Abstract
The microbial diversity in maritime meltwater pond sediments from Bratina Island, Ross Sea, Antarctica was investigated by 16S rDNA-dependent molecular phylogeny. Investigations of the vertical distribution, phylogenetic composition, and spatial variability of Bacteria and Archaea in the sediment were carried out. Results revealed the presence of a highly diverse bacterial population and a significantly depth-related composition. Assessment of 173 partial 16S rDNA clones analyzed by amplified rDNA restriction analysis (ARDRA) using tetrameric restriction enzymes (HinP1I 5′G∇CGC3′and Msp I. 5′C∇CGG3′, BioLabs) revealed 153 different bacterial OTUs (operational taxonomic units). However, only seven archaeal OTUs were detected, indicating low archaeal diversity. Based on ARDRA results, 30 bacterial clones were selected for sequencing and the sequenced clones fell into seven major lineages of the domain Bacteria; the α, γ, and δ subdivisions of Proteobacteria, the Cytophaga–Flavobacterium–Bacteroides, the Spirochaetaceae, and the Actinobacteria. All of the archaeal clones sequenced belonged to the group Crenarchaeota and phylogenetic analysis revealed close relationships with members of the deep-branching Group 1 Marine Archaea.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Benthic sediment represents one of the most complex microbial habitats on Earth (Torsvik et al. 1990; Priscu et al. 1998). The microorganisms in the sediment play a significant role in remineralization of organic matter within the aquatic ecosystem. For example, bacterial populations are the major contributors in the transformation of organic carbon, sulfur, nitrogenous compounds, and metals, with an important role in ecosystem food webs and nutrient cycling. In studies of the structure and function of aquatic ecosystems, reliable estimates of microbial numbers, diversity, and activity are critical. However, the accurate enumeration of the prokaryotic diversity has, until recently, been difficult because a very high proportion of the bacterial and archaeal species cannot be cultivated (Amann et al. 1995). The combination of various new molecular techniques, including FISH (fluorescent in situ hybridization); SSU (small subunit) rDNA PCR amplification, cloning, and sequencing; DGGE (denaturing gradient gel electrophoresis); SSCP (single-stranded conformation polymorphism); and TRFLP (terminal restriction fragment length polymorphism), now make it possible to describe the microbial diversity with a much higher level of accuracy (Stackebrandt et al. 1993; Amann et al. 1995; Muyzer et al. 1998), unveiling a new, previously unsuspected, world of microbial diversity.
The Antarctic continent provides a unique environment with extremely low temperatures and, in the Dry Valley regions of South Victoria Land, dramatic seasonal variation. Substantial portions of the McMurdo Ice Shelf, adjacent to the southern Dry Valley regions, are covered by gravels and mineral deposits originating from marine sediment (Hart 1990). Over decades, freezing, thawing, and evaporation have resulted in the development of meltwater ponds with different solute gradients, salinities, and biomass contents (Mountfort et al. 1999 and references therein). Studies of the meltwater ponds at McMurdo Ice Shelf show that leaching of local salt deposits and weathering of superficial sediments contribute to the enrichment of sulfate, sodium, calcium, and potassium in the meltwater (DeMora et al. 1994) that lower the freezing point of water and leave the pools essentially ice-free (Vincent 1988). The more saline McMurdo Ice Shelf meltwater ponds are dominated by cyanobacterial biomass, whereas sulfate reduction and methanogenesis predominate in ponds of lower salinity (Mountfort et al. 1999). Studies of psychrophiles (optimal growth temperature, <15°C; no growth occurring above 20°C) and their diversity in Antarctic sea ice show that the microbial community comprises many taxonomically novel members (Bowman et al. 1997; Christner et al. 2001; Gordon et al. 2000). Recently, interest in Antarctic pelagic microbiology has centered on the presence and significance of Archaea, now found to be widely abundant and constituting a significant part of Antarctic picoplankton assemblages (DeLong et al. 1994; Preston et al. 1996; Murray et al. 1998).
