan mountain ranges, India. Extremophiles 14, 377–395 (2010). https://doi.org/10.1007/s00792-010-0318-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-010-0318-3