To date, however, few cultivation-independent studies have been carried out on the microbial diversity of permanently cold Antarctic sediments (Knoblauch et al. 1999; Ravenschlag et al. 1999). Even less is known about the diversity and function of Archaea in the polar benthic sediments.
The potential benefits from the exploration of the microbial diversity of Antarctica derive from the future biotechnological exploitation of the Antarctic gene pool and from new insights into the biological mechanisms of adaptation to and tolerance of extreme environments by microorganisms.
In this study, we conducted a molecular phylogenetic investigation into the bacterial and archaeal community composition in benthic saline pond sediment from McMurdo Ice Shelf at Bratina Island, Ross Sea, Antarctica.
Materials and methods
Field sites and sampling procedures
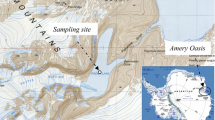
Sediment samples were collected from saline ponds in the area of Bratina Island (78°00.82′S, 165°33.05′E) in the Antarctic Ross Dependency region in January 1999. The area contains a large number of pools of different salinity, but isolated from the marine environment by a 50-m depth of sea ice. The pool sampled was approximately 12 m2 in area with a maximum of 1 m depth and a sediment depth of over 1 m. Sediment core samples were collected using a plastic tube (4 cm2 i.d.), sectioned into subsamples of different sediment depth after ejection, and aseptically transferred to sterile 90-ml polyethylene Kartell bottles for storage at −20°C during transport to the UK for subsequent analysis.
Environmental parameters
Water, soil, and atmospheric temperatures were monitored with a Solomat 520C digital thermometer (equipped with a 10-cm steel probe).
DNA extraction
Genomic DNA was extracted directly from 2 g dry weight of sediment sample (Bio 101 FastDNA spin protocol) from seven different sediment depths. DNA was also extracted from 2 ml of a sample from the water column above the sediment core. The concentration of dsDNA was determined fluorophotometrically using the sensitive fluorescent nucleic acid stain PicoGreen (Molecular Probes), excitation ~480 nm, emission ~520 nm (TD-700 Fluorimeter). Lambda DNA was used as a standard for preparation of calibration curves.
PCR amplification of 16S rDNA
The DNA purified from soil was used as a template for PCR. A eubacterial-specific 16S rDNA primer pair (Rudi et al. 1997) was used (forward primer CC342f, 5′-TCCAGACTCCTACGGGAGGCAGC-3′ and reverse primer, CD927r, 5′-CTTGTGCGGGCCCCCGTCAATTC-3′). For the amplification of Archaea, we used the specific primer pair [forward primer, ARCH21f, 5′-TTCCGGTTGATCCTGCCGGA-3′ and reverse primer, ARCH958r, 5′-CCCGGCGTTGAATCCAATT-3′ (DeLong 1992)]. Primers were synthesized by Pharmacia Biotech. The DNA was denatured at 94°C for 4 min prior to amplification with 35 PCR cycles (92°C 30 s, 42°C 45 s, 72°C 30 s) followed by an extension step at 72°C for 7 min for both primer pairs. Amplification reactions contained 30 pmol of each primer, 200 μM deoxynucleotides, 1 unit of DNA polymerase mix using the High Fidelity Expand PCR system (Boehringer), 1–4 ng of control DNA (E. coli, NCIMB 12210), or extracted genomic DNA, in a final volume of 50 μl. The mixture was overlaid with 20 μl mineral oil (Sigma Chemical).
PCR products were examined by gel electrophoresis on 0.8% agarose gels in 1×TAE buffer (4 mM Tris-acetate, pH 7.6, 1 mM EDTA) using λ DNA/HindIII molecular weight markers for size comparisons of the PCR products.
Cloning into pCR 2.1 TOPO vector and construction of clone libraries
Since the proofreading activity of the DNA polymerase Pwo removes the 3′A-overhangs necessary for TA cloning (Invitrogen), 3′A-overhangs were added to the blunt end fragments by amplification with 1 U Taq polymerase at 72°C for 20 min. Polymerases were removed by phenol–chloroform extraction and DNA precipitation with sodium acetate and ethanol. The PCR product was gel-purified (Qiagen) and then cloned into pCR 2.1 TOPO vector (TOPO TA cloning kit, Invitrogen). The cloning reaction was transformed into TOP10 One Shot cells, incubated overnight on selective plates, followed by blue–white selection.
White colonies were selected, amplified, and plasmid DNA isolated (QIAprep Spin miniprep kit, Qiagen). The plasmids were analyzed for inserts by restriction analysis using overnight EcoRI (5′GAATTC3′, BioLabs) digestion. The digested reactions were analyzed by agarose gel electrophoresis.
Amplified rDNA restriction analysis (ARDRA) and restriction fragment length polymorphism (RFLP) analysis
Cloned inserts were reamplified with the appropriate primers. The purity and size of each PCR product was checked on 1% agarose gels. The reamplified 16S rDNA product was used in restriction digests using HinP1I and MspI enzymes (in 10 mM Tris-HCl, 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol, pH 7.9). Digests were resolved on 2% agarose gels in TBE and were grouped manually based on the restriction patterns. RFLP clones were selected for sequencing (Oswel) based on ARDRA or RFLP patterns.
Phylogenetic analysis
The 16S rDNA sequences were compared with sequences from the BLAST database and the Ribosomal Database Program (RDP) and aligned. Phylogenetic trees were constructed from evolutionary distances using the neighbor-joining method implemented through NEIGHBOR (DNADIST) from the PHYLIP version 3.57c package (Felsenstein 1993) (with the Kimura two-parameter model with a transition/transversion ratio of 2). DNAPARS was used for maximum-parsimony and maximum-likelihood methods. A total of 100 bootstrapped replicate resampling data sets were generated (for each method) and a consensus tree was made (using CONSENS). Tree files generated by PHYLIP were analyzed with the TREEVIEW program (Page 1996). All sequences were analyzed for chimeras using the RDP program Check Chimera (Cole et al. 2003).
Results
Benthic sediment samples from saline ice ponds in Bratina Island, South Victoria Land, eastern Antarctica were analyzed for bacterial and archaeal diversity. Genomic DNA was extracted from sediment samples and amplified using universal bacteria- or archaea-specific 16S rDNA PCR primers. Physicochemical characterization showed that the salinity of the surface sediment was equivalent to 0.46% sodium chloride and the concentration of dissolved oxygen was 1.1 mg/l. The temperature of the pool water at 10 cm depth was 9°C at the point of sampling. Chlorophyll a concentrations in the top 2 cm sediment were less than 10 μg/g of dry weight sediment, indicating a low abundance of phototrophic organisms. The protein concentration in the top 2 cm of sediment was 5 μg/g dry weight.
The efficiency of direct genomic DNA extraction of the sediment samples was evaluated by re-extracting the samples. Electrophoretic analysis of primary and secondary DNA extracts on agarose gels showed that approximately 90–100% of the extractable DNA was obtained in the primary extraction. DNA fragments were predominantly >3 kb in size. The yield of extracted DNA in the different sediment samples varied from 50 to 10 μg/g dry weight sediment. The top 2 cm fraction and the 10–13 cm sediment layer gave the highest DNA yields.
Genomic library
Purified genomic DNA extracted from sediment and water samples of seven different depths was amplified using universal eubacterial 16S rDNA primers (Rudi et al. 1997). A 605-bp fragment, corresponding to variable regions (E. coli V3, V4, and V5, nucleotide 334–939 of 16S rDNA) was detected in each of the eight samples. This PCR fragment was cloned to produce a 16S rDNA library representing each sample. All libraries contained >103 clones. DNA of the sample from the depth interval of 10–13 cm was used to produce an archaeal 16S rDNA library using archaeal-specific primers for the PCR amplification.
Vertical profiling using ARDRA analysis
Tetrametric restriction enzymes have been shown to be useful tools for screening environmental clone libraries (Ravenschlag et al. 1999; Liu et al. 1997) and the common restriction fragment patterns, the so-called operational taxonomic units (OTUs) obtained from such analysis, assist in distinguishing between taxonomic groups. A summary of the ARDRA results using the restriction enzymes HinP1I and MspI is shown in Table 1. Of the 173 bacterial clones analyzed, 153 different ARDRA patterns were identified, indicating high bacterial diversity. However, digestion of 17 archaeal clones resulted in only seven different patterns, indicating relatively low archaeal diversity. The ARDRA results (Table 1) indicate that the bacterial library derived from the water column had a lower diversity of organisms (with a coverage of 54%) compared with the libraries derived from the sediment (where the coverage ranged from 40% to 0%). At several depths (1–2 cm, 4–6 cm, and 13–17 cm) every clone analyzed showed a unique pattern (Table 1), indicating very high diversity in these samples. A total rarefaction analysis of all the bacterial ARDRA patterns suggested that only a portion of the total diversity has been observed in the screening of 173 clones (Fig. 1).
Sequencing analysis of 16S rDNA clones
Based on restriction patterns derived from ARDRA, 36 clones (30 bacterial and 6 archaeal) were selected for sequencing. Between three and five bacterial nucleotide sequences corresponding approximately to positions 342–927 (E. coli numbering) were obtained per library (Table 2). Sequences suspected to be chimeric after examination by the CHECK_CHIMERA program were not included. Comparative 16S rDNA analysis showed that none of the clone sequences were identical to any of the known 16S rDNA sequences in the nucleotide databases (BLASTn). Most bacterial clone sequences showed >88% but <97% similarity to already known sequences, indicating the putative presence of new species. On the basis of complete 16S rDNA sequences, a binary similarity value of 97.5% indicates the presence of two distinct species (Stackebrandt and Goebel 1994). However, it has been recently shown that a single species (archaeal) may contain 16S rRNA genes that are up to 5% different (Amann et al. 2000). The use of 97.5% limit for species separation must therefore be treated with some caution.
The 16S rDNA sequences obtained from the different clones were subject to phylogenetic analysis. The sequenced clones were affiliated to seven major lineages of the domain Bacteria; the α-, γ-, and δ- subdivisions of the Proteobacteria, the Cytophaga–Flavobacterium–Bacteroides phylum, the Actinomycetaea, and the Spirochaeta (see Table 2, Fig. 2).
Phylogenetic tree of bacterial 16S rDNA clones derived from Bratina Island pond sediment (in bold) with reference species. The bar represents 10% estimated sequence divergence. Spirochaeta (Sp), Cytophagales–Flavobacterium–Bacteroides (CFB), α-Proteobacteria (α), γ-Proteobacteria (γ), δ- Proteobacteria (δ). The bar represents 10% estimated sequence divergence
Both the ARDRA and sequencing results showed that there were differences in the vertical distribution of bacteria. The top 10 cm of sediment was dominated by δ-Proteobacteria whereas the layer between 10 and 17 cm depth was dominated by Cytophagales and Bacteroidaceae. At 17–22 cm sediment depths the major fraction of bacteria consisted of γ-Proteobacteria. Four bacterial and one archaeal clone could not be affiliated to any known group.
Proteobacteria
38% of the sequenced clones were included in the Proteobacterial domain. The δ-Proteobacteria clones were exclusively detected within the upper 2 cm of the sediment profile, with the exception of one clone from 10–13 cm depth sample. Phylogenetic analysis revealed that the δ-Proteobacteria clone sequences clustered into three groups (not shown). One of the groups consisted of BIs1–2.5 and BIs1–2.22 (similarity >0.90 and bootstrap values above 97%), which formed a common lineage with an unusual sulfate-reducing, taurine-fermenting Desulforhopalus and an uncultured Arctic sediment clone (Sva0999) affiliated with Arctic Desulfotalea sp. (Ravenschlag et al. 1999). The other upper-sediment clone grouped together with an unidentified Xanthomonas-related clone (rj15) from phenol-degrading activated sludge (Watanabe et al. 1999). The clone from the deeper sediment (BIs10–13.6) grouped together with Pelobacter and Desulfuromonas sp.
Other proteobacteria clones belonged to the subdivisions of α- or γ-Proteobacteria. The α-Proteobacteria was related to a marine uncultured Rhodobacter (BIs10–13.2, similarity 0.96). The clones belonging to the subclass of γ-Proteobacteria were derived from the deepest sediment sample (17–22 cm) except for three E. coli clones that were derived from the water-column sample (BIs1.1, 1.9, and 1.22, which were probably the result of contamination of the library host, as the clones were 99% identical to the laboratory strain E. coli K12). Clone BIs17–22.9 was related to a Legionella-like pathogen (similarity 0.97), whereas clone BIs17–22.4 showed the closest relation to the deep-branching marine Curacaobacter baltica (similarity 0.90), a Pseudomonas-related chemoheterotrophic aerobe isolated from the Baltic Sea (NCBI AJ002006).
Cytophaga–Flavobacterium–Bacteroides
The majority of clones that clustered with the CFB division derived from libraries of sediment samples of 7–17 cm depth. The fermentative group Bacteroidaceae was represented by several clones (BIs7–10.2, BIs7–10.7, BIs7–10.8) with different ARDRA patterns but all related to an uncultured bacteria from a low-temperature hydrocarbon- and chlorine-contaminated aquifer undergoing intrinsic bioremediation (Dojka et al. 1998). This group included clone BIs13–17.2, related to a benzene-mineralizing sulfate reducer (Phelps et al. 1998). Clones in the Cytophagales group (BIs4–6.6, BIs7–10.6, BIs10–13.3, BIs13–17.9, and BIs17–22.5) were related to uncultured marine or stone sublithic psychrotrophs (NCBI AF207849, AB015532, AF170754, AF001373) (see Table 2, Fig. 2).
Other bacterial divisions
Clone BIs4–6.0 was most closely related (similarity 0.93) to an uncultured environmental Spirochaeta sp. identified in a hydrocarbon- and chlorinated solvent-contaminated aquifer (Dojka et al. 1998). From the library representing the 13–17 cm depth sample, one clone (BIs13–17.6) clustered with the Gloeobacter, a member of the Chroococcales cyanobacteria (similarity 0.89). Actinomycetes were represented by two sequenced clones from deeper sediment (BIs13–17.10 and BIs17–22.1) that were affiliated with an uncultured deep-sea sediment actinomycete and Clavibacter michiganesis, respectively.
Archaea
In contrast to the bacterial library, ARDRA patterns showed that the archaeal library contained few phylotypes. Seven ARDRA patterns were detected. All of the analyzed archaeal clones were associated with Crenarchaeota except for two that could not be classified as they showed low sequence similarity to any known archaeon.
Phylogenetic analysis indicated that clones BIsArch 2, 3, and 7 (and BIsArch 4, which was not included in the final tree because the sequence was too short) clustered with high bootstrap values to the Group I marine archaeal sequences (Fig. 3), a deep-branching group with members of the pelagic planktonic division in the Crenarchaeota (DeLong 1992; DeLong et al. 1994). A group of terrestrial Archaea (SCA) was also affiliated to this group (Bintrim et al. 1997).
Discussion
Prokaryotic diversity analysis based on 16S rDNA techniques provides a means for understanding ecologically complex microbial community structures through the identification of otherwise obscure (e.g., unculturable) populations. Several investigators have raised concerns about whether the DNA extract obtained is representative of the indigenous microflora and about the problems associated with PCR of 16S rDNA gene for the phylogenetic analysis, and therefore this will not be discussed here (Chandler et al. 1997; Urakawa et al. 2000). With the limitations of culture techniques, however, the sequenced-based phylogenetic techniques provide a less biased picture of the community composition than would any cultivation technique (Amann et al. 1995).
Our results show that the prokaryotic diversity is comparable to, if not higher than, that shown in other studies of Antarctic meltwater lake sediments (Bowman et al. 2000), microbial mats in East Antarctic lakes (Brambilla et al. 2001), and the Arctic Ocean (Ravenschlag et al. 1999), each of which report a high level of complexity in the prokaryotic community structure. The linearity of the rarefaction curve (Fig. 1) suggests that the diversity of prokaryotes in the Bratina Island meltwater pond is probably higher than has been determined experimentally. The results also demonstrate the complexity of the microbial community in the cold sediments, where most of the sequenced clones represent novel taxa. Putative strains are related to species detected either in cold marine or lake waters and sediment or in bioremediation processes, with several heterotrophs involved in the cycling of CO2 and inorganic nutrients and decomposition. Some of the more metabolically 'specialized' bacteria, such as the Fe (III)-reducing Pelobacter, have also been detected in microbial mats in Lake Fryxell, Antarctic Dry Valleys (Brambilla et al. 2001).
In addition, our results reveal a vertical stratification of bacterial species, probably reflecting a vertical gradient of temperature, oxygen content, and redox potential of the sediment. We note, however, that the limited sample sizes and numbers (one clone library from 8 g of sediment for each depth) may generate results which do not fully reflect the spatial distribution of the sediment communities. Sulfate-reducing δ-Proteobacteria were abundant in the top centimeter of sediment, indicating low redox potential. These obligate anaerobic bacteria may also thrive in anaerobic micro-niches, such as flocs of organic material, in an otherwise aerobic environment (Brune et al. 2000).
Cytophagales and microaerophilic Bacteroidaceae, which are important in mineralization of organic matter, were the most prevalent groups between 4 and 17 cm depth. Interestingly, two phototrophs, the uncultured Rhodobacter-related α-Proteobacteria, which is prevalent in aquatic environments with abundant organic matter and which grows photoautotrophically under anaerobic conditions in the presence of sulfide, and a Chroococcales were also present in this sample, at a greater depth than might be expected. Deeper in the sediment (below 13 cm depth) Actinobacter and microaerophilic γ-Proteobacteria were detected. Several authors have reported the isolation of high GC-containing Gram-positive bacteria from Antarctic soil and sea ice. However, there is a disagreement on whether these are the prevalent microbiota or not. Few genera have actually been isolated (Nocardia, Nocardiopsis, and Streptomyces). Interestingly, it has been suggested that Actinobacter species that have disappeared from specific biotopes due to changing conditions or competition can survive (as spores) for thousands of years in polar ice (Abyzov 1993 and references therein).
Phylogenetic analysis revealed that the archaeal clones (only analyzed from a depth of 17–22 cm) were closely clustered with the deeply branching Marine Group 1 Crenarchaeota (Fig. 3). This group appears to be ubiquitous (DeLong 1992; Preston et al. 1996; Bintrim et al. 1997) and has recently been reported in a wide variety of temperate and cold environments including agricultural and forest soils, fresh water lake sediments, marine picoplankton, and deep-sea locations (Preston et al. 1996; Bintrim et al. 1997; Buckley et al. 1998 and references therein). The abundance of nonthermophilic crenarchaeotal rRNA found in cold Antarctic waters and sediments suggests that these organisms are ecologically relevant members of the marine microbial community (DeLong et al. 1994; MacGregor et al. 1997; Buckley et al. 1998; this study).
We detected no methanogenic archaea but several sulfate reducers were identified, suggesting that a low redox potential and high SO4 2− concentration (DeMora et al. 1994, 1996) in the sediment may favor sulfate reducers over methanogens in competition for acetate (Vincent 1988; Mountfort et al. 1999). Studies of sediment from Antarctic lakes using most-probable-number analysis showed that methanogens represented only a small proportion of the total microbial population (Bowman et al. 1997). The detection of methanogens by use of specific PCR primers may, however, cause this view to be revised.
Interestingly, our results indicate the presence of a number of aromatic hydrocarbon degraders which could be assumed to be cold-temperature active. With improved cultivation techniques or multigenomic expression, cloning these bacteria and their enzymes will be an attractive source of catalysts for biotransformations and bioremediation.
References
Abyzov SS (1993) Microorganisms in the Antarctic ice. In: Friedmann EI (ed) Antarctic microbiology. Wiley-Liss, New York, pp 265–295
Amann G, Stetter OK, Llobet-Brossa E, Amann R, Antón J (2000) Direct proof for the presence and expression of two 5% different 16Sr RNA genes in individual cells of Haloarcula marismortui. Extremophiles 4:373–376
Amann R, Ludwig W, Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59:143–169
Bintrim SB, Donohue TJ, Handelsman J, Roberts GP, Goodman RM (1997) Molecular phylogeny of Archaea from soil. Proc Natl Acad Sci USA 94:277–282
Bowman JP, McCammon SA, Brown MV, Nichols DS, McMeekin TA (1997) Diversity and association of pschychrophilic bacteria in Antarctic sea ice. Appl Environ Microbiol 63:3068–3078
Bowman JP, Rea SM, McCammon SA, McMeekin TA (2000) Diversity and community structure within anoxic sediment from marine salinity meromitic lakes and a coastal meromitic marine basin, Vestfold Hills, Eastern Antarctica. Environ Microbiol 2:227–237
Brambilla E, Hippe H, Hagelstein A, Tindall BJ, Stackebrandt E (2001) 16S rDNA diversity of cultured and uncultured prokaryotes of mat sample from Lake Fryxell, McMurdo Dry Valleys, Antarctica. Extremophiles 5:23–33
Brune A, Frenzel P, Cypionka H (2000) Life at the oxic–anoxic interface: microbial activities and adaptations. FEMS Microbiol Rev 24:691–710
Buckley DH, Graber JR, Schmidt TM (1998) Phylogenetic analysis of nonthermophilic members of the kingdom Crenarchaeota and their diversity and abundance in soils Appl Environ Microbiol 64:4333–4339
Chandler DP, Fredrickson JK, Brockman FJ (1997) Effect of PCR template concentration on the composition and distribution of total community 16S rDNA clone libraries. Mol Ecol 6:475–482
Christner BC, Mosley-Thompson E, Thompson LG, Reeve JN (2001) Isolation of bacteria and 16S rDNAs from Lake Vostok accretion ice. Environ Microbiol 3:570–577
Cole JR, Chai B, Marsh TL, Farris RJ, Wang Q, Kulam SA, Chandra S, McGarrell DM, Schmidt TM, Garrity GM, Tiedje JM (2003) The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res 31:442–443
DeLong EF (1992) Archaea in coastal marine environments. Proc Natl Acad Sci USA 89:5685–5689
DeLong EF, Wu KY, Prézelin BB, Jovine RM (1994) High abundance of Archaea in Antarctic marine picoplankton. Nature 37:695–697
DeMora SJ, Whitehead RF, Gregory M (1994) The chemical composition of glacial melt water ponds and streams on the McMurdo Ice Shelf, Antarctica. Antarct Sci 6:17–27
DeMora SJ, Lee PA, Grout A, Schall C, Heumann KG (1996) Aspects of the biogeochemistry of sulphur in glacial melt water ponds on the McMurdo Ice Shelf, Antarctica. Antarct Sci 8:15–22
Dojka MA, Hugenholtz P, Haack SK, Pace NR (1998) Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol 64:3869–3877
Felsenstein J (1993) PHYLIP (phylogenetic inference package) version 3.57c. University of Washington, Seattle, USA
Gordon DA, Priscu J, Giovannoni S (2000) Origin and phylogeny of microbes living in permanent Antarctic lake ice. Microb Ecol 39:197–202
Hart CP (1990) Holocene megafauna in the McMurdo Ice Shelf sediments: fossilization and implications for glacial processes. Antarct J 25:11–14
Knoblauch C, Jørgensen BB, Harder J (1999) Community size and metabolic rates of psychrophilic sulphate-reducing bacteria in Arctic sediments. Mar Ecol Prog Ser 180:2–21
Liu WT, Marsh TL, Cheng H, Forney LJ (1997) Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl Environ Microbiol 63:4516–4522
MacGregor BJ, Moser DP, Alm EW, Nealson KH, Stahl DA (1997) Crenarchaeota in Lake Michigan sediment. Appl Environ Microbiol 63:1178–1181
Mountfort DO, Kaspar HF, Downes M, Asher RA (1999) Partitioning effects during terminal carbon and electron flow in sediments of a low-salinity meltwater pond near Bratina Island, McMurdo Ice Shelf, Antarctica. Appl Environ Microbiol 65:5493–5499
Murray AE, Preston DA, Massana L, Taylor T, Blakis A, Wu K, De Long EF (1998) Seasonal and spatial variability of bacterial and archaeal assemblages in the coastal waters near Anvers Island, Antarctica. Appl Environ Microbiol 64:2582–2595
Muyzer G, Smalla K (1998) Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie van Leeuwenhoek Int J Gen Mol Microbiol 73:127–141
Page RD (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Phelps CD, Kerkhof LJ, Young LY (1998) Molecular characterization of a sulfate-reducing consortium which mineralizes benzene. FEMS Microbiol Ecol 27:269–279
Preston CM, Wu KY, Molinski TF, De Long EF (1996) A psychrophilic crenarchaeon in habits a marine sponge: Crenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA 93:6241–626
Priscu JC, Fritsen CH, Adams EE, Giovannoni SJ, Paerl HW, McKay CP, Doran PT, Gordon DA, Lanoil BD, Pickney JL (1998) Perennial Antarctic Lake Ice: an oasis for life in a polar desert. Science 280:2095–2098
Ravenschlag K, Sahm K, Pernthaler J, Amann R (1999) High bacterial diversity in permanently cold marine sediments. Appl Environ Microbiol 65:3982–3989
Rudi KO, Skulber M, Laarsen F, Jakobsen KS (1997) Strain characterization and classification of oxyphotobacteria in clone cultures on the basis of 16S rRNA sequences from the variable regions V6, V7 and V8. Appl Environ Microbiol 63:2593–2599
Stackebrandt E, Goebel BM (1994) A place for DNA–DNA reassociation and 16S ribosomal-RNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol 44:846–849
Stackebrandt E, Liesack W, Goebel BM (1993) Bacterial diversity in a soil sample from a subtropical Australian environment as determined by 16S rRNA analysis. FASEB J 7:232–236
Torsvik V, Goksoyr J, Daae FL (1990) High diversity in DNA of soil bacteria. Appl Environ Microbiol 56:782–787
Urakawa H, Yoshida T, Nishimura M, Ohwada K (2000) Characterization of depth-related population variation in microbial communities of a coastal marine sediment using 16S rDNA-based approaches and quinone profiling. Environ Microbiol 2:542–554
Vincent WF (1988) Microbial ecosystems of Antarctica. Cambridge University Press, Cambridge, UK
Watanabe K, Teramoto M, Harayama S (1999) An outbreak of nonflocculating catabolic populations caused the breakdown of a phenol-digesting activated-sludge process. Appl Environ Microbiol 65:2813–2819
Acknowledgments
This research was carried out under the auspices of the University of Waikato Antarctic Terrestrial Biology Research program. We gratefully acknowledge financial support from the University of Waikato and Antarctica New Zealand (D.A.C.), the Carl Tryggers Foundation (S.S.) and the Wenner-Gren foundations (S.S.). We thank Dr. M. Thomas for providing useful information on phylogenetic analysis.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by K. Horikoshi
Rights and permissions
About this article
Cite this article
Sjöling, S., Cowan, D.A. High 16S rDNA bacterial diversity in glacial meltwater lake sediment, Bratina Island, Antarctica. Extremophiles 7, 275–282 (2003). https://doi.org/10.1007/s00792-003-0321-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00792-003-0321-z